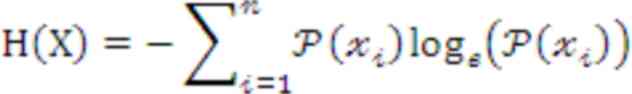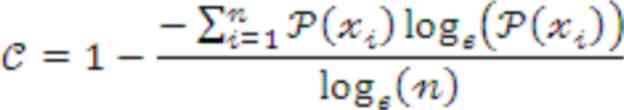Cancer develops due to a series of genetic changes
in normal cells, which causes them to become malignant (1) with the ability to invade surrounding
normal tissues and metastasize. Immunotherapy is a promising
treatment method for tumors. The most studied immunotherapy
involves immune checkpoint inhibitors (ICIs). ICIs function by
targeting specific proteins expressed on immune or cancer cells,
thereby alleviating inhibitory signals that prevent the immune
system from attacking cancer cells and augmenting the immune
response against these cells (2).
Key checkpoint proteins, including cytotoxic
T-lymphocyte-associated protein 4 (CTLA-4), programmed cell death
protein 1 (PD-1) and programmed cell death ligands (PD-Ls), serve
as primary targets for ICIs. By inhibiting these checkpoint
proteins, ICIs enhance the immune-mediated attack on malignant
cells (3). Additionally, various
other immunotherapies, including sipuleucel-T, have been clinically
employed for cancer treatment. Sipuleucel-T is an immunotherapy
that was approved in 2010 for the treatment of advanced prostate
cancer. It is a personalized treatment designed to stimulate the
immune system of the patient, causing it to target and attack
prostate cancer cells (4).
It is important to determine the effect of treatment
and the prognosis after treatment, and biomarkers can help to
achieve these goals. Due to some patients with cancer being unable
to undergo surgery or biopsy, the identification of non-invasive
cancer biomarkers is urgently necessary.
A number of studies have focused on the
characteristics of T cell receptor (TCR) repertoires, and have
demonstrated that they differ among cancer tissues, adjacent
tissues and peripheral blood (5)
and may be different after treatment than they were before
(6). As the characteristics of
certain TCR repertoires are associated with prognosis (7), TCR repertoires have potential as
biomarkers. Therefore, it is important to understand the
characteristics of the TCR repertoire in different locations in the
body and at different time points in patients with cancer.
The immune system comprises adaptive and innate
components. The innate immune system protects the body from general
pathogenic factors, whereas the adaptive immune system targets and
is able to remember specific pathogens (8).
T cells are key components of the adaptive immune
system that recognize pathogens or abnormal peptides and
specifically initiate adaptive immunity. T cells function by either
directly killing infected cells (9)
or releasing cytokines to attract other immune cells. This activity
is triggered when T cells recognize foreign antigens displayed on
the surface of antigen-presenting cells through the major
histocompatibility complex (MHC) (10). This display allows TCRs on T cells
to identify specific peptides or antigen epitopes, which initiates
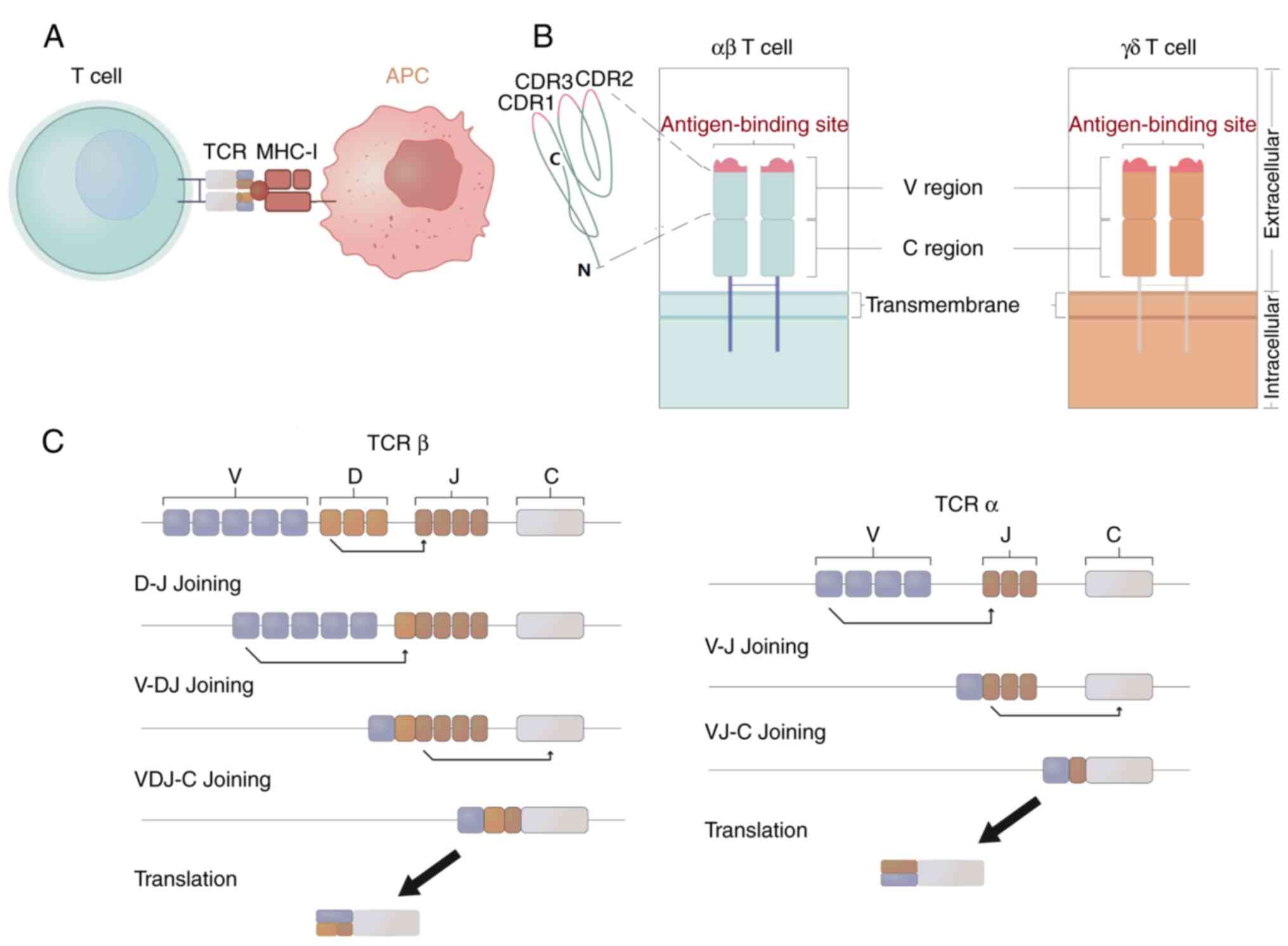
the T-cell response (Fig. 1A). TCRs
are highly diverse heterodimers composed of α and β chains, or γ
and δ chains, encoded by TRA, TRB, TRG and TRD genes, respectively
(Fig. 1B). The ab heterodimers
constitute the majority of these TCRs, accounting for ~95% of the
total number of TCRs, and can identify MHC-presented antigen
peptides or antigen epitopes. By contrast, the γδ TCRs are less
abundant, with only 1–5% of T cells expressing them; moreover, they
are involved in the innate immune response and are not restricted
to antigens presented by MHC molecules. However, the ligands to
which γδ T cells bind are unclear (11). TCR genes contain variable regions (V
regions) and constant regions. The variable region is assembled by
variable (V), diversity (D) and joining (J) gene fragments through
an orderly process known as V(D)J recombination, in which one
allele of each gene fragment is randomly recombined with other gene
fragment alleles to form a functional antigen recognition region
(12,13) (Fig.
