Introduction
The development of cancer immunotherapy,
particularly immune checkpoint blockades, has revolutionized cancer
treatment (1–3). Immunotherapy activates the natural
defense system of the host, which identifies and eliminates tumor
cells. This strategy has emerged as an effective treatment with
unparalleled and synergistic survival benefits in multiple cancer
types, such as melanoma and non-small cell lung carcinoma (4–6). At
present, 11 immune checkpoint inhibitors (ICIs) have been
clinically approved for the treatment of 16 malignant diseases
(7,8). However, overcoming treatment
resistance is becoming increasingly challenging, and fewer than
one-third of patients with cancer achieve significant and long-term
responses to cancer immunotherapy (9–11).
Thus, there is an urgent need to identify predictive biomarkers of
immunotherapy responses.
The histone lysine N-methyltransferase 2 (KMT2)
family of proteins regulates the expression of specific regions of
the genome by methylating histone H3 lysine K4 (H3K4). This
modification leads to chromatin remodeling and DNA accessibility,
which are involved in the occurrence, progression and immune
tolerance of a number of cancer type, such as breast and prostate
cancer (12,13). The KMT2 family of proteins includes
KMT2A, KMT2B, KMT2C and KMT2D. KMT2A and KMT2B dimethylate and
trimethylate chromatin at the promoter regions and polycomb
response elements of genes, while KMT2C and KMT2D monomethylate
H3K4 at the gene enhancer regions (14–17).
KMT2C (also known as MLL3) regulates DNA damage
response and repair by directly binding to sites of DNA damage and
mediating histone methylation. This histone modification process
relaxes the chromosomal structure near the damaged DNA, enabling
other key proteins to access the damaged sites and repair the
damage (18,19). KMT2-encoding genes are frequently
mutated in a number of cancer types, such as small cell lung and
breast cancer (20–22). These genes are closely related to
the occurrence and development of multiple tumors, and
significantly affect the clinical eradication of tumors,
particularly with immunotherapy (20–22).
Zhang and Huang (23) reported that
mutations targeting the KMT2 family of proteins may be predictive
biomarkers of a favorable response to treatment with ICIs in
multiple cancer types, such as melanoma, bladder, uterine, and lung
carcinomas. Compared with patients harboring wild-type
KMT2A/C, those with KMT2A/C mutations achieved higher
overall survival (OS), progression-free survival, objective
response rate (ORR) and durable clinical benefits upon ICI
treatment (24). However, the
relationship between KMT2C expression in tumor immune infiltration
and the predicted immunotherapeutic response remains unclear due to
the publication of few comprehensive pan-cancer studies to
date.
In the present study, the relationship between KMT2C
expression and various tumor-associated parameters were evaluated
in a pan-cancer setting. The present study will contribute to the
understanding of the role of KMT2C in tumor progression and
immunotherapy.
