Introduction
Despite advancements in treatment, the prognosis of
patients with epithelial ovarian cancer (EOC), an aggressive
malignant form of cancer, is poor; which may be attributed to the
combined effect of metastasis and multidrug resistance (1–3). The
development of understanding of the underlying mechanism of tumor
growth and development in EOC is important to identify new
diagnostic markers and therapeutic targets.
Non-coding RNAs (ncRNAs) including microRNAs
(miRNAs/miRs) and long non-coding RNAs (lncRNAs), do not have
protein-coding potential, and serve vital roles in the regulation
of cellular processes, such as cellular growth, differentiation and
apoptosis (4–6). Previous studies have reported that
miRNAs and lncRNAs serve crucial roles in tumorigenesis in certain
types of cancer including lung cancer (7) and liver cancer (8) and can function as oncogenes or tumor
suppressor genes (7,8). Certain miRNAs and lncRNAs have also
been reported to be involved in tumor development in EOC (9,10).
One such lncRNA, HOXA-AS2, is located between the
homeobox protein Hox-A3 and homeobox protein Hox-A4 genes in the
homeobox A (HOXA) cluster, and has been reported to promote tumor
progression in prostate cancer (11), bladder cancer (12), papillary thyroid cancer (13), colorectal cancer (14), breast cancer (15), hepatocellular carcinoma (16) and pancreatic cancer (17). However, the expression status,
cellular function and molecular mechanism underlying the role of
HOXA-AS2 in the progression of EOC remains unclear. In the present
study, the role of HOXA-AS2 in the development of EOC was assessed
by a series of molecular and cellular biology methods.
Materials and methods
Tissue samples
EOC tissue samples were collected from patients who
had undergone curative surgical treatment between March 2013 and
March 2014 at the First Hospital of Jilin University (Changchun,
China). All patients provided written informed consent for
participation in the present study. Paired cancerous and
noncancerous tissues (2 cm from the EOC tissues; n=52) were
collected and stored in liquid nitrogen until further use. Two
pathologists independently confirmed the EOC diagnosis. None of the
enrolled patients had received any perioperative treatment, such as
radio-chemotherapy. The clinicopathologic data of the patients with
EOC that participated in the present study are presented (Table I). The present study was compliant
with the Declaration of Helsinki and was approved by the Ethics
Committee of the First Hospital of the Jilin University
(Jlu20210121-1; Changchun, China).
 | Table I.Association of HOXA-AS2 expression
with clinicopathologic factors of patients with epithelial ovarian
cancer. |
Table I.
Association of HOXA-AS2 expression
with clinicopathologic factors of patients with epithelial ovarian
cancer.
|
|
| HOXA-AS2
expression |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
feature | No. of cases | High, n | Low, n | P-value |
|---|
| Age, years |
|
|
| 0.5751 |
|
<60 | 22 | 10 | 12 |
|
|
≥60 | 30 | 17 | 13 |
|
| FIGO stage |
|
|
| 0.0001 |
|
I–II | 37 | 13 | 24 |
|
|
III–IV | 15 | 14 | 1 |
|
| Tumor size |
|
|
| 0.5737 |
| ≤
2 | 33 | 16 | 17 |
|
|
>2 | 19 | 11 | 8 |
|
| Lymph node
metastasis |
|
|
| 0.0067 |
| No | 36 | 14 | 22 |
|
|
Yes | 16 | 13 | 3 |
|
Cell culture and transfection
The human EOC SKOV3 and A2780 cell lines, and human
ovarian surface epithelial cells (HOSEpiCs) were purchased from the
American Type Culture Collection and were grown in RPMI-1640 medium
containing 10% fetal bovine serum (FBS; Gibco; Thermo Fisher
Scientific, Inc.) in a 5% CO2 incubator at 37°C.
Lentivirus-mediated HOXA-AS2 short hairpin RNAs
(sh-HOXA-AS2s) and a non-targeting control shRNA (sh-NC) were
constructed and packaged by Guangzhou Geneseed Biotech. Co., Ltd.
with sequences as follows: sh-HOXA-AS2#1,
5′-GCTTACCTAGAAAGATGTTTCAAGAGAACATCTTTCTAGGTAAGCG-3′;
sh-HOXA-AS2#2,
5′-TTTGCGTCTACAGACCTATCTTCAAGAGAGATAGGTCTGTAGACGCAAAG-3′;
sh-HOXA-AS2#3,
5′-AGTTCAGCTCAAGTTGAACATTCAAGAGATGTTCAACTTGAGCTGAACTC-3′; and
sh-NC, 5′-TTCTCCGAACGTGTCACGTCAAGAGATTACGTGACACGTTCGGAGAA-3′.
SKOV3 cells were plated in 6-well plates and grown
to a cell density of ~60%, and then transfected with sh-HOXA-AS2 or
sh-NC using Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's
instructions.miR-372-3p inhibitor (anti-miR;
5′-GCUCAAAUGUCGCAGCACUUUUU-3′), inhibitor-negative control
(anti-miR-NC; 5′-UUCUCCGAACGUGUCACGUTT-3′), miR-372-3p mimic
(5′-AAAGUGCUGCGACAUUUGAGCGUGCUCAAAUGUCGCAGCACUUUUU-3′) and miR-NC
(5′-ACGUGACACGUUCGGAGAATT-3′) were purchased from MedChemExpress.
Transfection, of 100 nM miR-372 mimics, miR-NC, anti-miR-372 and
anti-miR-NC into SKOV3 cells using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) was performed
according to the manufacturer's protocol. Transfection efficiency
was examined after 48 h using reverse transcription-quantitative
PCR (RT-qPCR).
RT-qPCR
RNA was extracted from EOC samples using TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and reverse
transcribed using the PrimeScript RT reagent kit (Takara Bio, Inc.)
at 25°C for 10 min, 50°C for 45 min and 85°C for 5 min.