1C). The V region of each TCR chain consists of three highly
variable complementary determining regions (CDRs), namely CDR1,
CDR2 and CDR3, CDR3 has the highest variability of these CDRs and
is the most important region for specific antigen recognition
(14) (Fig. 1B).
The CDR3 region is responsible for binding antigenic
peptides presented by MHC molecules (15,16).
Due to its direct interaction with antigenic peptides and high
variability, which enables the recognition of diverse antigens, the
CDR3 region provides a wealth of knowledge about TCR
specificity.
The development of next-generation sequencing (NGS)
has revolutionized the characterization of immune libraries,
allowing for large-scale parallel TCR sequencing. A wide range of
computing and mathematical tools has been created to model and
describe the diversity of these libraries. NGS is advantageous in
that it has greater sequencing depth and quantifies TCR clonal
abundance with markedly higher accuracy than is possible by
spectratyping, in which the number of clonotypes is determined
based on the number of different CDR3 lengths (17). Compared with single-cell TCR
sequencing, population TCR sequencing is more commonly used in the
study of TCR diversity, and facilitates the analysis and comparison
of different repertoires in tumors.
gDNA and RNA are both suitable starting materials
for multiplex PCR, and can be used for multiple rounds of PCR.
These multiple rounds of PCR may introduce sequencing bias and
error, potentially leading to some alleles being more easily
amplified, thus affecting the accuracy of the results (20,21).
However, this can be corrected, e.g., by changing primer
concentrations (22). RNA can also
be analyzed using 5′-RACE (23),
which uses a reverse transcriptase with terminal transferase
activity to reverse transcribe RNA while untemplated sequences,
mainly including deoxycytidine triphosphate are added at the 3′
end. A template switch oligonucleotide containing a complementary
poly(G) strand binds to the 3′-terminal sequence of the first
strand and initiates the chain reaction (24). Almost all the cDNA fragments that
are obtained via this method remained intact. Therefore, only a
pair of primers are required for subsequent template amplification
to achieve complete amplification of the possible V gene.
In the analysis of the TCR diversity index, metrics
such as Shannon entropy and clonality are commonly used to quantify
TCR diversity and the expansion of specific TCR clones within a
sample. Shannon entropy effectively quantifies the complexity and
breadth of the TCR repertoire by evaluating how uniformly the
different TCR variants are distributed within the T-cell
population. The formula introduces the TCR diversity index, H(X),
as a measure of TCR diversity, as follows:
In this formula, the variable C is a metric for the
assessment of TCR clonality, which is confined to the range of 0 to
1. Essentially, TCR clonality is the extent of proliferation
exhibited by specific TCR variants within a sample relative to
others. A higher value of C signifies elevated clonality, which is
concomitant with diminished diversity (27).
Numerous studies have demonstrated that the TCR
repertoire diversity in the peripheral blood or tumor tissues of
patients with cancer has its own specific characteristics compared
with that in healthy individuals, and may be useful in the
diagnosis of various cancers.
Through the TCRβ sequencing and analysis of cancer
tissues and distant noncancerous tissues from 15 patients with lung
cancer, Wang et al (30)
demonstrated that the TCR repertoire diversity in lung cancer
tissues was significantly greater than that in distant normal lung
tissues. However, in patients with non-small cell lung cancer
(NSCLC), Song et al (31)
found that the diversity of the TCR repertoire revealed by
high-throughput sequencing was not different from that of adjacent
non-tumor tissues. The reason for these inconsistent results may be
that the tumor microenvironment (TME) in lung cancer tissues
differs among individuals, thus resulting in differences in the TCR
repertoire diversity among cancerous tissues.
Changes in the TCR repertoire diversity in tumorous
or normal tissues have also been observed in other types of
carcinoma. In a recent study, high-throughput TCR sequencing
demonstrated that the TCR repertoire diversity of HCC tumor tissues
and peritumoral tissues was similar (5). This differed from the results of one
previous study, in which the TCR repertoire diversity in HCC tumor
tissues was higher than that in adjacent non-tumor tissues
(32), but consistent with another,
in which no significant difference in TCR repertoire diversity was
identified between HCC tumor tissues and adjacent normal tissues
(33). Sherwood et al
(34) found that the diversity of
the TCR repertoire in tumor tissues was much lower than that in
peripheral blood in a study of patients with colorectal cancer.
Similarly, the TCR repertoire diversity in bladder cancer tissues
(35) and in penile squamous cell
carcinoma (PSCC) (36) was found to
be lower than that in adjacent normal tissues. By contrast, the
diversity of the TCR repertoire in breast cancer tissues has been
demonstrated to be greater than that in adjacent normal tissues
(37). Moreover, the TCR repertoire
diversity in oesophageal squamous cell carcinoma was revealed to be
not significantly different from that in peripheral blood and
adjacent normal tissues (38).
Information on the changes in TCR repertoire observed in different
cancers are summarized in Tables I
and II.
The TCR repertoire diversity in the peripheral blood
or tumor tissues of patients with tumors has been shown to be
inconsistent compared with that in healthy individuals or adjacent
normal tissues. Whether these characteristics of TCR repertoire
diversity in tumors have the potential to be used as markers for
tumor diagnosis requires further validation.
Tumor immunotherapy has profoundly advanced cancer
research and effectively improved the prognosis of patients with
cancer (39,40), and the success of ICIs underscores
the importance of the anticancer immune response in patients. The
expression of biomarkers, such as PD-Ls, in cancer tissues has been
shown to have an marked effect on the clinical response to ICIs.
However, as tumor tissue cannot be biopsied during treatment, new
biomarkers of the response to ICIs are necessary to determine the
efficacy of clinical treatment. Blood samples are becoming more
widely used because they are relatively easy and non-invasive to
obtain compared with biopsies of tumor tissue. In addition, as
numerous studies have shown that the TCR repertoire in the
peripheral blood can change significantly during tumor treatment,
such changes may be used as a biomarker of ICI response.