Materials and methods
Data collection
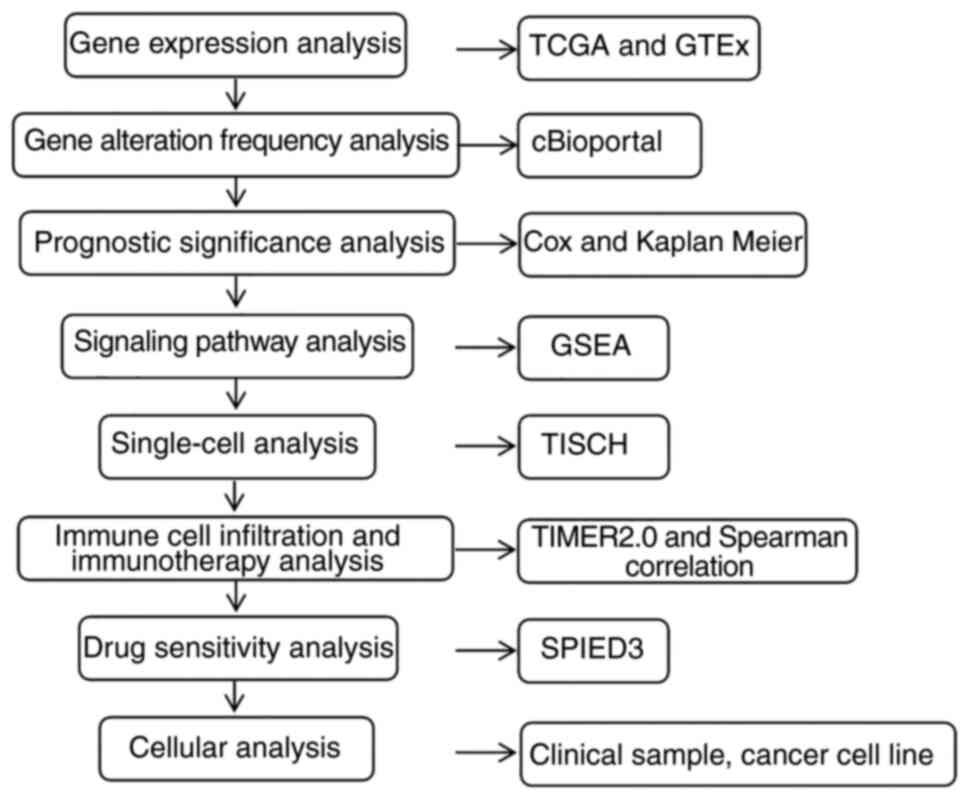
The workflow of the present study is shown in
Fig. 1. The Cancer Genome Atlas
(TCGA) pan-cancer dataset were downloaded from the Genotype-Tissue
Expression project (https://xenabrowser.net/datapages/) and the UCSC Xena
database (https://xenabrowser.net/datapages/). Data on the
following cancer types were included in the present study:
Adrenocortical carcinoma (ACC), bladder urothelial carcinoma
(BLCA), breast invasive carcinoma (BRCA), cervical squamous cell
carcinoma and endocervical adenocarcinoma (CESC),
cholangiocarcinoma (CHOL), colon adenocarcinoma (COAD), large B
cell lymphoma (DLBC), esophageal cancer (ESCA), glioblastoma (GBM),
head and neck squamous cell carcinoma (HNSC), kidney chromophobe
(KICH), kidney renal clear cell carcinoma (KIRC), kidney renal
papillary cell carcinoma, acute myeloid leukemia (LAML), low grade
glioma (LGG), liver hepatocellular carcinoma (LIHC), lung
adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC),
mesothelioma, ovarian serous cystadenocarcinoma (OV), pancreatic
adenocarcinoma (PAAD), pheochromocytoma and paraganglioma (PCPG),
prostate adenocarcinoma (PRAD), rectal adenocarcinoma (READ),
sarcoma (SARC), skin cutaneous melanoma (SKCM), stomach
adenocarcinoma (STAD), testicular germ cell tumors (TGCT), thyroid
carcinoma (THCA), thymoma (THYM), uterine corpus endometrial
carcinoma (UCEC), uterine carcinosarcoma (UCS) and uveal melanoma
(UVM). The transcriptomic, CRISPR-Cas9 and small interfering RNA
data generated using cancer cell lines were downloaded from the
Cancer Cell Line Encyclopedia (CCLE) website (https://sites.broadinstitute.org/ccle/).
Immunotherapy cohort data were downloaded from the Gene Expression
Omnibus (https://www.ncbi.nlm.nih.gov/geo/).
Prognostic analysis
The prognosis data including OS, disease-specific
survival (DSS), disease-free interval (DFI) and progression-free
interval (PFI) were downloaded from the UCSC Xena database
(xenabrowser.net/datapages/). Kaplan-Meier and univariate Cox
regression analyses were conducted to calculate the association
between KMT2C expression and the pan-cancer OS, DSS, DFI and PFI.