Complimentary DNA was quantified using an ABI 7900 qPCR System with
SYBR Green Real-time PCR Master Mix (Takara Bio, Inc.). The
thermocycling conditions used were as follows: Denaturation at 94°C
for 3 min, followed by 40 cycles of denaturation at 94°C for 15
sec, annealing at 60°C for 30 sec and extension at 72°C for 30 sec.
All experiments were performed using previously described primers
(14,18). The primers used were as follows:
miR-372 forward (F), 5′-ACACTCCAGCTGGGAAAGTGCTGCGACATTT-3′ and
reverse (R), 5′-GTGCAGGGTCCGAGGT-3′; HOXA-AS2 F,
5′-CCCGTAGGAAGAACCGATGA-3′ and R, 5′-TTTAGGCCTTCGCAGACAGC-3′; U6 F,
5′-CTCGCTTCGGCAGCACATATACT-3′ and R, 5′-ACGCTTCACGAATTTGCGTGTC-3′;
and GAPDH F, 5′-GGGAAACTGTGGCGTGAT-3′ and R,
5′-GAGTGGGTGTCGCTGTTGA-3′. Relative gene expression levels were
calculated from the data of three independent experiments using the
2–∆∆Cq method (19). U6
was used as the internal reference for miR-372 and GAPDH was used
as the internal reference for HOXA-AS2.
Subcellular fractionation
A PARIS Kit (Thermo Fisher Scientific, Inc.) was
used to separate the nuclear and cytoplasmic fractions of SKOV3
cells according to the manufacturer's protocols. RT-qPCR was then
used to assess HOXA-AS2 expression in these fractions, with U6
serving as a nuclear control and GAPDH as a cytoplasmic control,
according to the aforementioned method.
Cell proliferation assay
Transfected EOC cells (5×103 cells/well)
were seeded and incubated in a 96-well plate for up to 72 h, with
samples being isolated at 3 time points (24, 48 and 72 h) followed
by the addition of CCK-8 solution (10 µl/well, Takara Bio, Inc.)
for 28 h. A spectrophotometer (BioTek Instruments; Agilent
Technologies, Inc.) was then used to measure the absorbance at 450
nm for each time point.
Cell apoptosis assay
The apoptosis of HOXA-AS2-depleted SKOV3 cells was
assessed using Annexin V-FITC/PI apoptosis detection kits (cat. no.
A211; Vazyme Biotech Co., Ltd.) for fluorescence activated cell
sorting on a BD FACSCanto™ II flow cytometer (BD Biosciences)
according to the manufacturer's protocols. The apoptotic rate was
calculated and analyzed using FlowJo 6.10 (FlowJo LLC).
Cell invasion assay
A Transwell invasion plate with a pore size of 8 mm,
which was precoated with Matrigel was used to investigate the
invasion ability of SKOV3 cells. Briefly, the transfected cells
(5×104 cells/well) were added to the top chamber of the
Transwell plate in serum-free medium, while the bottom chamber was
filled with RPMI1640 medium containing 20% FBS (Gibco; Thermo
Fisher Scientific, Inc.). After 24 h, the cells fixed with 4%
paraformaldehyde at 25°C for 30 min and stained with 0.1% crystal
violet (Sigma-Aldrich; Merck KGaA) at 25°C for 10 min. Cells were
imaged and counted in five different fields of view using an X71
inverted light microscope (Olympus Corporation).
Wound healing assay
The transfected cells (5×104 cells/well)
were seeded and cultured in a 6-well plate until confluent. The
monolayer was then scratched with a pipette tip and cultured in a
serum-free medium for 24 h. Baseline (0 h) and 24 h images were
acquired using an X71 inverted light microscope (Olympus
Corporation). Migration rate was calculated by dividing the change
in wound width by the time spent in migration as previously
described by Grada et al (20). ImageJ software (V1.8.0; National
Institutes of Health) was used to measure the size of the
wound.
Bioinformatics and luciferase reporter
assays
The ENCORI database (http://starbase.sysu.edu.cn) was used to analyze the
HOXA-AS2 binding interaction with miRNAs. Among the miRNAs
identified, miR-372, which is known to serve tumor-suppressive
roles in several cancers (18,21,22),
was selected for further study. Wild-type and mutant HOXA-AS2
fragments with/without the binding sites for miR-372 were
synthesized and placed into the psiCHECK2 vector (Promega
Corporation) and were referred to as WT-HOXA-AS2 or MT-HOXA-AS2,
respectively. The SKOV3 cells were cultured in a 12-well plate util
~80% confluent. Then, cells were co-transfected with a luciferase
plasmid and either miR-372 mimics or miR-NC using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). After 24 h, activity was measured using a
Dual-Luciferase Reporter Assay System (Promega Corporation).
Firefly luciferase activity was normalized to Renilla luciferase
activity.
Biotinylated RNA pull-down assay
Biotinylated derivatives of wild-type, mutant or
control miR-372 (Bio-miR-372-WT, Bio-miR-372-MT or Bio-NC,
respectively) were purchased from Guangzhou RiboBio Co., Ltd. The
biotinylated RNA was transfected into SKOV3 cells using
Lipofectamine® 2000 and cultured at 37°C with 5%
CO2 for 48 h based on the manufacturer's instructions.
SKOV3 cells (1×107) were lysed in the soft lysis buffer
plus 80 U/ml RNasin (Promega Corporation). The cell lysate (100 µl)
was then precipitated with M-280 streptavidin beads (Sigma-Aldrich;
Merck KGaA) at 4°C for 12 h. The beads were harvested by
centrifugation at 13,000 × g for 10 min at 4°C. The bound RNAs were
purified using the RNeasy Mini kit (Qiagen GmbH). HOXA-AS2
expression was assessed in the purified RNA using RT-qPCR performed
to the aforementioned method
Statistical analyses
SPSS v19.0 (IBM Corp.) was used to analyze data,
which was presented as the mean ± SD. The χ2 test was
used to assess the relationship between HOXA-AS2 expression and the
clinicopathologic features of patients with EOC. An unpaired
Student's t-test (two-tailed) or ANOVA followed by Bonferroni's
post hoc test were used to assess the statistical significance of
differences. Kaplan-Meier analysis, coupled with the log-rank test,
was performed to investigate the overall survival rate. Pearson's
correlation coefficient was used to assess the correlation between
HOXA-AS2 and miR-372. P<0.05 was considered to indicate a
statistically significant difference.
