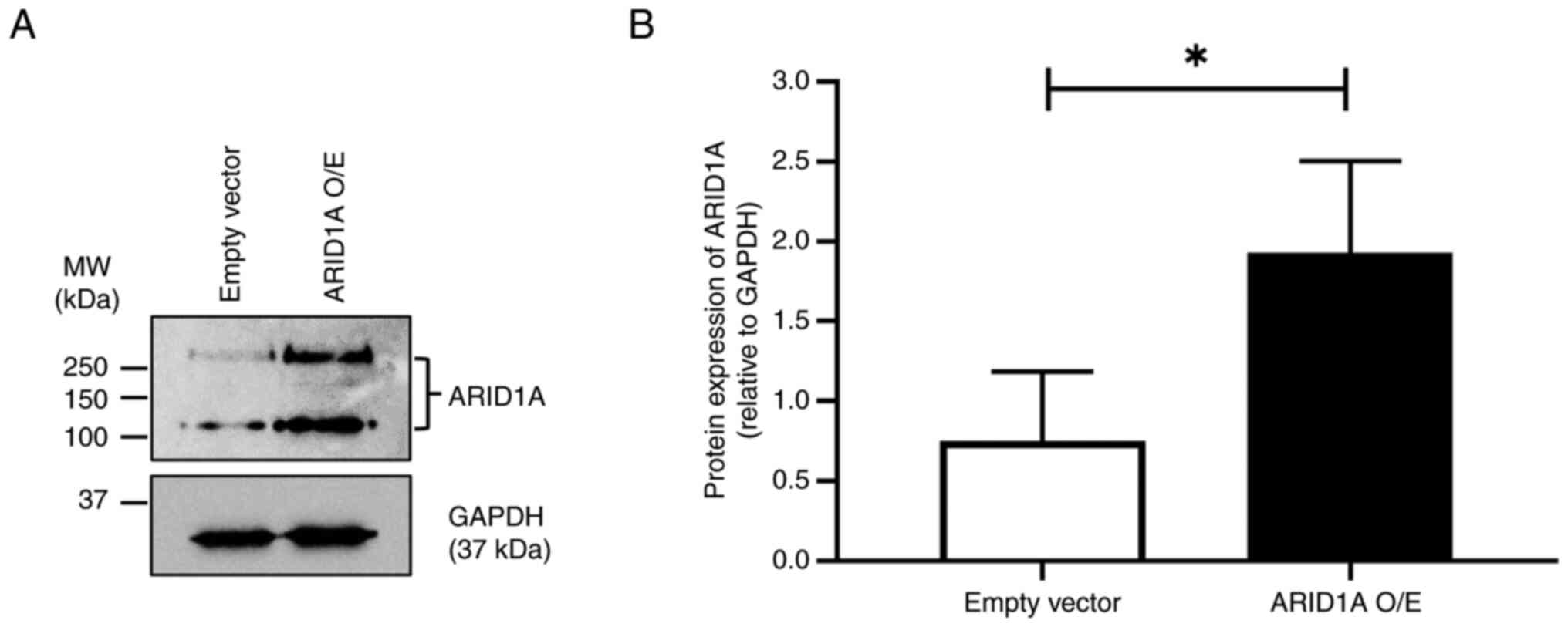
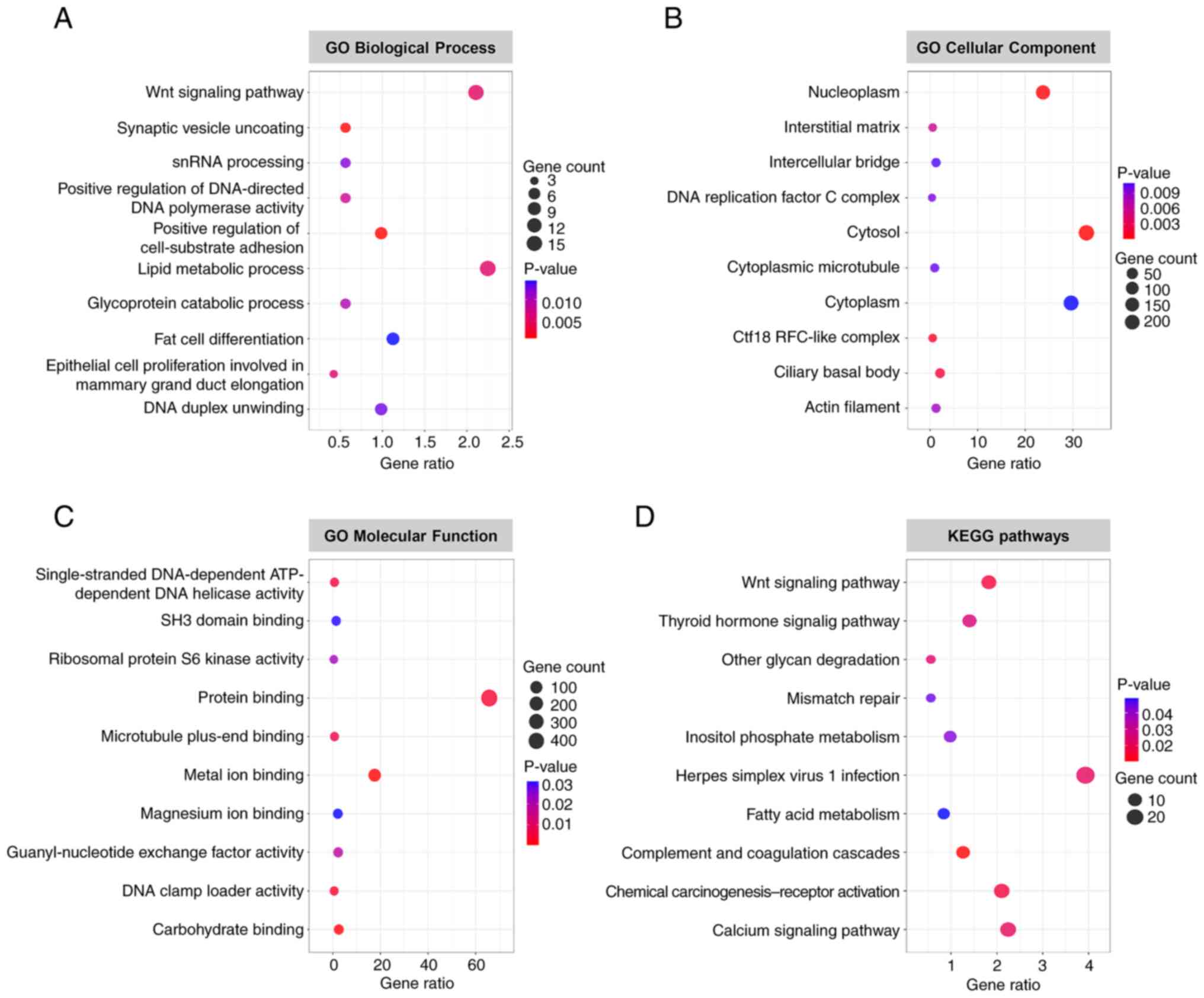
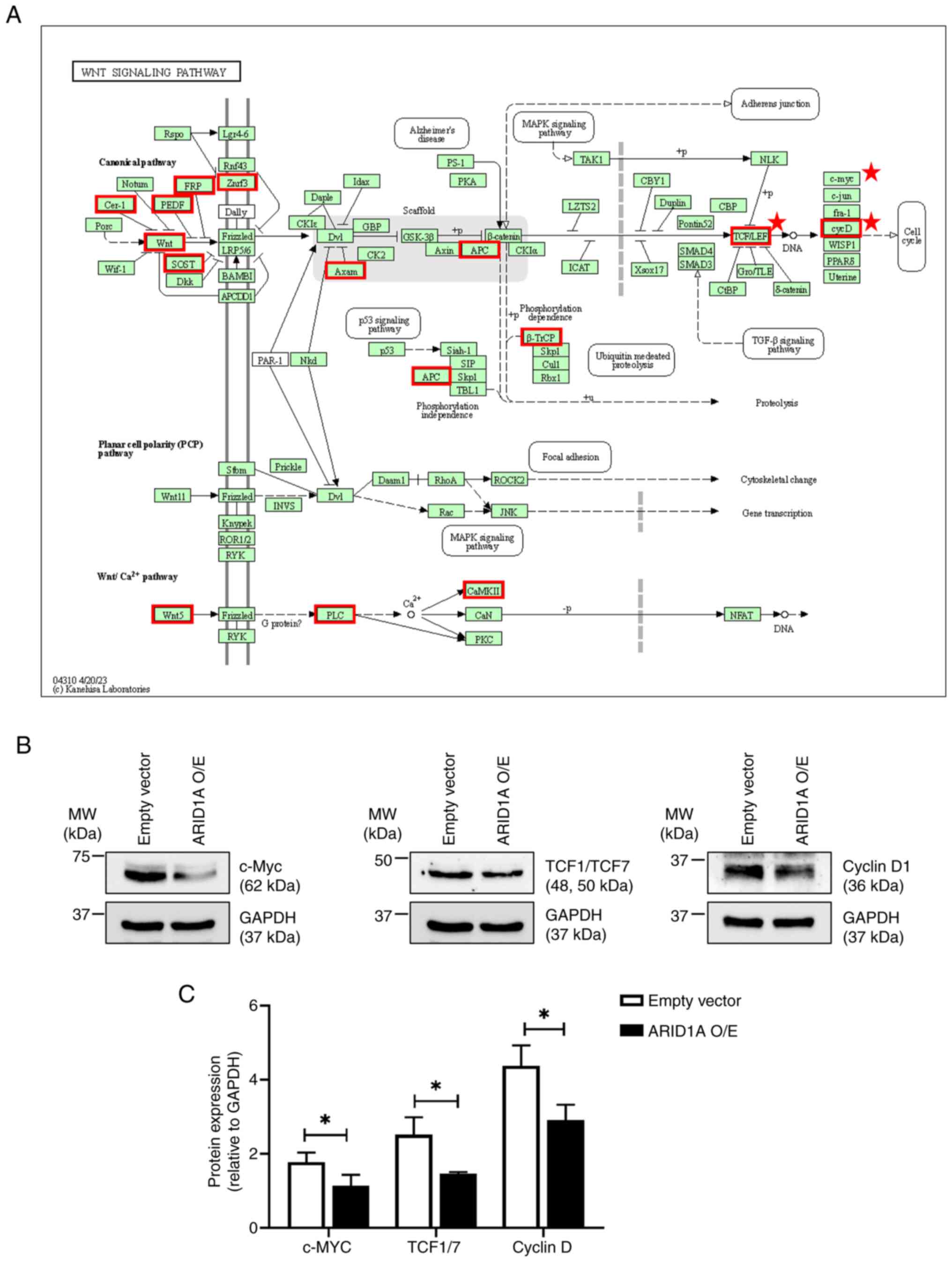
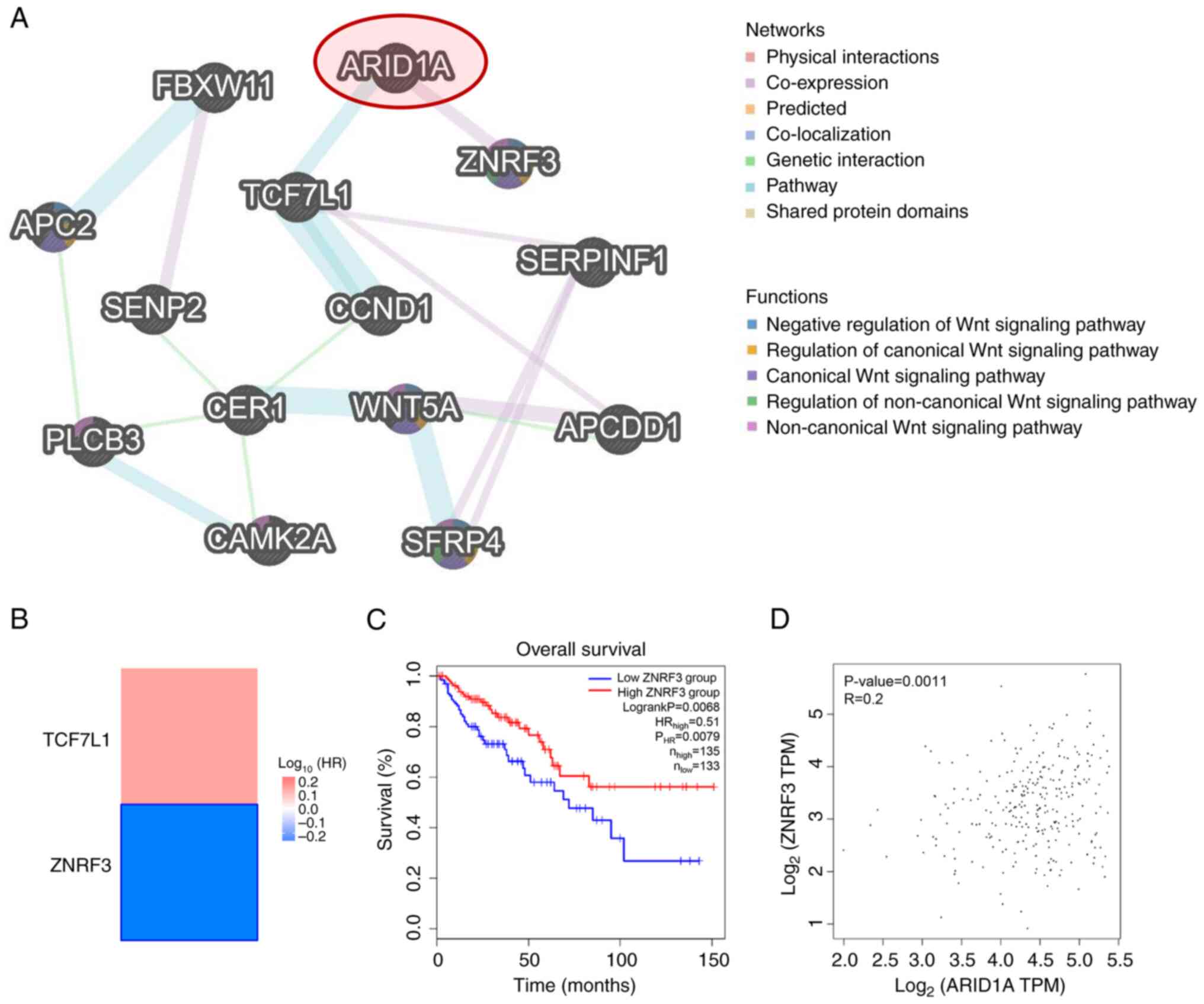
|
1
|
Zhao S, Wu W, Jiang Z, Tang F, Ding L, Xu
W and Ruan L: Roles of ARID1A variations in colorectal cancer: A
collaborative review. Mol Med. 28:422022. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wilsker D, Probst L, Wain HM, Maltais L,
Tucker PW and Moran E: Nomenclature of the ARID family of