The samples were divided into high and low expression according to
the optimal cut-off point. Multivariate Cox regression analyses
were also conducted to calculate the association between KMT2C
expression and the pan-cancer OS. Univariate Cox analysis was
performed using the UCSC-Xena-Shiny website (https://shiny.hiplot-academic.com/ucsc-xena-shiny/).
Multivariate Cox analyses were performed using the R package,
‘survival’. Kaplan-Meier analyses were performed using the R
packages, ‘survival’ and ‘survminer’ (R 4.2.0; r-project.org/).
Gene set enrichment analysis
(GSEA)
TCGA patient samples were divided into two groups
based on the KMT2C expression levels. The samples were arranged in
expression order and the top 30% of the samples were defined as the
high KMT2C expression group and the bottom 30% as the low KMT2C
expression group, with the remainder excluded from analysis
(25). The gene sets were
downloaded from the MsigDB database (https://www.gsea-msigdb.org/gsea/msigdb). GSEA was
performed using the R package, ‘ClusterProfiler’.
Single-cell analysis
Single-cell analysis was performed using Tumor
Immune Single-cell Hub (TISCH) web tool (26). Results were visualized using the R
package, ‘ggplot2’. The single-cell hepatocellular carcinoma
dataset (dataset ID, SCDS0000020) was also analyzed using the
Cell-omics Data Coordinate Platform (CDCP; http://db.cngb.org/cdcp/dataset/SCDS0000020/).
Chemotherapy sensitivity analysis
The correlation between KMT2C expression and the
sensitivity to small molecule inhibitors was investigated using the
CMap (https://portals.broadinstitute.org/cmap/) and SPIED3
(92.205.225.222/HGNC-SPIED3-QF.py) web tools (27). Drug sensitivity and gene expression
data were obtained from experiments with cancer cell lines and
downloaded from the GDSC (https://www.cancerrxgene.org/) database (28). The correlation between drug
sensitivity and gene expression was analyzed using the Spearman's
test.
Analysis of the tumor microenvironment
(TME)
Single-sample (ss)GSEA was conducted to calculate
the tumor immune cell infiltration scores (29) using R 4.2.0. A Spearman's
correlation analysis was performed to investigate the correlation
between tumor immune cell infiltration scores, immune
checkpoint-related genes and KMT2C expression (R 4.2.0). The TIMER2
webtool (http://timer.cistrome.org/) was used
to evaluate the relationship between KMT2C expression and tumor
immune cell infiltration.
Immunotherapy response prediction
analyses
Immunotherapy response prediction analyses of KMT2C
expression were performed using the Tumor Immune Dysfunction and
Exclusion (TIDE) computational method (http://tide.dfci.harvard.edu) (30). Correlations between KMT2C expression
and tumor mutational burden (TMB) and microsatellite instability
(MSI) were analyzed using Spearman's correlation test using R
4.2.0. IMvigor210 (31), PRJEB23709
(32), PRJNA482620 (33) and PRJEB25780 (34) immunotherapy cohort datasets were
used to analyze and identify the predictive value of KMT2C.
Immunohistochemistry
Paraffin-embedded kidney tumor tissue samples were
obtained from patients treated at the Second Affiliated Hospital of
Anhui Medical University (Hefei, China) with informed consent from
May 2019 to March 2022. The inclusion criteria were as follows:
diagnosed with renal cancer, not receiving treatment, and the
patient is willing to provide pathology sections for study. Tumors
were staged according to the 8th edition of the INM classification
of malignant tumors (35). The
basic patient clinical information is provided in Table SI. All experiments were approved by
the Medical Ethics Committee of the Second Affiliated Hospital of
Anhui Medical University (approval no. 81220282).