In an early study, it was reported that the TCR
repertoire diversity increased after anti-CTLA-4 treatment and
improved the survival of patients with metastatic
castration-resistant prostate cancer or metastatic melanoma
(41). Consistent with this, an
increase in TCR repertoire diversity was also observed after
anti-CTLA-4 treatment in another study of patients with metastatic
melanoma (6). This may be
attributed to the anti-CTLA-4 treatment promoting reconstruction of
the TCR lineage and increasing its diversity (42). By contrast, the diversity of the TCR
repertoire in peripheral blood has not been found to change
significantly after anti-PD-1 therapy (43–45);
however, increased TCR clonality has been observed in the
peripheral blood of patients with melanoma treated with anti-PD-1
therapy (44,45). In addition, Kato et al
(46) reported that after anti-PD-1
treatment, the clonality of the TCR repertoire in the peripheral
blood of patients with advanced renal cell carcinoma (RCC)
increased; however, the diversity of the TCR repertoire
decreased.
Changes in the TCR repertoire have been observed
after other treatments, including radiotherapy and chemotherapy.
Liu et al (47) reported
that the diversity of the TCR repertoire in the peripheral blood of
patients with lung cancer was significantly lower than that in
healthy individuals, with the TCR repertoire diversity decreasing
after chemotherapy, radiotherapy, tyrosine kinase inhibitor therapy
and/or antiangiogenic therapy. Similarly, the TCR repertoire
diversity in the peripheral blood was found to decrease after
FOLFIRI chemotherapy with bevacizumab or cetuximab in most patients
with metastatic colorectal cancer (48), and in patients with prostate cancer
after sipuleucel-T therapy (49).
The TCR repertoire of tumor-infiltrating lymphocytes
(TILs) also undergoes changes. TILs are immune cells located in the
TME that are crucial in the immune response against tumors
(50,51). In tumors, high infiltration levels
of TILs, particularly cytotoxic CD8+ T cells which have
the ability to kill tumor cells, are often associated with an
improved prognosis (52). TILs can
be used in the assessment of tumor response to immunotherapy.
Tumors rich in TILs, known as ‘hot’ tumors, are more likely to
respond to immunotherapy than are ‘cold’ tumors in which TILs are
scarce (53). In a study of
patients with rectal cancer who responded well to radiotherapy and
chemotherapy, the TCR repertoire diversity of TILs was observed to
increase after treatment (54).
However, in a study of patients with head and neck squamous cell
carcinoma, the TCR repertoire diversity of TILs was shown to be
reduced, while the TCR clonotypes in TILs were expanded after
cetuximab treatment (Table III)
(55). Therefore, changes in the
TCR repertoire of TILs with regard to diversity, clonality or
clonotypes may serve as predictive therapeutic markers.
A high TCR repertoire diversity in peripheral blood
is associated with an improved immune response in numerous types of
tumors (56). Hopkins et al
(57) reported that the diversity
of the TCR repertoire in the peripheral blood of patients with PDAC
increased after treatment. Moreover, in patients receiving
anti-CTLA-4 treatment, patients whose TCR repertoire diversity
increased after treatment exhibited an improved therapeutic
response; however, this was not observed in patients receiving
anti-PD-1 treatment. In patients with advanced melanoma, a highly
diverse TCR repertoire in the peripheral blood before treatment was
found to be associated with an improved therapeutic effect of
anti-CTLA-4 treatment (58,59). Notably, similar findings have also
been reported for patients with other types of cancer treated with
anti-PD-1 therapy. For example, in a study of gastrointestinal
tumors, anti-PD-1 treatment exhibited improved therapeutic effects
in patients with a higher baseline TCR repertoire diversity in the
peripheral blood (43). In
addition, patients with NSCLC with high TCR repertoire diversity in
the peripheral blood exhibited a superior therapeutic response to
anti-PD-1 treatment (60,61). Also, patients with lung cancer who
had a greater TCR repertoire diversity in the peripheral blood
before treatment exhibited a more favorable response to
radiotherapy and chemotherapy (Table
IV) (47).
The aforementioned studies demonstrate that a wider
range of TCR repertoires before ICI immunotherapy is largely
associated with improved clinical outcomes in patients with cancer.
This may indicate that increasing the diversity of the TCR
repertoire is beneficial to patients receiving ICI immunotherapy.
This may be due to the increase in the diversity or clonality of
the TCR repertoire, or due to the expansion of the TCR repertoire
induced after ICI immunotherapy improving existing immunity, thus
guaranteeing the effectiveness of immunotherapy (62). However, TCR repertoire
characteristics alone are not sufficient to determine the response
to tumor immunotherapy. First, tumor cells are genetically
heterogeneous and undergo changes as the cancer progresses
(63), which results in inability
of the TCR repertoire to recognize all tumor cell variants. In
addition, high-affinity TCRs may also recognize antigens expressed
in healthy tissues, trigger non-targeted effects and cause
autoimmune diseases (64). Second,
in addition to the TCR repertoire, PD-1, PD-Ls, tumor mutational
burden (65,66) and microsatellite instability
(66,67) have also been shown to serve as
biomarkers for tumor immunotherapy, including ICIs (68,69),
chimeric antigen receptor (CAR) T-cell therapies (70), or CAR T cells combined with
chemotherapy, radiotherapy or angiogenesis inhibitors (71). A comprehensive treatment strategy
may combine the complementary effects of different immunotherapy
methods to obtain an improved therapeutic effect. Therefore,
consideration of the characteristics of the TCR repertoire in
combination with those ICIs or other tumor immunotherapies may
promote the further development of tumor immunotherapy.
There is evidence to suggest that the diversity of
the TCR repertoire can be used as a prognostic biomarker of the
immune response to tumors. For example, an increased TCR repertoire
diversity in PDAC tissue was found to be associated with an
improved prognosis (72). However,
different findings were reported for muscle-invasive bladder cancer
(MIBC); specifically, patients with low TCRβ chain diversity,
associated with oligoclonal TIL expansion, had longer
recurrence-free survival (73).
Although the patients with MIBC had low TCRβ chain diversity, they
also exhibited oligoclonal TIL expansion and high number of
neoantigens, which improved their prognosis (73). In patients with nasopharyngeal
carcinoma, a lower TCR repertoire diversity in tumor tissues than
in paired adjacent normal tissues was found to be associated with a
poor prognosis. Lower diversity in the tumor may indicate that T
cells were hindered from infiltrating or inducing apoptosis in the
TME, thus suggesting that the T-cell immune response was
insufficient, and that the patient may benefit from checkpoint
blockade treatment (74). In a
study conducted by Valpione et al (75), an increase in the baseline TCR
repertoire diversity of tumor-infiltrating T cells was found to be
associated with an improved prognosis in patients with various
cancers, including breast cancer, melanoma, lung cancer and
RCC.