DNA-binding proteins. Genomics. 86:242–251. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang X, Nagl NG, Wilsker D, Van Scoy M,
Pacchione S, Yaciuk P, Dallas PB and Moran E: Two related ARID
family proteins are alternative subunits of human SWI/SNF
complexes. Biochem J. 383:319–325. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wu RC, Wang TL and Shih IeM: The emerging
roles of ARID1A in tumor suppression. Cancer Biol Ther. 15:655–664.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liu X, Li Z, Wang Z, Liu F, Zhang L, Ke J,
Xu X, Zhang Y, Yuan Y, Wei T, et al: Chromatin remodeling induced
by ARID1A loss in lung cancer promotes glycolysis and confers JQ1
vulnerability. Cancer Res. 82:791–804. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wilson MR, Reske JJ, Holladay J, Neupane
S, Ngo J, Cuthrell N, Wegener M, Rhodes M, Adams M, Sheridan R, et
al: ARID1A mutations promote P300-dependent endometrial invasion
through super-enhancer hyperacetylation. Cell Rep. 33:1083662020.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jiang T, Chen X, Su C, Ren S and Zhou C:
Pan-cancer analysis of ARID1A alterations as biomarkers for
immunotherapy outcomes. J Cancer. 11:776–780. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fontana B, Gallerani G, Salamon I, Pace I,
Roncarati R and Ferracin M: ARID1A in cancer: Friend or foe? Front
Oncol. 13:11362482023. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Guan B, Gao M, Wu CH, Wang TL and Shih
IeM: Functional analysis of in-frame indel ARID1A mutations reveals
new regulatory mechanisms of its tumor suppressor functions.