Immunohistochemistry was performed by Servicebio Co., Ltd. 4%
Paraformaldehyde Fix Solution (Beyotime Institute of Biotechnology;
cat. no. P0099) fixed (room temperature, 24 h) and
paraffin-embedded kidney tumor tissue were cut into 5-µm-thick
sections, dried, deparaffinized and dehydrated in a graded ethanol
series. The antigen was retrieved by microwave method using sodium
citrate (10 mM, pH 6.0) for 20 min, and then washed by phosphate
buffered saline (PBS) 3 times. The tissue sections were treated
with 1% hydrogen peroxide (Beyotime, P0100A) for 10 min to block
endogenous tissue peroxidase activity and treated with goat serum
(Beyotime, C0265) for 1 h at room temperature to block non-specific
protein binding. The slides were incubated with rabbit monoclonal
anti-human PD-L1 antibody (Abcam, ab205921, 1:500) or rabbit
polyclonal anti-human KMT2C antibody (absin, abs113638 1:200)
overnight at 4°C. Then, the slides were incubated with the
Universal kits (ZSGB-BIO; cat. no. PV-6000) at room temperature for
20 min. The slides were washed with PBS and colored with
3,3′-diaminobenzidine substrate kit (ZSGB-BIO, ZLI-9017) for 5 min.
Then, the slides were counterstained with hematoxylin staining
solution (ZSGB-BIO, ZLI-9610) at room temperature for 1 min, and
visualized with a light microscope (Nikon, ECLIPSE Ti2). The tumor
proportion score (TPS) was used to evaluate the expression of
programmed death-ligand 1 (PD-L1) in tumor samples. The TPS was
calculated as follows: TPS=(number of tumor cells with positive
PD-L1 membrane staining/total number of tumor cells) ×100%. The
average optical density (representing KMT2C expression) was
calculated using ImageJ1.53t software (National Institutes of
Health).
In vitro cellular assays
The A549 and H1975 human lung cancer cell lines was
purchased from Nanjing CoBioer Biotechnology Co., Ltd. A549 cells
were cultured in DMEM (Corning, Inc.) supplemented with 10% (v/v)
fetal bovine serum (FBS; ExCell, Inc.) and 1% (v/v)
penicillin/streptomycin. H1975 cells were cultured in RPMI-1640
(Corning, Inc.) supplemented with 10% (v/v) FBS and 1% (v/v)
penicillin/streptomycin. The immortalized human umbilical vein
endothelial cell (HUVEC) line was purchased from Xiamen Immocell
Biotechnology Co., Ltd (IM-H205). and cultured in human umbilical
vein endothelial cell medium (HUVEC-90011; Cyagen Biosciences,
Inc.).
KMT2C knockdown cell line
construction
KMT2C knockdown pLKO.1 plasmids were
purchased from Shanghai GenePharma Co., Ltd. Plasmids
(pLKO.1:pSPAX.2: pMD2G=2:2:1) were transfected into 293T with
polyethylenimine (YEASEN, 40815ES03, 1 µg/ml). After 48 h, the
lentivirus was collected. When the cells were in the logarithmic
phase, 1×105 cells were seeded into a 6-well plate and 1
ml lentivirus (MOI=10) and 1.5 µl 10 µg/µl polybrene(absin) were
added at 37°C for 24 h. The medium was replaced with fresh medium
after 24 h. After 48 h, 1 µg/µl puromycin was added to the medium
for screening. The transfection efficiency was examined by
immunoblotting analysis. The short hairpin (sh)RNA sequences used
were as follows: shNC, 5′-TTCTCCGAACGTGTCACGT-3′; sh1,
5′-CGATCTCCTCAGCAGAATATA-3′; and sh2,
5′-CTGAGCTCACTACAGATTATA-3′.