A number of studies have shown that the diversity of
the TCR repertoire in peripheral blood is able to predict tumor
prognosis. In a study of patients with lung cancer, a lower
diversity of the TCR repertoire in the peripheral blood after
treatment was found to be associated with a poor prognosis
(47). However, in other studies of
lung cancer, those patients with a high TCR repertoire diversity in
the peripheral blood had an improved prognosis (76–79).
In patients with gastrointestinal tumors, a high diversity of the
TCR repertoire in the peripheral blood after anti-PD-1 treatment
was also found to be associated with a good prognosis (43). Similarly, in metastatic colorectal
cancer, a high TCR repertoire diversity in the peripheral blood
exhibited an association with a good prognosis (48). Also, in patients with melanoma, a
low diversity of the peripheral blood TCR repertoire was associated
with a poor prognosis (7). Similar
findings have been reported in patients with breast cancer
(80), where a low TCR repertoire
diversity in the peripheral blood was associated with poor overall
survival. In addition, Yan et al (81) reported that for patients with
esophageal squamous cell carcinoma, a greater TCR repertoire
diversity in the peripheral blood was associated with an improved
prognosis, while high TCR repertoire clonality was associated with
a poor prognosis (Table V).
TCR repertoire diversity has been found to be
associated with prognosis in other tumors. In patients with
high-grade serous ovarian cancer (HGSOC), the diversity of the TCR
repertoire was low in patients who experienced recurrence.
Therefore, it was speculated that patients with high TCR repertoire
diversity have an improved prognosis (82). In MIBC, basal/squamous-like and
stroma-rich subtypes exhibited high TCR richness and diversity,
which were associated with a good prognosis. However, no changes in
TCR richness or diversity were observed for the luminal subtype
(83).
In addition to diversity, other characteristics of
the TCR repertoire have been found to be prognostically useful. For
example, in a study of HCC, similarity of the TCR repertoire
between tumor and adjacent normal tissues was indicated to be
associated with an improved prognosis (33). However, in patients with NSCLC, high
TCR repertoire clonality in the tumor compared with that in
adjacent normal tissues was found to be associated with a good
prognosis (60,77). A similar finding was reported in a
study of HGSOC (84). In patients
with advanced RCC, those with elevated TCR repertoire clonality
after anti-PD-1 treatment exhibited a good prognosis (46), while in patients with CC, fewer
clonal types in the sentinel lymph node TCR repertoire was
associated with a poor prognosis (28) (Table
VI). Notably, TCR convergence, where T cells share identical
TCRs with an identical amino acid sequence yet have different DNA
sequences due to codon degeneracy, also has an association with
tumor prognosis, with a greater TCR convergence corresponding to an
improved prognosis (85).
Therefore, TCR repertoire diversity is a promising tumor prognostic
marker. However, the relationships between other characteristics of
the TCR repertoire and prognosis require verification in studies
involving large numbers of patients.
The diversity distribution of TCR repertoires in
peripheral blood mononuclear cells (PBMCs) and tumor infiltrates,
and their association with prognosis is disparate for tumors of
different types. The diversity of a TCR repertoire is determined by
the number of different clonotypes (richness) and the relative
abundance of these clonotypes (evenness) in a sample (86). All clonotypes derived from clonal
expansion carry an identical CDR3 in which the V-region used during
V(D)J recombination which depends on subnuclear relocation of the
rearranging TCR loci (tr), DNA methylation status,
recruitment of chromatin remodelling enzymes, histone modification
and germline transcription as well as spatial and temporal
regulation in different subtypes of cancers, and then influence the
number and abundance of clonotypes (87). Additionally, exposure to certain
antigens influences the expansion of specific TCR clonotypes.
Therefore, the TCR repertoires of different tissues and tumors are
dynamic, and dependent on factors such as age and antigen exposure
(87). Tumor antigens include
tumor-associated and tumor-specific antigens; the latter are only
expressed in tumor tissues and activate specific T cells, which
conditions TCR repertoire diversity. Peripheral blood TCR diversity
is closely associated with the global transcriptomes of peripheral
blood and the intratumor microenvironment mainly referred to the
number of tumor-specific lymphocytes which spread to peripheral
blood (88). Additionally,
peripheral blood TCR diversity decreases while the clonality
increases with age (89). This may
explain the discrepancy between TCR repertoires in PBMCs and tumor
infiltrates.
The importance of the TCR repertoire in tumor
immunity has become increasingly apparent. In the present review,
TCR structures are summarized. TCR repertoire characteristics are
potentially useful as tools for the diagnosis of tumors, the
enhancement of tumor immunity and the prediction of tumor
prognosis. The TCR repertoire holds promise as a biomarker, and
patients are expected to benefit from research into this
repertoire. However, compared with the immunohistochemical
detection of PD-Ls and targeted sequencing of liquid biopsies,
which allows for the detection of tumor-derived DNA or circulating
tumor cells that are present in the bloodstream or other bodily
fluids, TCR repertoire analysis is complex and expensive. It
comprises multiple steps, including the preparation of blood or
tissue samples, TCR sequencing, data analysis and interpretation,
which require professional and technical personnel, high-throughput
sequencing platforms and bioinformatics tools. Moreover, the volume
of data generated by TCR sequencing is substantial, and the
identification of specific TCRs for cancer therapy is time
consuming (19). Novel prediction
models have been developed to identify epitope-specific TCRs
(90). In addition, more sensitive
and cost-effective sequencing tools have been developed, including
characterizing TCR repertoires (91), spatially resolved TCR sequencing
(92), and formalin-fixed
paraffin-embedded-suitable unique molecular identifier-based-TCR
sequencing (93). Tools such as
these are expected to further utilize and maximize the value of TCR
repertoire analysis. Although TCR analysis indicators, such as
Shannon entropy and clonality, can help in understanding the
relationships between TCR clonality and solid tumors, the
associations between TCR repertoires and clinical benefits require
further investigation. Nevertheless, the TCR repertoire can be used
as a tumor immune biomarker, with potential clinical significance
in the prediction of patient prognosis and the monitoring of
therapeutic efficacy. In the future, more advanced sequencing,
database construction techniques and comprehensive analysis
algorithms are likely to be developed to further simplify and
clarify the evaluation and comparison of TCR repertoires, reduce
the cost of TCR repertoire analysis, and allow more people to
benefit.
In addition, despite the extreme diversity of TCR
chain pairs, the specific antigens recognized by the TCRs of γδ T
cells remain largely unknown (94).