Neoplasia. 14:986–993. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mullen J, Kato S, Sicklick JK and Kurzrock
R: Targeting ARID1A mutations in cancer. Cancer Treat Rev.
100:1022872021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Somsuan K, Peerapen P, Boonmark W,
Plumworasawat S, Samol R, Sakulsak N and Thongboonkerd V:
ARID1A knockdown triggers epithelial-mesenchymal transition
and carcinogenesis features of renal cells: Role in renal cell
carcinoma. FASEB J. 33:12226–12239. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xu S and Tang C: The role of ARID1A
in tumors: Tumor initiation or tumor suppression? Front Oncol.
11:7451872021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wei XL, Wang DS, Xi SY, Wu WJ, Chen DL,
Zeng ZL, Wang RY, Huang YX, Jin Y, Wang F, et al: Clinicopathologic
and prognostic relevance of ARID1A protein loss in colorectal
cancer. World J Gastroenterol. 20:18404–18412. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ye J, Zhou Y, Weiser MR, Gönen M, Zhang L,
Samdani T, Bacares R, DeLair D, Ivelja S, Vakiani E, et al:
Immunohistochemical detection of ARID1A in colorectal carcinoma:
Loss of staining is associated with sporadic microsatellite
unstable tumors with medullary histology and high TNM stage. Hum
Pathol. 45:2430–2436. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Guan X, Cui L, Ruan Y, Fang L, Dang T,
Zhang Y and Liu C: Heterogeneous expression of ARID1A in colorectal
cancer indicates distinguish immune landscape and efficacy of
immunotherapy. Discov Oncol. 15:922024. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xie C, Fu L, Han Y, Li Q and Wang E:
Decreased ARID1A expression facilitates cell proliferation and
inhibits 5-fluorouracil-induced apoptosis in colorectal carcinoma.
Tumour Biol. 35:7921–7927. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Peerapen P, Sueksakit K, Boonmark W,
Yoodee S and Thongboonkerd V: ARID1A knockdown enhances
carcinogenesis features and aggressiveness of Caco-2 colon cancer
cells: An in vitro cellular mechanism study. J Cancer. 13:373–384.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Baldi S, Zhang Q, Zhang Z, Safi M, Khamgan
H, Wu H, Zhang M, Qian Y, Gao Y, Shopit A, et al: ARID1A
downregulation promotes cell proliferation and migration of colon
cancer via VIM activation and CDH1 suppression. J Cell Mol Med.
26:5984–5997. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Erfani M, Zamani M, Hosseini SY,
Mostafavi-Pour Z, Shafiee SM, Saeidnia M and Mokarram P: ARID1A
regulates E-cadherin expression in colorectal cancer cells: A
promising candidate therapeutic target. Mol Biol Rep. 48:6749–6756.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kwon YW, Jo HS, Bae S, Seo Y, Song P, Song
M and Yoon JH: Application of proteomics in cancer: Recent trends
and approaches for biomarkers discovery. Front Med (Lausanne).