Immunoblotting analysis
Proteins from the cells extracted using RIPA buffer
(Beyotime, P0013B) and quantified by the Pierce BCA Protein assay
(Thermo, 23225), and 40 ug of each sample was loaded on
Nitrocellulose (0.45 µm) membrane (Bio-Rad, 1620115), and was
blocked by 5% skim milk 1 h, then TBST (1% TWEEN 20) wash 3 times
for 15 min. Next, the membrane was incubated with the primary
antibody KMT2C (absin, abs113664, dilution: 1:1,000) and
anti-β-Actin Monoclonal Antibody (absin, abs830031, dilution:
1:1,000) at 4°C overnight and followed by peroxidase-conjugated
secondary antibody (HUABIO, HA1008, dilution: 1:5,000) for 1 h. at
37°C. Protein bands were visualized by an enhanced chemiluminescent
detection kit (Thermo, A38554) with ChemiDoc XRS+ System (Bio-Rad,
USA).
Reverse transcription-quantitative
polymerase chain reaction
The RNA extraction kit (AG21101; Accurate Biology)
was used to extract the total cellular RNA and the reverse
transcription kit (AG11706; Accurate Biology) to reverse
transcription. RT-qPCR) reactions were run on a Bio-Rad IQ 5 RT-PCR
detection system using SYBR Green Premix Pro Taq HS qPCR Kit
(AG11701; Accurate Biology). The primers were ordered from Biosune
company, having the following sequences: Forward (F)_KMT2C,
5′-CCTCCCTCCCAACACCACCTC-3′; Reverse(R)_KMT2C,
5′-TCTGGATACCTGCTCACTTCTACCC-3′;F_GAPDH,
5′-GAGAAGTATGACAACAGCCTCAA-3′; and R_GAPDH,
5′-GCCATCACGCCACAGTTT-3′. Use the reverse transcription kit AG11706
reverse recording reaction: instantaneous separation after mixing;
the procedure is 37°C/15 min, 85°C 5 sec, 4°C. Then the target
primer and template cDNA were then diluted 40 times with the
AG11701 SYBRGreen ProTaq HS pre-mixed qPCR kit (10 µl system) with
3 complex wells for each sample. GAPDH was used as the internal
reference gene. The thermocycling conditions were as follows:
samples were incubated at 95°C for 30 sec, followed by 40 cycles at
95°C for 5 sec and 60°C for 30 sec. The results were analyzed by
the 2-ΔΔCq method (36).
Colony formation and wound healing
assays
Colony formation and wound healing assays were
conducted as previously reported (37). For the colony formation assay,
1×104 cells were seeded into a 6-well plate with 2 ml
complete medium and cultured for ~2 weeks. Then, cells fixed with
4% paraformaldehyde 20 min at room temperature and stained by 0.1%
crystal violet 20 min at room temperature. The number of colonies
were calculated using ImageJ software. The number of cells over 60
was defined as a clone. For the cell migration assay, cells were
grown to 100% confluency and a wound gap was created by a scratch
instrument. The medium was replaced with medium containing 0.1%
FBS, and images were collected using a light microscope EVOS XL
Core Cell Imaging System (Thermo Fisher Scientific, Inc.) on days 0
and 7 after scratch formation. The gap closure was calculated using
ImageJ software 1.53t.
Anti-proliferation assay
Small molecular inhibitors SN-38 (HY-13704),
vorinostat (HY-10221), UNC0224 (HY-10929), SGC0946 (HY-15650) were
purchased from MedChemExpress (Shanghai, China). The cells were
plated in 96-well plate (3,000 cells/well) and medium is replaced
with DMEM (Corning, Inc.) containing different concentrations (10,
3, 1, 0.3, 0.1, 0.03, 0.010, 0.003, 0.001 µM) of small molecule
inhibitors, and incubated in 37°C and 5% CO2. After 72
h, 10 µl CCK8 (C0038, Beyotime Institute of Biotechnology) were
added in per well and incubate at 37°C for 30 min. Detection (450
nm) was performed with a microplate reader (Thermo Fisher
Scientific, Inc.).
Statistical analysis
All data were statistically analyzed using R
software 4.2.0 (https://www.r-project.org/). Kaplan-Meier and
univariate Cox regression analyses were conducted for survival
analysis. Statistically significant correlations was determined
using the Spearman's test. Analyses of KMT2C expression in cancer
and normal tissues were conducted using unpaired Student's t-test.