Therefore, more research into γδ T cells is necessary. Studies have
shown that γδ T cells play an important role in tumor immunotherapy
(95–98) and can directly attack tumor cells
without relying on MHC for antigen presentation. Therefore, in the
future, in addition to further study of αβ T cells, more attention
should be focused on γδT cells. The assessment of these cells may
promote the further development of tumor immunotherapy.
Not applicable.
This study was supported by a fund from the Science and
Technology Cooperation and Exchange Special Project of Shanxi
Province (grant no. 202104041101012).
Not applicable.
ALH drafted the manuscript and was responsible for
drawing the figure and preparing the tables. YZH and YY were
responsible for the acquisition, analysis and interpretation of
data. GPZ, HLW and MP reviewed and revised the manuscript. All
authors read and approved the final version of the manuscript. Data
authentication is not applicable.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Pasetto A and Lu YC: Single-cell TCR and
transcriptome analysis: An indispensable tool for studying T-cell
biology and cancer immunotherapy. Front Immunol. 12:6890912021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Papadakis M, Karniadakis I, Mazonakis N,
Akinosoglou K, Tsioutis C and Spernovasilis N: Immune checkpoint
inhibitors and infection: What is the interplay? In Vivo.
37:2409–2420. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kong X, Zhang J, Chen S, Wang X, Xi Q,
Shen H and Zhang R: Immune checkpoint inhibitors: Breakthroughs in
cancer treatment. Cancer Biol Med. j.issn.2095-3941.2024.0055.
2024.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kantoff PW, Higano CS, Shore ND, Berger
ER, Small EJ, Penson DF, Redfern CH, Ferrari AC, Dreicer R, Sims
RB, et al: Sipuleucel-T immunotherapy for castration-resistant
prostate cancer. N Engl J Med. 363:411–422. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang Z, Zhong Y, Zhang Z, Zhou K, Huang Z,
Yu H, Liu L, Liu S, Yang H, Zhou J, et al: Characteristics and
clinical significance of T-cell receptor repertoire in
hepatocellular carcinoma. Front Immunol. 13:8472632022. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Robert L, Tsoi J, Wang X, Emerson R, Homet
B, Chodon T, Mok S, Huang RR, Cochran AJ, Comin-Anduix B, et al:
CTLA4 blockade broadens the peripheral T-cell receptor repertoire.
Clin Cancer Res. 20:2424–2432. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Charles J, Mouret S, Challende I, Leccia
MT, De Fraipont F, Perez S, Plantier N, Plumas J, Manuel M,
Chaperot L and Aspord C: T-cell receptor diversity as a prognostic
biomarker in melanoma patients. Pigment Cell Melanoma Res.
33:612–624. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chaplin DD: Overview of the immune
response. J Allergy Clin Immunol. 125 (2 Suppl 2):S3–S23. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bonilla FA and Oettgen HC: Adaptive
immunity. J Allergy Clin Immunol. 125 (2 Suppl 2):S33–S40. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gielis S, Moris P, Bittremieux W, De
Neuter N, Ogunjimi B, Laukens K and Meysman P: Detection of
enriched T cell epitope specificity in full T cell receptor
sequence repertoires. Front Immunol. 10:28202019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lo Presti E, Dieli F and Meraviglia S:
Tumor-infiltrating γδ T lymphocytes: pathogenic role, clinical
significance, and differential programing in the tumor
microenvironment. Front Immunol. 5:6072014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Roth DB: V(D)J recombination: Mechanism,
errors, and fidelity. Microbiol Spectr.
2:10.1128/microbiolspec.MDNA3. 0041–2014. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Schatz DG and Ji Y: Recombination centres
and the orchestration of V(D)J recombination. Nat Rev Immunol.
11:251–263. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mahe E, Pugh T and Kamel-Reid S: T cell
clonality assessment: Past, present and future. J Clin Pathol.
71:195–200. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Rosati E, Dowds CM, Liaskou E, Henriksen
EKK, Karlsen TH and Franke A: Overview of methodologies for T-cell
receptor repertoire analysis. BMC Biotechnol. 17:612017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Rossjohn J, Gras S, Miles JJ, Turner SJ,
Godfrey DI and McCluskey J: T cell antigen receptor recognition of
antigen-presenting molecules. Annu Rev Immunol. 33:169–200. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Laydon DJ, Bangham CRM and Asquith B:
Estimating T-cell repertoire diversity: Limitations of classical
estimators and a new approach. Philos Trans R Soc Lond B Biol Sci.
370:201402912015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Woodsworth DJ, Castellarin M and Holt RA:
Sequence analysis of T-cell repertoires in health and disease.
Genome Med. 5:982013. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Pai JA and Satpathy AT: High-throughput
and single-cell T cell receptor sequencing technologies. Nat
Methods. 18:881–892. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shugay M, Britanova OV, Merzlyak EM,
Turchaninova MA, Mamedov IZ, Tuganbaev TR, Bolotin DA, Staroverov
DB, Putintseva EV, Plevova K, et al: Towards error-free profiling
of immune repertoires. Nat Methods. 11:653–655. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu X, Zhang W, Zeng X, Zhang R, Du Y,
Hong X, Cao H, Su Z, Wang C, Wu J, et al: Systematic comparative
evaluation of methods for investigating the TCRβ repertoire. PLoS
One. 11:e01524642016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Carlson CS, Emerson RO, Sherwood AM,
Desmarais C, Chung MW, Parsons JM, Steen MS,
LaMadrid-Herrmannsfeldt MA, Williamson DW, Livingston RJ, et al:
Using synthetic templates to design an unbiased multiplex PCR
assay. Nat Commun. 4:26802013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
No authors listed. Rapid amplification of
5′ complementary DNA ends (5′ RACE). Nat Methods. 2:629–630. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhu YY, Machleder EM, Chenchik A, Li R and
Siebert PD: Reverse transcriptase template switching: A SMART
approach for full-length cDNA library construction. Biotechniques.
30:892–897. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Stewart JJ, Lee CY, Ibrahim S, Watts P,
Shlomchik M, Weigert M and Litwin S: A Shannon entropy analysis of
immunoglobulin and T cell receptor. Mol Immunol. 34:1067–1082.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shannon CE: A mathematical theory of
communication. Bell Syst Tech J. 27:379–423. 1948. View Article : Google Scholar
|
|
27
|
Pielou EC: The measurement of diversity in
different types of biological collections. J Theor Biol.