Results
Elevated HOXA-AS2 in EOC samples is
correlated with poor prognosis
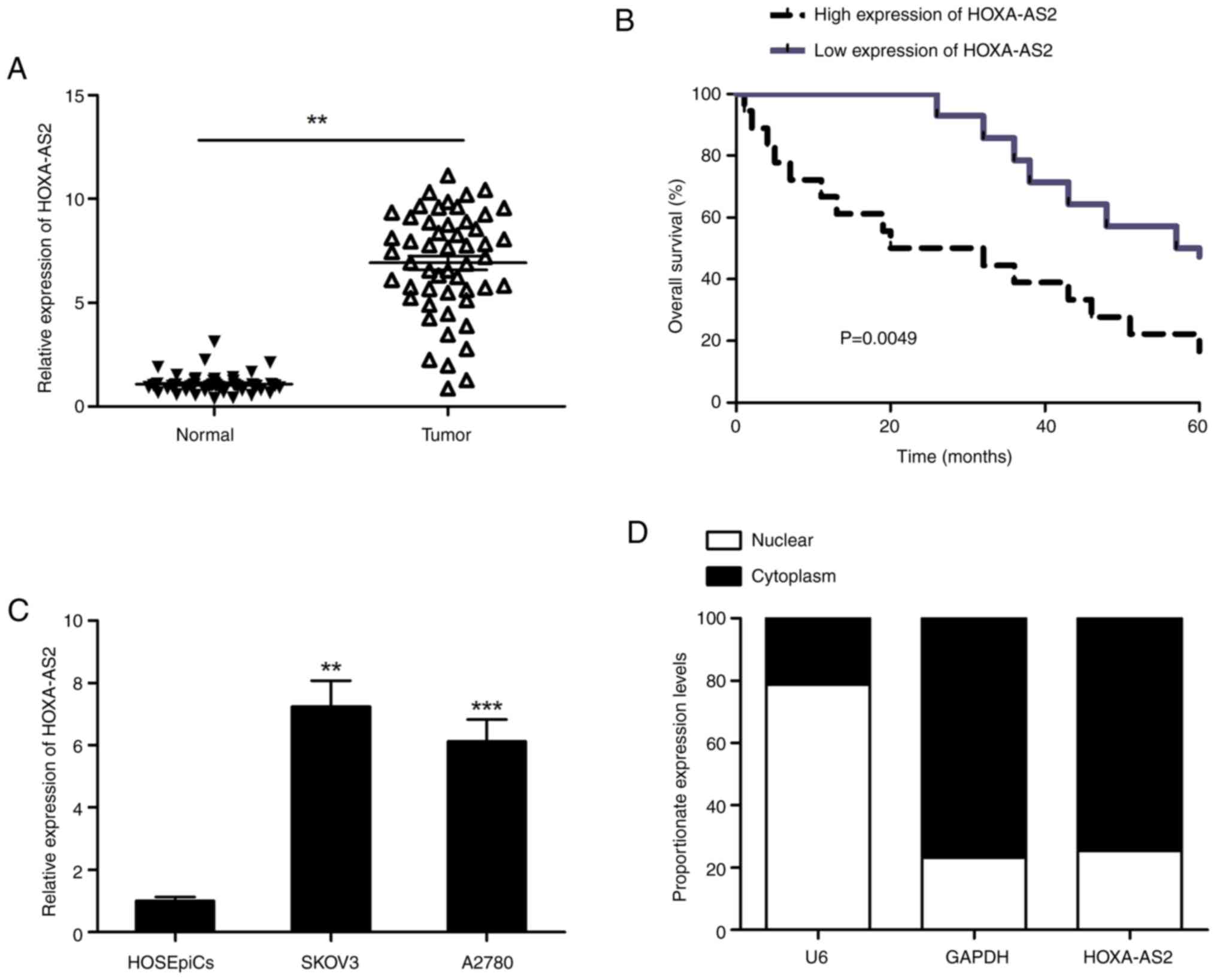
Expression levels of HOXA-AS2 in the EOC tissues
were significantly higher compared with those in the noncancerous
tissues (Fig. 1A). All 52 patients
with EOC were classified into two groups based on the mean
expression level of HOXA-AS2, a high expression level (n=27) and a
low expression level (n=25) group, to assess the relationship
between levels of HOXA-AS2 and clinicopathological features. There
was no significant association between the levels of HOXA-AS2 and,
age or tumor size in the patients with EOC; however, a significant
association was demonstrated between the expression level of
HOXA-AS2 and lymph node metastasis and advanced Federation
International of Gynecology and Obstetrics (FIGO) stage (Table I). Furthermore, Kaplan-Meier
analysis demonstrated that elevated levels of HOXA-AS2 was
associated with reduced overall survival in patients with EOC
(Fig. 1B). Moreover, both EOC cell
lines demonstrated significantly higher HOXA-AS2 expression levels
compared with HOSEpiCs (Fig. 1C).
The SKOV3 cell line showed a notably higher HOXA-AS2 expression
level compared with the A2780 cell line, and was used in subsequent
experiments. The localization of HOXA-AS2 in SKOV3 cells was
assessed, which indicated that HOXA-AS2 was mainly located in the
cytoplasm of SKOV3 cells (Fig.
1D).