One way ANOVA and Bonferroni was used for multi-group data
comparisons. All P-values were two-sided, and P<0.05 was
considered to indicate a statistically significant difference.
Results
Analysis of pan-cancer KMT2C
expression
KMT2C expression in The Cancer Genome Atlas (TCGA)
pan-cancer dataset was evaluated. KMT2C was highly expressed in
BRCA, CHOL, ESCA, GBM, KICH, LAML, LGG, PAAD, PRAD and STAD tumor
samples. By contrast, low KMT2C expression was observed in ACC,
BLCA, CESC, COAD, LUAD, LUSC, OV, SKCM, TGCT, THCA, THYM, UCEC and
UCS (Fig. 2A). Analysis of KMT2C
expression in paired tumor and normal pan-cancer tissues revealed
that KMT2C was highly expressed in CHOL, LIHC and KICH tumor
tissues, whereas its expression was low in COAD and THCA tumor
tissues (Figs. 2B-E and S1A). The expression of KMT2C at different
tumor stages was further investigated, and it was demonstrated that
the expression varied significantly at the different stages of
COAD, KIRC, OV and STAD (Fig.
S1B-E).
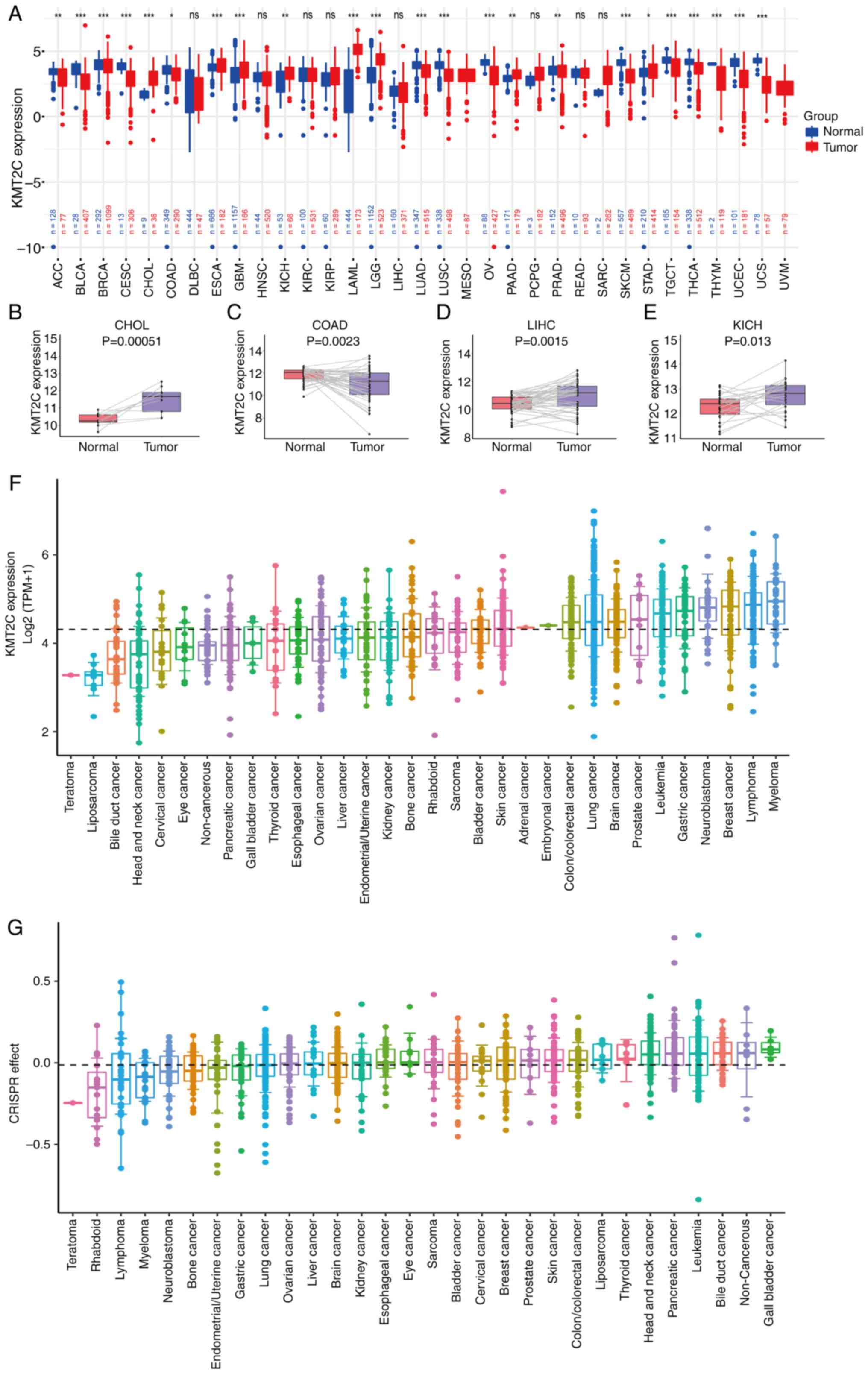 | Figure 2.Expression analysis of KMT2C in
pan-cancer. (A) Pan-cancer analysis of KMT2C expression in tumor
and normal tissues from TCGA and Genotype-Tissue Expression
databases. (B) Analysis of KMT2C expression in paired CHOL tumor
and adjacent normal tissues. (C) Analysis of KMT2C expression in
paired COAD tumor and adjacent normal tissues. (D) Analysis of
KMT2C expression in paired LIHC tumor and adjacent normal tissues.
(E) Analysis of KMT2C expression in paired KICH tumor and adjacent
normal tissues. (F) Pan-cancer analysis of KMT2C expression in
cancer cell lines from the CCLE. The threshold lines represent the
mean KMT2C expression in all cells. (G) Effect of CRISPR/Cas9