8:7473332021. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Aluksanasuwan S, Somsuan K, Ngoenkam J,
Chiangjong W, Rongjumnong A, Morchang A, Chutipongtanate S and
Pongcharoen S: Knockdown of heat shock protein family D member 1
(HSPD1) in lung cancer cell altered secretome profile and
cancer-associated fibroblast induction. Biochimica Biophys Acta Mol
Cell Res. 1871:1197362024. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Guan B, Wang TL and Shih IeM: ARID1A, a
factor that promotes formation of SWI/SNF-mediated chromatin
remodeling, is a tumor suppressor in gynecologic cancers. Cancer
Res. 71:6718–6727. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Stewart SA, Dykxhoorn DM, Palliser D,
Mizuno H, Yu EY, An DS, Sabatini DM, Chen IS, Hahn WC, Sharp PA, et
al: Lentivirus-delivered stable gene silencing by RNAi in primary
cells. RNA. 9:493–501. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lowry OH, Rosebrough NJ, Farr AL and
Randall RJ: Protein measurement with the Folin phenol reagent. J
Biol Chem. 193:265–275. 1951. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhu W, Smith JW and Huang CM: Mass
spectrometry-based label-free quantitative proteomics. J Biomed
Biotechnol. 2010:8405182010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Neilson KA, Ali NA, Muralidharan S,
Mirzaei M, Mariani M, Assadourian G, Lee A, van Sluyter SC and
Haynes PA: Less label, more free: Approaches in label-free
quantitative mass spectrometry. Proteomics. 11:535–553. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tyanova S, Temu T and Cox J: The MaxQuant
computational platform for mass spectrometry-based shotgun
proteomics. Nat Protoc. 11:2301–2319. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tyanova S, Temu T, Sinitcyn P, Carlson A,
Hein MY, Geiger T, Mann M and Cox J: The perseus computational
platform for comprehensive analysis of (prote)omics data. Nat
Methods. 13:731–740. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Huang da W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhao T and Wang Z: GraphBio: A shiny web
app to easily perform popular visualization analysis for omics
data. Front Genet. 13:9573172022. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Warde-Farley D, Donaldson SL, Comes O,
Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT,
et al: The GeneMANIA prediction server: Biological network
integration for gene prioritization and predicting gene function.
Nucleic Acids Res(Web Server issue). 38:W214–W220. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cancer Genome Atlas Research Network, .
Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA,
Ellrott K, Shmulevich I, Sander C and Stuart JM: The cancer genome
atlas pan-cancer analysis project. Nat Genet. 10:1113–1120. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tang Z, Kang B, Li C, Chen T and Zhang Z:
GEPIA2: An enhanced web server for large-scale expression profiling
and interactive analysis. Nucleic Acids Res. 47:W556–W560. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Koo BK, Spit M, Jordens I, Low TY, Stange
DE, van de Wetering M, van Es JH, Mohammed S, Heck AJ, Maurice MM
and Clevers H: Tumour suppressor RNF43 is a stem-cell E3 ligase
that induces endocytosis of Wnt receptors. Nature. 488:665–669.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sun X, Wang SC, Wei Y, Luo X, Jia Y, Li L,
Gopal P, Zhu M, Nassour I, Chuang JC, et al: Arid1a has
context-dependent oncogenic and tumor suppressor functions in liver
cancer. Cancer Cell. 32:574–589.e6. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Shen J, Ju Z, Zhao W, Wang L, Peng Y, Ge
Z, Nagel ZD, Zou J, Wang C, Kapoor P, et al: ARID1A deficiency
promotes mutability and potentiates therapeutic antitumor immunity
unleashed by immune checkpoint blockade. Nat Med. 24:556–562. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liu X, Dai SK, Liu PP and Liu CM: Arid1a
regulates neural stem/progenitor cell proliferation and
differentiation during cortical development. Cell Prolif.