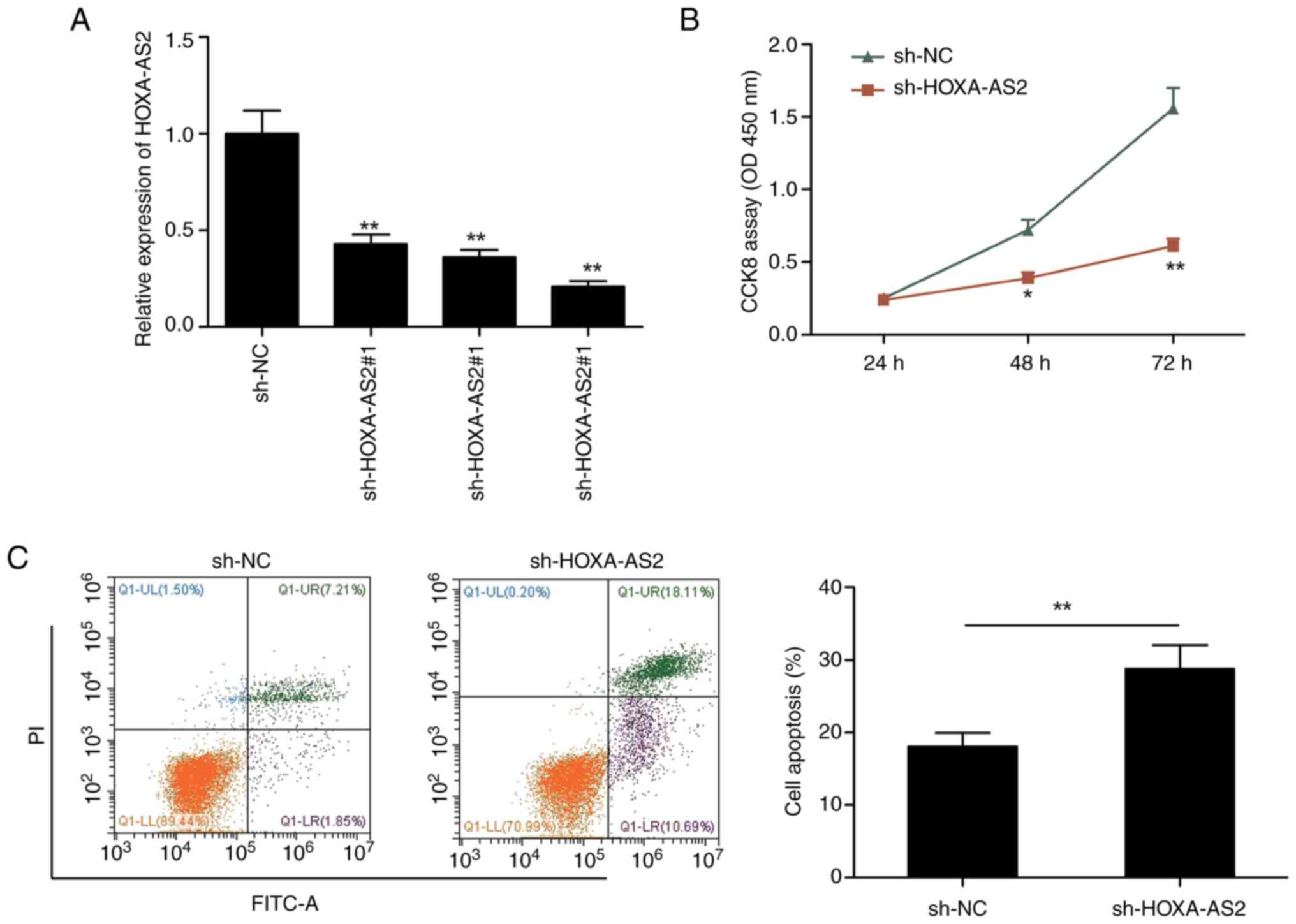
HOXA-AS2 knockdown inhibits EOC cell
proliferation and induces apoptosis
To evaluate the biological role of HOXA-AS2, three
lentivirus-mediated HOXA-AS2 shRNAs (sh-HOXA-AS2#1, sh-HOXA-AS2#2
and sh-HOXA-AS2#3) and a sh-NC were transfected into SKOV3 cells.
Compared with sh-NC, all three shRNAs significantly reduced the
expression of HOXA-AS2 in SKOV3 (Fig.
2A). sh-HOXA-AS2#3 showed the maximum reduction and was used in
all subsequent experiments, where it was labeled as sh-HOXA-AS2.
Furthermore, the CCK-8 assay indicated that the knockdown of
HOXA-AS2 significantly decreased SKOV3 cells proliferation
(Fig. 2B) and significantly induced
cell apoptosis (Fig. 2C) compared
with the sh-NC.