13:131–144. 1966. View Article : Google Scholar
|
|
28
|
Cui JH, Lin KR, Yuan SH, Jin YB, Chen XP,
Su XK, Jiang J, Pan YM, Mao SL, Mao XF and Luo W: TCR repertoire as
a novel indicator for immune monitoring and prognosis assessment of
patients with cervical cancer. Front Immunol. 9:27292018.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Pan Y, Gao J, Lin J, Ma Y, Hou Z, Lin Y,
Wen S, Pan M, Lu F and Huang H: High-dimensional single-cell
analysis unveils distinct immune signatures of peripheral blood in
patients with pancreatic ductal adenocarcinoma. Front Endocrinol
(Lausanne). 14:11815382023. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang X, Zhang B, Yang Y, Zhu J, Cheng S,
Mao Y, Feng L and Xiao T: Characterization of distinct T cell
receptor repertoires in tumor and distant non-tumor tissues from
lung cancer patients. Genomics Proteomics Bioinformatics.
17:287–296. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Song Z, Chen X, Shi Y, Huang R, Wang W,
Zhu K, Lin S, Wang M, Tian G, Yang J and Chen G: Evaluating the
potential of t cell receptor repertoires in predicting the
prognosis of resectable non-small cell lung cancers. Mol Ther
Methods Clin Dev. 18:73–83. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chen Y, Xu Y, Zhao M, Liu Y, Gong M, Xie
C, Wu H and Wang Z: High-throughput T cell receptor sequencing
reveals distinct repertoires between tumor and adjacent non-tumor
tissues in HBV-associated HCC. Oncoimmunology. 5:e12190102016.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lin KR, Deng FW, Jin YB, Chen XP, Pan YM,
Cui JH, You ZX, Chen HW and Luo W: T cell receptor repertoire
profiling predicts the prognosis of HBV-associated hepatocellular
carcinoma. Cancer Med. 7:3755–3762. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sherwood AM, Emerson RO, Scherer D,
Habermann N, Buck K, Staffa J, Desmarais C, Halama N, Jaeger D,
Schirmacher P, et al: Tumor-infiltrating lymphocytes in colorectal
tumors display a diversity of T cell receptor sequences that differ
from the T cells in adjacent mucosal tissue. Cancer Immunol
Immunother. 62:1453–1461. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ma J, Sun G, Zhu P, Liu S, Ou M, Chen Z,
Zou C, Chan FL, Dai Y and Sui W: Determination of the complexity
and diversity of the TCR β-chain CDR3 repertoire in bladder cancer
using high-throughput sequencing. Oncol Lett. 17:3808–3816.
2019.PubMed/NCBI
|
|
36
|
Zhang J, Wang Y, Huang Y, Tan X, Xu J, Yan
Q, Tan J, Zhang Y, Zhang J, Ma Q, et al: Characterization of T cell
receptor repertoire in penile cancer. Cancer Immunol Immunother.
73:242024. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang T, Wang C, Wu J, He C, Zhang W, Liu
J, Zhang R, Lv Y, Li Y, Zeng X, et al: The different T-cell
receptor repertoires in breast cancer tumors, draining lymph nodes,
and adjacent tissues. Cancer Immunol Res. 5:148–156. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chen Z, Zhang C, Pan Y, Xu R, Xu C, Chen
Z, Lu Z and Ke Y: T cell receptor β-chain repertoire analysis
reveals intratumour heterogeneity of tumour-infiltrating
lymphocytes in oesophageal squamous cell carcinoma. J Pathol.
239:450–458. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Rizvi NA, Hellmann MD, Snyder A, Kvistborg
P, Makarov V, Havel JJ, Lee W, Yuan J, Wong P, Ho TS, et al: Cancer
immunology. Mutational landscape determines sensitivity to PD-1
blockade in non-small cell lung cancer. Science. 348:124–128. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Motzer RJ, Escudier B, McDermott DF,
George S, Hammers HJ, Srinivas S, Tykodi SS, Sosman JA, Procopio G,
Plimack ER, et al: Nivolumab versus everolimus in advanced
renal-cell carcinoma. N Engl J Med. 373:1803–1813. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Cha E, Klinger M, Hou Y, Cummings C, Ribas
A, Faham M and Fong L: Improved survival with T cell clonotype
stability after anti-CTLA-4 treatment in cancer patients. Sci
Transl Med. 6:238ra702014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhu Y, Zhao F, Li Z and Yu J: Current
landscape and future directions of biomarkers for predicting
responses to immune checkpoint inhibitors. Cancer Manag Res.
10:2475–2488. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ji S, Li J, Chang L, Zhao C, Jia R, Tan Z,
Liu R, Zhang Y, Li Y, Yin G, et al: Peripheral blood T-cell
receptor repertoire as a predictor of clinical outcomes in
gastrointestinal cancer patients treated with PD-1 inhibitor. Clin
Transl Oncol. 23:1646–1656. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Tumeh PC, Harview CL, Yearley JH, Shintaku
IP, Taylor EJ, Robert L, Chmielowski B, Spasic M, Henry G, Ciobanu
V, et al: PD-1 blockade induces responses by inhibiting adaptive
immune resistance. Nature. 515:568–571. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Roh W, Chen PL, Reuben A, Spencer CN,
Prieto PA, Miller JP, Gopalakrishnan V, Wang F, Cooper ZA, Reddy
SM, et al: Integrated molecular analysis of tumor biopsies on
sequential CTLA-4 and PD-1 blockade reveals markers of response and
resistance. Sci Transl Med. 9:eaah35602017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kato T, Kiyotani K, Tomiyama E, Koh Y,
Matsushita M, Hayashi Y, Nakano K, Ishizuya Y, Wang C, Hatano K, et
al: Peripheral T cell receptor repertoire features predict durable
responses to anti-PD-1 inhibitor monotherapy in advanced renal cell
carcinoma. Oncoimmunology. 10:18629482021. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Liu YY, Yang QF, Yang JS, Cao RB, Liang
JY, Liu YT, Zeng YL, Chen S, Xia XF, Zhang K and Liu L:
Characteristics and prognostic significance of profiling the
peripheral blood T-cell receptor repertoire in patients with
advanced lung cancer. Int J Cancer. 145:1423–1431. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Chen YT, Hsu HC, Lee YS, Liu H, Tan BC,
Chin CY, Chang IY and Yang CY: Longitudinal high-throughput
sequencing of the T-cell receptor repertoire reveals dynamic change
and prognostic significance of peripheral blood TCR diversity in
metastatic colorectal cancer during chemotherapy. Front Immunol.
12:7434482022. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Sheikh N, Cham J, Zhang L, DeVries T,
Letarte S, Pufnock J, Hamm D, Trager J and Fong L: Clonotypic
diversification of intratumoral T cells following sipuleucel-T
treatment in prostate cancer subjects. Cancer Res. 76:3711–3718.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Frankowska K, Zarobkiewicz M, Dąbrowska I
and Bojarska-Junak A: Tumor infiltrating lymphocytes and
radiological picture of the tumor. Med Oncol. 40:1762023.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Kazemi MH, Sadri M, Najafi A, Rahimi A,
Baghernejadan Z, Khorramdelazad H and Falak R: Tumor-infiltrating
lymphocytes for treatment of solid tumors: It takes two to tango?