54:e131242021. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yoodee S, Peerapen P, Plumworasawat S and
Thongboonkerd V: ARID1A knockdown in human endothelial cells
directly induces angiogenesis by regulating angiopoietin-2
secretion and endothelial cell activity. Int J Biol Macromol.
180:1–13. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yoodee S, Peerapen P, Plumworasawat S,
Malaitad T and Thongboonkerd V: Identification and characterization
of ARID1A-interacting proteins in renal tubular cells and their
molecular regulation of angiogenesis. J Transl Med. 21:8622023.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhao H, Ming T, Tang S, Ren S, Yang H, Liu
M, Tao Q and Xu H: Wnt signaling in colorectal cancer: Pathogenic
role and therapeutic target. Mol Cancer. 21:1442022. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lo YH, Kolahi KS, Du Y, Chang CY,
Krokhotin A, Nair A, Sobba WD, Karlsson K, Jones SJ, Longacre TA,
et al: A CRISPR/Cas9-engineered ARID1A-deficient human
gastric cancer organoid model reveals essential and nonessential
modes of oncogenic transformation. Cancer Discov. 11:1562–1581.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Vaicekauskaitė I, Dabkevičienė D, Šimienė
J, Žilovič D, Čiurlienė R, Jarmalaitė S and Sabaliauskaitė R:
ARID1A, NOTCH and WNT signature in gynaecological tumours.
Int J Mol Sci. 24:58542023. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Hiramatsu Y, Fukuda A, Ogawa S, Goto N,
Ikuta K, Tsuda M, Matsumoto Y, Kimura Y, Yoshioka T, Takada Y, et
al: Arid1a is essential for intestinal stem cells through Sox9
regulation. Proc Natl Acad Sci USA. 116:1704–1713. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Heunis T, Lamoliatte F, Marín-Rubio JL,
Dannoura A and Trost M: Technical report: Targeted proteomic
analysis reveals enrichment of atypical ubiquitin chains in
contractile murine tissues. J Proteomics. 229:1039632020.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Hong X, Wang T, Du J, Hong Y, Yang CP,
Xiao W, Li Y, Wang M, Sun H and Deng ZH: ITRAQ-based quantitative
proteomic analysis reveals that VPS35 promotes the expression of
MCM2-7 genes in HeLa cells. Sci Rep. 12:97002022. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Pino JMV, Nishiduka ES, da Luz MHM, Silva
VF, Antunes HKM, Tashima AK, Guedes PLR, de Souza AAL and Lee KS:
Iron-deficient diet induces distinct protein profile related to
energy metabolism in the striatum and hippocampus of adult rats.
Nutr Neurosci. 25:207–218. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhang X, Morikawa K, Mori Y, Zong C, Zhang
L, Garner E, Huang C, Wu W, Chang J, Nagashima D, et al: Proteomic
analysis of liver proteins of mice exposed to 1,2-dichloropropane.
Arch Toxicol. 94:2691–2705. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Van Dross R, Browning PJ and Pelling JC:
Do truncated cyclins contribute to aberrant cyclin expression in
cancer? Cell Cycle. 5:472–477. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wang J, Su W, Zhang T, Zhang S, Lei H, Ma
F, Shi M, Shi W, Xie X and Di C: Aberrant cyclin D1 splicing in
cancer: From molecular mechanism to therapeutic modulation. Cell
Death Dis. 14:2442023. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yu N, Zhu H, Tao Y, Huang Y, Song X, Zhou
Y, Li Y, Pei Q, Tan Q and Pei H: Association between prognostic
survival of human colorectal carcinoma and ZNRF3 expression. Onco
Targets Ther. 9:6679–6687. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Okuda S, Watanabe Y, Moriya Y, Kawano S,
Yamamoto T, Matsumoto M, Takami T, Kobayashi D, Araki N, Yoshizawa
AC, et al: jPOSTrepo: An international standard data repository for
proteomes. Nucleic Acids Res. 45:D1107–D1111. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|