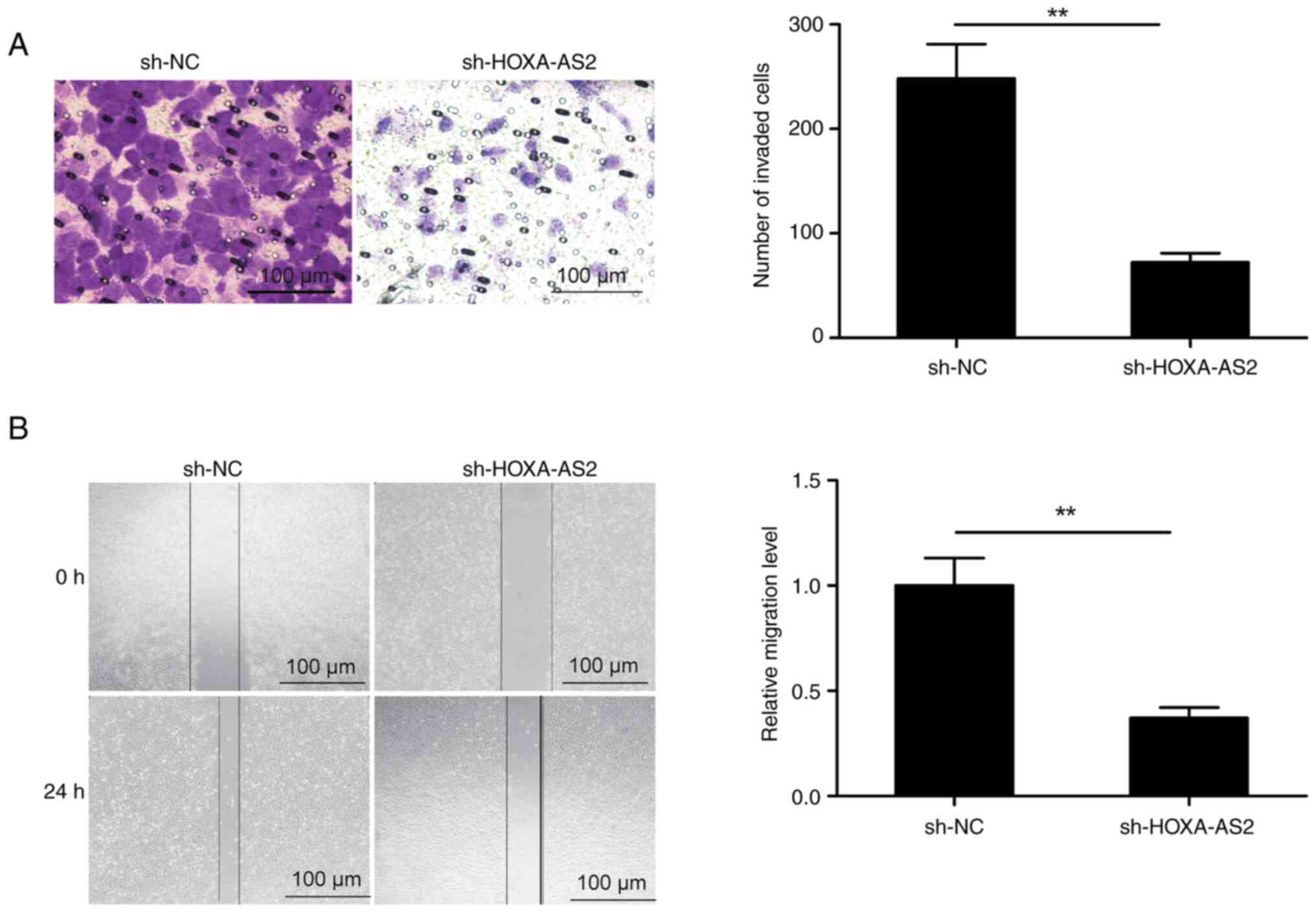
The migration and invasion of the EOC
cells is suppressed by the knockdown of HOXA-AS2
Transwell and wound healing assays were performed to
assess the effect of the knockdown of HOXA-AS2 on the invasion and
migration of SKOV3 cells. The results demonstrated that knockdown
of HOXA-AS2 significantly decreased both cell invasion and
migration in SKOV3 cells compared with the control (Fig. 3).
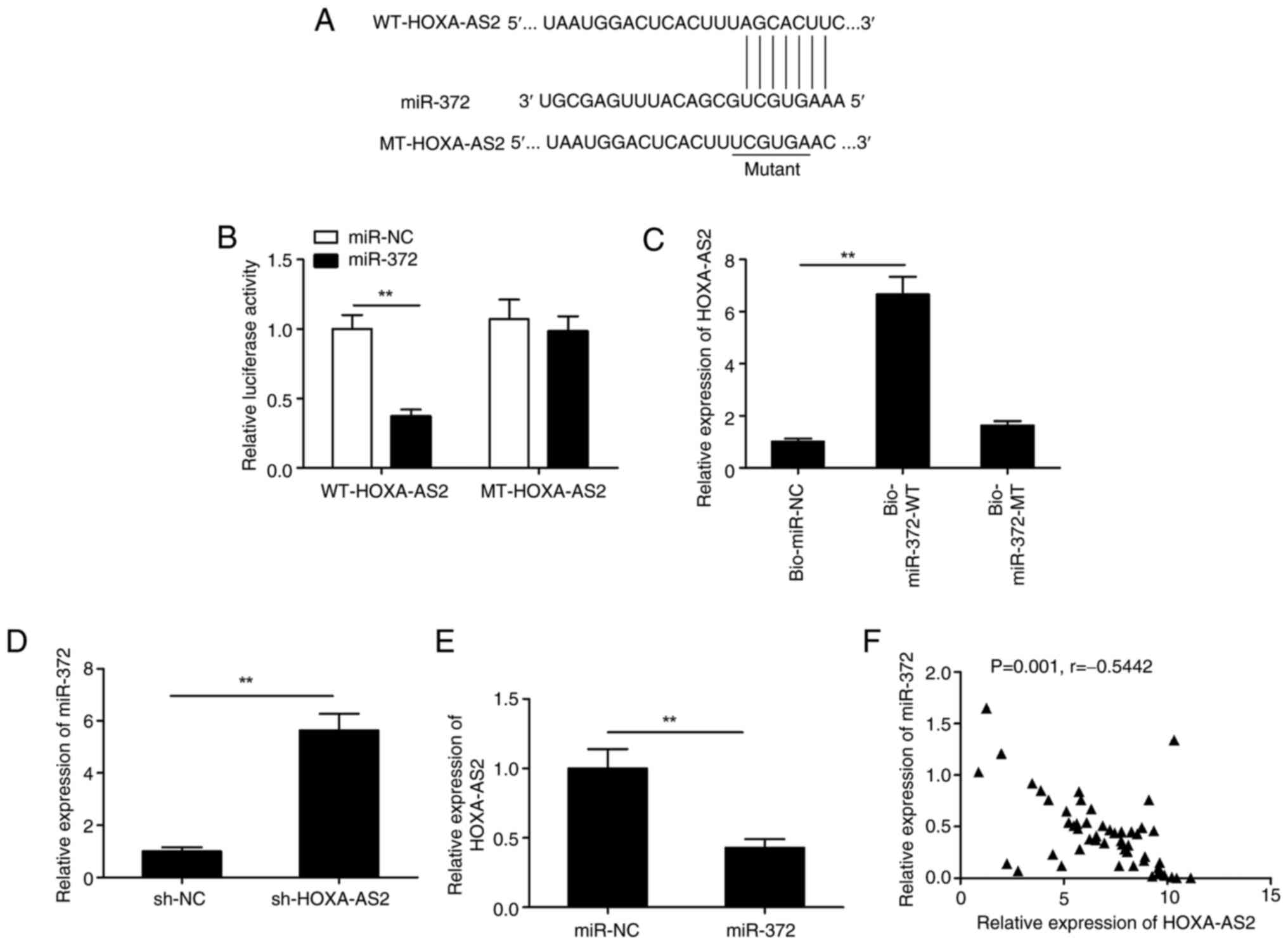
HOXA-AS2 functions as a competing
endogenous RNA (ceRNA) which directly interacts with miR-372 in EOC
cells
It has been previously reported that lncRNAs can
regulate specific miRNAs involved in carcinogenesis by functioning
as ceRNAs (23,24). The ENCORI database was used to
predict the miR-372-HOXA-AS2 interaction. The results indicated
that miR-372 contained a complementary binding sequence to HOXA-AS2
and thus, this was the potential target of HOXA-AS2 (Fig. 4A). This hypothesis was tested using
a luciferase activity assay. Overexpression of miR-372
significantly reduced the luciferase activity of WT-HOXA-AS2
exclusively, with no significant difference demonstrated in
MT-HOXA-AS2 (Fig. 4B). Moreover,
the biotinylated RNA pull-down assay demonstrated that the
bio-miR-372-WT group had significantly higher levels of HOXA-AS2
compared with both the control (bio-NC) and bio-miR-372-MT groups
(Fig. 4C). HOXA-AS2 knockdown
significantly increased the expression of miR-372 in SKOV3 cells
compared with the negative control (Fig. 4D). However, a significant reduction
in the expression of HOXA-AS2 was caused by the overexpression of
miR-372 (Fig. 4E). There was a
significant negative correlation between miR-372 expression and
HOXA-AS2 in EOC tissues (P=0.001; r=−0.5442; Fig. 4F). This indicated that there was a
direct interaction between HOXA-AS2 and miR-372, and thus that
miR-372 functioned as a ceRNA.
Knockdown of HOXA-AS2 inhibits the
progression of EOC through the sponging of miR-372
Since HOXA-AS2 functions as a ceRNA by sponging
miR-372, it was hypothesized that HOXA-AS2 might affect the
progression of EOC by regulating miR-372. Rescue experiments in
SKOV3 cells demonstrated that the knockdown of HOXA-AS2
significantly increased the levels of miR-372 in SKOV3 cells, while
transfection with the miR-372 inhibitor significantly reversed this
effect (Fig. 5A). Furthermore,
downregulation of miR-372 significantly reversed the inhibitory
effects of HOXA-AS2 depletion on cellular proliferation, apoptosis,
invasion and migration in SKOV3 cells (Fig. 5B-E). Thus, HOXA-AS2 depletion
inhibited the progression of EOC through the regulation of