Front Immunol. 13:10189622022. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Whiteside TL: Tumor-infiltrating
lymphocytes and their role in solid tumor progression. Exp Suppl.
113:89–106. 2022.PubMed/NCBI
|
|
53
|
Giraldo NA, Becht E, Remark R, Damotte D,
Sautès-Fridman C and Fridman WH: The immune contexture of primary
and metastatic human tumours. Curr Opin Immunol. 27:8–15. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Akiyoshi T, Gotoh O, Tanaka N, Kiyotani K,
Yamamoto N, Ueno M, Fukunaga Y and Mori S: T-cell complexity and
density are associated with sensitivity to neoadjuvant
chemoradiotherapy in patients with rectal cancer. Cancer Immunol
Immunother. 70:509–518. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Ge H, Ferris RL and Wang JH: Cetuximab
responses in patients with HNSCC correlate to clonal expansion
feature of peripheral and tumor-infiltrating T cells with top
T-cell receptor clonotypes. Clin Cancer Res. 29:647–658. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Liu XS and Mardis ER: Applications of
immunogenomics to cancer. Cell. 168:600–612. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Hopkins AC, Yarchoan M, Durham JN, Yusko
EC, Rytlewski JA, Robins HS, Laheru DA, Le DT, Lutz ER and Jaffee
EM: T cell receptor repertoire features associated with survival in
immunotherapy-treated pancreatic ductal adenocarcinoma. JCI
Insight. 3:e1220922018. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Postow MA, Manuel M, Wong P, Yuan J, Dong
Z, Liu C, Perez S, Tanneau I, Noel M, Courtier A, et al: Peripheral
T cell receptor diversity is associated with clinical outcomes
following ipilimumab treatment in metastatic melanoma. J Immunother
Cancer. 3:232015. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Arakawa A, Vollmer S, Tietze J, Galinski
A, Heppt MV, Bürdek M, Berking C and Prinz JC: Clonality of
CD4+ blood T cells predicts longer survival with CTLA4
or PD-1 checkpoint inhibition in advanced melanoma. Front Immunol.
10:13362019. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Han J, Duan J, Bai H, Wang Y, Wan R, Wang
X, Chen S, Tian Y, Wang D, Fei K, et al: TCR repertoire diversity
of peripheral PD-1+CD8+ T cells predicts
clinical outcomes after immunotherapy in patients with non-small
cell lung cancer. Cancer Immunol Res. 8:146–154. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Dong N, Moreno-Manuel A, Calabuig-Fariñas
S, Gallach S, Zhang F, Blasco A, Aparisi F, Meri-Abad M, Guijarro
R, Sirera R, et al: Characterization of circulating T cell receptor
repertoire provides information about clinical outcome after PD-1
blockade in advanced non-small cell lung cancer patients. Cancers
(Basel). 13:29502021. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Joshi K, Milighetti M and Chain BM:
Application of T cell receptor (TCR) repertoire analysis for the
advancement of cancer immunotherapy. Curr Opin Immunol. 74:1–8.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
McGranahan N and Swanton C: Clonal
heterogeneity and tumor evolution: Past, present, and the future.
Cell. 168:613–628. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Bendle GM, Linnemann C, Hooijkaas AI, Bies
L, de Witte MA, Jorritsma A, Kaiser AD, Pouw N, Debets R, Kieback
E, et al: Lethal graft-versus-host disease in mouse models of T
cell receptor gene therapy. Nat Med. 16:565–570. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Samstein RM, Lee CH, Shoushtari AN,
Hellmann MD, Shen R, Janjigian YY, Barron DA, Zehir A, Jordan EJ,
Omuro A, et al: Tumor mutational load predicts survival after
immunotherapy across multiple cancer types. Nat Genet. 51:202–206.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Marabelle A, Fakih M, Lopez J, Shah M,
Shapira-Frommer R, Nakagawa K, Chung HC, Kindler HL, Lopez-Martin
JA, Miller WH Jr, et al: Association of tumour mutational burden
with outcomes in patients with advanced solid tumours treated with
pembrolizumab: Prospective biomarker analysis of the multicohort,
open-label, phase 2 KEYNOTE-158 study. Lancet Oncol. 21:1353–1365.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Overman MJ, McDermott R, Leach JL, Lonardi
S, Lenz HJ, Morse MA, Desai J, Hill A, Axelson M, Moss RA, et al:
Nivolumab in patients with metastatic DNA mismatch repair-deficient
or microsatellite instability-high colorectal cancer (CheckMate
142): An open-label, multicentre, phase 2 study. Lancet Oncol.
18:1182–1191. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Cercek A, Lumish M, Sinopoli J, Weiss J,
Shia J, Lamendola-Essel M, El Dika IH, Segal N, Shcherba M,
Sugarman R, et al: PD-1 blockade in mismatch repair-deficient,
locally advanced rectal cancer. N Engl J Med. 386:2363–2376. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
O'Malley DM, Neffa M, Monk BJ, Melkadze T,
Huang M, Kryzhanivska A, Bulat I, Meniawy TM, Bagameri A, Wang EW,
et al: Dual PD-1 and CTLA-4 checkpoint blockade using balstilimab
and zalifrelimab combination as second-line treatment for advanced
cervical cancer: An open-label phase II study. J Clin Oncol.
40:762–771. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Hu Y, Feng J, Gu T, Wang L, Wang Y, Zhou
L, Hong R, Tan Su Yin E, Zhang M, Lu P and Huang H: CAR T-cell
therapies in China: Rapid evolution and a bright future. Lancet
Haematol. 9:e930–e941. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Grosser R, Cherkassky L, Chintala N and
Adusumilli PS: Combination immunotherapy with CAR T cells and
checkpoint blockade for the treatment of solid tumors. Cancer Cell.