miR-372.
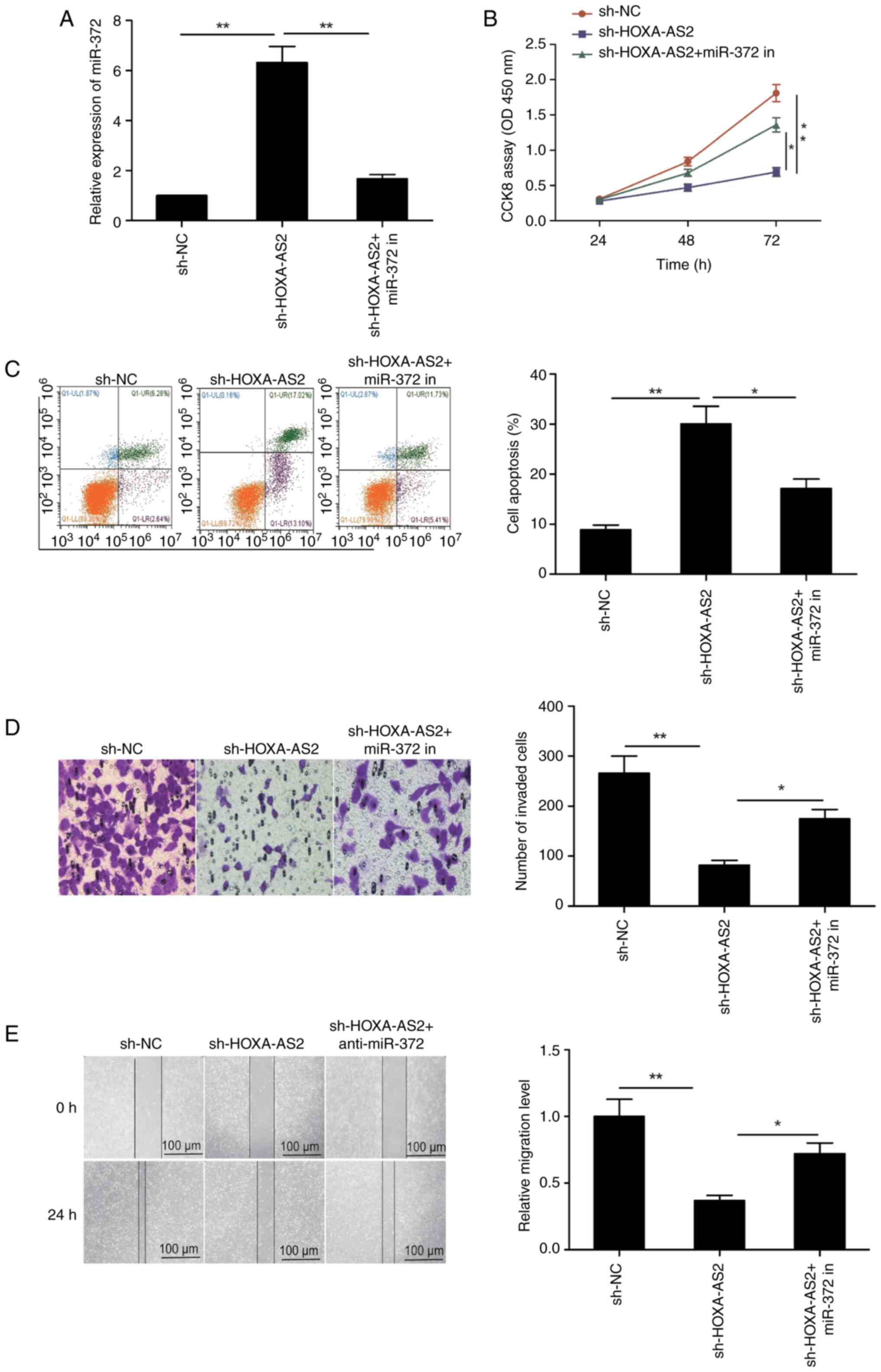 | Figure 5.HOXA-AS2 regulates EOC progression by
sponging miR-372. (A) The expression of miR-372 was examined in
SKOV3 cells transfected with sh-NC, sh-HOXA-AS2 and sh-HOXA-AS2 +
miR-372 in. (B) Cell proliferation, (C) apoptosis, (D) invasion and
(E) migration were assessed in SKOV3 cells transfected with sh-NC,
sh-HOXA-AS2 and sh-HOXA-AS2 + miR-372 in. All results are presented
as mean ± standard deviation from at least three independent
experiments. *P<0.05 and **P<0.01. miR-372 in, miR-372
inhibitor; HOXA-AS2, homeobox A cluster antisense RNA2; sh, short
hairpin RNA; NC, negative control; miR, micro RNA; PI, propidium
iodide. |
Discussion
LncRNAs function as oncogenic genes in tumor
development and are used as diagnostic markers for EOC (9,10).
Wang et al (25) reported
that the lncRNA TP73-AS1 promoted EOC cell proliferation and
metastasis via regulation of MMP2 and MMP9. Similarly, Fang and Xia
(26) reported that the interaction
between lncRNA HLA-F-AS1 and the miR-21-3p/PEG3 axis promoted
cellular growth in EOC, both in vitro and in vivo.
Another study reported that SNHG17 promoted EOC proliferation and
metastasis by regulating the transcription of Forkhead box A1
(27). In the present study, it was
demonstrated that compared with noncancerous tissues and HOSEpiCs,
HOXA-AS2 levels were upregulated in EOC tissues and cell lines, and
that HOX-AS2 levels were associated with advanced FIGO grade and
poor prognosis. It was also demonstrated that HOXA-AS2 served a
tumorigenic role in EOC development through sponging of
miR-372.
The association between abnormal expression of
HOXA-AS2 and carcinogenesis has been extensively studied (11–17).
However, the biological role, especially in migration and invasion,
and regulatory mechanism of HOXA-AS2 in EOC remain unclear. To the
best of our knowledge, the present study is the first to identify
upregulated expression of HOXA-AS2 in EOC samples, which was linked
to its poor prognosis. Moreover, a loss-of-function assay
demonstrated that knockdown of HOXA-AS2-resulted in a decrease in
EOC proliferation and invasion, which indicated that HOXA-AS2
promoted the progression of EOC. LncRNAs are known to exert their
biology role through the ceRNA mechanism by acting as a ‘sponge’
for miRNAs, to regulate their expression and function (23,24).
The role of miRNAs has been reported in the tumorigenesis and
progression of numerous cancers, where they function as oncogenes
and tumor suppressors (28,29). Numerous miRNAs have been reported to
serve key roles in the initiation and development of EOC, and might