36:471–482. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Pothuri VS, Hogg GD, Conant L, Borcherding
N, James CA, Mudd J, Williams G, Seo YD, Hawkins WG, Pillarisetty
VG, et al: Intratumoral T-cell receptor repertoire composition
predicts overall survival in patients with pancreatic ductal
adenocarcinoma. Oncoimmunology. 13:23204112024. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Choudhury NJ, Kiyotani K, Yap KL,
Campanile A, Antic T, Yew PY, Steinberg G, Park JH, Nakamura Y and
O'Donnell PH: Low T-cell receptor diversity, high somatic mutation
burden, and high neoantigen load as predictors of clinical outcome
in muscle-invasive bladder cancer. Eur Urol Focus. 2:445–452. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Jin YB, Luo W, Zhang GY, Lin KR, Cui JH,
Chen XP, Pan YM, Mao XF, Tang J and Wang YJ: TCR repertoire
profiling of tumors, adjacent normal tissues, and peripheral blood
predicts survival in nasopharyngeal carcinoma. Cancer Immunol
Immunother. 67:1719–1730. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Valpione S, Mundra PA, Galvani E, Campana
LG, Lorigan P, De Rosa F, Gupta A, Weightman J, Mills S, Dhomen N
and Marais R: The T cell receptor repertoire of tumor infiltrating
T cells is predictive and prognostic for cancer survival. Nat
Commun. 12:40982021. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Reuben A, Zhang J, Chiou SH, Gittelman RM,
Li J, Lee WC, Fujimoto J, Behrens C, Liu X, Wang F, et al:
Comprehensive T cell repertoire characterization of non-small cell
lung cancer. Nat Commun. 11:6032020. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Chen C, Liu SYM, Chen Y, Ou Q, Bao H, Xu
L, Zhang Y, Zhong W, Zhou Q, Yang XN, et al: Predictive value of
TCR Vβ-Jβ profile for adjuvant gefitinib in EGFR mutant NSCLC from
ADJUVANT-CTONG 1104 trial. JCI Insight. 7:e1526312022. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Wang J, Bie Z, Zhang Y, Li L, Zhu Y, Zhang
Y, Nie X, Zhang P, Cheng G, Di X, et al: Prognostic value of the
baseline circulating T cell receptor β chain diversity in advanced
lung cancer. Oncoimmunology. 10:18996092021. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Abed A, Beasley AB, Reid AL, Law N,
Calapre L, Millward M, Lo J and Gray ES: Circulating pre-treatment
T-cell receptor repertoire as a predictive biomarker in advanced or
metastatic non-small-cell lung cancer patients treated with
pembrolizumab alone or in combination with chemotherapy. ESMO Open.
8:1020662023. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Manuel M, Tredan O, Bachelot T, Clapisson
G, Courtier A, Parmentier G, Rabeony T, Grives A, Perez S, Mouret
JF, et al: Lymphopenia combined with low TCR diversity (divpenia)
predicts poor overall survival in metastatic breast cancer
patients. OncoImmunology. 1:432–440. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Yan C, Ma X, Guo Z, Wei X, Han D, Zhang T,
Chen X, Cao F, Dong J, Zhao G, et al: Time-spatial analysis of T
cell receptor repertoire in esophageal squamous cell carcinoma
patients treated with combined radiotherapy and PD-1 blockade.
Oncoimmunology. 11:20256682022. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Kim JW, Kim S, Yang SY, Joung JG and Hwang
S: T-cell receptor repertoire characteristics associated with
prognostic significance in high-grade serous ovarian carcinoma.
Genes (Basel). 14:7852023. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Benítez R, Yu K, Sirota M, Malats N and
Pineda S: Characterization of the tumor-infiltrating immune
repertoire in muscle invasive bladder cancer. Front Immunol.
14:9865982023. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Lecuelle J, Boidot R, Mananet H, Derangère
V, Albuisson J, Goussot V, Arnould L, Tharin Z, Ray Coquard I,
Ghiringhelli F, et al: TCR clonality and genomic instability
signatures as prognostic biomarkers in high grade serous ovarian
cancer. Cancers (Basel). 13:43942021. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Pan M and Li B: T cell receptor
convergence is an indicator of antigen-specific T cell response in
cancer immunotherapies. Elife. 11:e819522022. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Aran A, Garrigós L, Curigliano G, Cortés J
and Martí M: Evaluation of the TCR repertoire as a predictive and
prognostic biomarker in cancer: Diversity or clonality? Cancers
(Basel). 14:17712022. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Attaf M, Huseby E and Sewell AK: αβ T cell
receptors as predictors of health and disease. Cell Mol Immunol.
12:391–399. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Nishida J, Cristea S, Bodapati S, Puleo J,
Bai G, Patel A, Hughes M, Snow C, Borges V, Ruddy KJ, et al:
Peripheral blood TCR clonotype diversity as an age-associated
marker of breast cancer progression. Proc Natl Acad Sci USA.
120:e23167631202023. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Hiam-Galvez KJ, Allen BM and Spitzer MH:
Systemic immunity in cancer. Nat Rev Cancer. 21:345–359. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Flumens D, Gielis S, Bartholomeus E,
Campillo-Davo D, van der Heijden S, Versteven M, De Reu H, Smits E,
Ogunjimi B, Laukens K, et al: Training of epitope-TCR prediction
models with healthy donor-derived cancer-specific T cells. Methods
Cell Biol. 183:143–160. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Chen SY, Liu CJ, Zhang Q and Guo AY: An
ultra-sensitive T-cell receptor detection method for TCR-Seq and
RNA-Seq data. Bioinformatics. 36:4255–4262. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Benotmane JK, Kueckelhaus J, Will P, Zhang
J, Ravi VM, Joseph K, Sankowski R, Beck J, Lee-Chang C, Schnell O
and Heiland DH: High-sensitive spatially resolved T cell receptor
sequencing with SPTCR-seq. Nat Commun. 14:74322023. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Baker AM, Nageswaran G, Nenclares P, Ronel
T, Smith K, Kimberley C, Laclé MM, Bhide S, Harrington KJ, Melcher
A, et al: FUME-TCRseq enables sensitive and accurate sequencing of
the T-cell receptor from limited input of degraded RNA. Cancer Res.
84:1560–1569. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Park JH and Lee HK: Function of γδ T cells
in tumor immunology and their application to cancer therapy. Exp
Mol Med. 53:318–327. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Liu Z, Eltoum IEA, Guo B, Beck BH, Cloud
GA and Lopez RD: Protective immunosurveillance and therapeutic
antitumor activity of gammadelta T cells demonstrated in a mouse
model of prostate cancer. J Immunol. 180:6044–6053. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
96
|
He W, Hao J, Dong S, Gao Y, Tao J, Chi H,
Flavell R, O'Brien RL, Born WK, Craft J, et al: Naturally activated
V gamma 4 gamma delta T cells play a protective role in tumor
immunity through expression of eomesodermin. J Immunol.
185:126–133. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Chauvin C, Joalland N, Perroteau J, Jarry
U, Lafrance L, Willem C, Retière C, Oliver L, Gratas C,
Gautreau-Rolland L, et al: NKG2D controls natural reactivity of
Vγ9Vδ2 T lymphocytes against mesenchymal glioblastoma cells. Clin
Cancer Res. 25:7218–7228. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Hao Q, Li R, Li H, Rui S, You L, Zhang L,
Zhao Y, Li P, Li Y, Kong X, et al: Dynamics of the Γδtcr
repertoires during the dedifferentiation process and pilot
implications for immunotherapy of thyroid cancer. Adv Sci (Weinh).
11:e23063642024. View Article : Google Scholar : PubMed/NCBI
|