act as novel therapeutic targets and clinical biomarkers for EOC
(30,31). HOXA-AS2 acts via the ceRNA mechanism
for certain types of miRNAs, including miR-855-5p (11), miR-509-3p (32), miR-125b (12), miR-15a-3p (13), miR-106a (15) and miR-520c-3p (16). In EOC, miR-372 has been reported to
be downregulated and to function as a tumor suppressor (18). The luciferase reporter assay,
RT-qPCR, and RNA pull-down assays confirmed the binding of miR-372
and HOXA-AS2 in EOC cells. Moreover, there was a negative
correlation between miR-372 levels and HOXA-AS2 in samples from
patients with EOC. Downregulation of HOXA-AS2 mediated the
inhibition of cell growth which was effectively reversed by the
inhibition of miR-372. These results indicated that HOXA-AS2 acted
as an endogenous sponge RNA to inhibit the action of miR-372 in
human EOC cells.
There are two main limitations in the present study.
First, at least one additional cell line should have been used to
assess HOXA-AS2 function in EOC. Second, the molecular mechanism of
HOXA-AS2 in EOC require further investigation
In conclusion, the present study demonstrated that
elevated levels of HOXA-AS2 in EOC tissues and cell lines was
associated with poor prognosis. Mechanistically, HOXA-AS2
facilitated cellular growth in the SKOV3 cells by regulating
miR-372. Further experiments using different EOC cell lines are
required to examine the efficiency of HOXA-AS2 as a novel
therapeutic target for EOC.
Acknowledgements
Not applicable.
Funding
This work was supported by the Program of Science and Technology
Development Plan of Jilin Province (grant no. 20200201466JC).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
WS conceived the study. YW performed the experiments
and wrote the manuscript. WS analyzed the data. WS and YW confirm
the authenticity of all the raw data. Both authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the First Hospital of the Jilin University (approval
no. Jlu20210121-1; Changchun, China) and was in accordance with the
Declaration of Helsinki.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sambasivan S: Epithelial ovarian cancer:
Review article. Cancer Treat Res Commun. 33:1006292022. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lheureux S, Braunstein M and Oza AM:
Epithelial ovarian cancer: Evolution of management in the era of
precision medicine. CA Cancer J Clin. 69:280–304. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nougaret S, McCague C, Tibermacine H,
Vargas HA, Rizzo S and Sala E: Radiomics and radiogenomics in
ovarian cancer: A literature review. Abdom Radiol (NY).
46:2308–2322. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang X, Arai S, Song X, Reichart D, Du K,
Pascual G, Tempst P, Rosenfeld MG, Glass CK and Kurokawa R: Induced
ncRNAs allosterically modify RNA-binding proteins in cis to inhibit
transcription. Nature. 454:126–130. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cech TR and Steitz JA: The noncoding RNA
revolution-trashing old rules to forge new ones. Cell. 157:77–94.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sun T: Long noncoding RNAs act as
regulators of autophagy in cancer. Pharmacol Res. 129:151–155.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Huarte M: The emerging role of lncRNAs in
cancer. Nat Med. 21:1253–1261. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Braga EA, Fridman MV, Moscovtsev AA,
Filippova EA, Dmitriev AA and Kushlinskii NE: LncRNAs in ovarian
cancer progression, metastasis, and main pathways: ceRNA and
alternative mechanisms. Int J Mol Sci. 21:88552020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang JY, Lu AQ and Chen LJ: LncRNAs in
ovarian cancer. Clin Chim Acta. 490:17–27. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang Z, Zhang F, Cai K and Xu J: LncRNA
HOXA-AS2 facilitates prostate cancer progression by inhibiting
miR-885-5p to upregulate KDM5B. Kidney Blood Press Res. 48:45–55.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang F, Wu D, Chen J, Chen S, He F, Fu H,
Wu Q, Liu S, Li X and Wang W: Long non-coding RNA HOXA-AS2 promotes
the migration, invasion and stemness of bladder cancer via
regulating miR-125b/Smad2 axis. Exp Cell Res. 375:1–10. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jiang L, Wu Z, Meng X, Chu X, Huang H and
Xu C: LncRNA HOXA-AS2 facilitates tumorigenesis and progression of
papillary thyroid cancer by modulating the miR-15a-5p/HOXA3 Axis.
Hum Gene Ther. 30:618–631. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ding J, Xie M, Lian Y, Zhu Y, Peng P, Wang
J, Wang L and Wang K: Long noncoding RNA HOXA-AS2 represses P21 and
KLF2 expression transcription by binding with EZH2, LSD1 in
colorectal cancer. Oncogenesis. 6:e2882017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wu J, Li M and Zhang Y: Long noncoding RNA
HOXA-AS2 regulates the expression of SCN3A by sponging miR-106a in
breast cancer. J Cell Biochem. 120:14465–14475. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang Y, Xu J, Zhang S, An J, Zhang J,
Huang J and Jin Y: HOXA-AS2 promotes proliferation and induces
epithelial-mesenchymal transition via the miR-520c-3p/GPC3 Axis in
Hepatocellular Carcinoma. Cell Physiol Biochem. 50:2124–2138. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lian Y, Li Z, Fan Y, Huang Q, Chen J, Liu
W, Xiao C and Xu H: The lncRNA-HOXA-AS2/EZH2/LSD1 oncogene complex
promotes cell proliferation in pancreatic cancer. Am J Transl Res.
9:5496–5506. 2017.PubMed/NCBI
|
|
18
|
Guan X, Zong ZH, Chen S, Sang XB, Wu DD,
Wang LL, Liu Y and Zhao Y: The role of miR-372 in ovarian carcinoma
cell proliferation. Gene. 624:14–20. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta DeltaC(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Grada A, Otero-Vinas M, Prieto-Castrillo
F, Obagi Z and Falanga V: Research techniques made simple: Analysis
of collective cell migration using the wound healing assay. J
Invest Dermatol. 137:e11–e16. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Huang X, Huang M, Kong L and Li Y: miR-372
suppresses tumour proliferation and invasion by targeting IGF2BP1
in renal cell carcinoma. Cell Prolif. 48:593–599. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li Y, Li F, Feng C, Wu T, Chen Y, Shah JA,
Wang F, Cai Y, Wang J and Jin J: MiR-372-3p functions as a tumor
suppressor in colon cancer by targeting MAP3K2. Front Genet.
13:8362562022. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chan JJ and Tay Y: Noncoding RNA:RNA
regulatory networks in cancer. Int J Mol Sci. 19:13102018.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang X, Yang B, She Y and Ye Y: The lncRNA
TP73-AS1 promotes ovarian cancer cell proliferation and metastasis
via modulation of MMP2 and MMP9. J Cell Biochem. 119:7790–7799.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Fang W and Xia Y: LncRNA HLA-F-AS1
attenuates the ovarian cancer development by targeting
miR-21-3p/PEG3 axis. Anticancer Drugs. 33:671–681. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zheng ZJ, Liu Y, Wang HJ, Pang WW and Wang
Y: LncRNA SNHG17 promotes proliferation and invasion of ovarian
cancer cells through up-regulating FOXA1. Eur Rev Med Pharmacol
Sci. 24:9282–9289. 2020.PubMed/NCBI
|
|
28
|
Croce CM and Calin GA: miRNAs, cancer, and
stem cell division. Cell. 122:6–7. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kasinski AL and Slack FJ: Epigenetics and
genetics MicroRNAs en route to the clinic: Progress in validating
and targeting microRNAs for cancer therapy. Nat Rev Cancer.
11:849–864. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
Di Leva G and Croce CM: The role of
microRNAs in the tumori-genesis of ovarian cancer. Front Oncol.
3:1532013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Dahiya N, Sherman-Baust CA, Wang TL,
Davidson B, Shih IeM, Zhang Y, Wood W III, Becker KG and Morin PJ:
MicroRNA expression and identification of putative miRNA targets in
ovarian cancer. PLoS One. 3:e24362008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chen R and He P: Long noncoding RNA
HOXA-AS2 accelerates cervical cancer by the miR-509-3p/BTN3A1 axis.
J Pharm Pharmacol. 73:1387–1396. 2021. View Article : Google Scholar : PubMed/NCBI
|