knockout on KMT2C expression on cancer cell lines (data from CCLE).
*P<0.05, **P<0.01, ***P<0.001. CCLE, Cancer Cell Line
Encyclopedia; KMT2C, histone lysine N-methyltransferase 2C; ns, not
significant; TCGA, The Cancer Genome Atlas; TMP, transcripts per
million; CHOL, cholangiocarcinoma; COAD, colon adenocarcinoma;
LIHC, liver hepatocellular carcinoma; KICH, kidney chromophobe. |
Next, the expression of KMT2C in different cancer
cell lines was investigated using CCLE datasets. KMT2C was highly
expressed in myeloma, lymphoma, breast cancer, gastric cancer and
leukemia cell lines, whereas low expression was observed in
teratoma, liposarcoma, bile duct cancer, head and neck cancer and
cervical cancer cell lines (Fig.
2F). CRISPR/Cas9 data showed that KMT2C knock-out
inhibited the proliferation of teratoma, rhabdoid, lymphoma and
myeloma cells (Fig. 2G). Similarly,
RNA interference analyses indicated that silencing of KMT2C
expression inhibited the proliferation of myeloma and rhabdoid
cells (Fig. S1F). The genomic
alterations in KMT2C in pan-cancer samples were also
analyzed. The results indicated that the cancer types with
frequencies >20% were bladder cancer and melanoma (Fig. S2A). The effect of KMT2C
mutations on its protein expression were further investigated. The
results showed that the presence or absence of a KMT2C
mutation did not affect its protein expression (Fig. S2B).
Prognostic significance of KMT2C
The pan-cancer prognostic value of KMT2C was
investigated using univariate Cox regression analysis. The OS and
PFI results indicated that KMT2C acted as a protective factor for
patients with KIRC and OV but was a risk factor for patients with
LUSC and UVM. The DSS results indicated that KMT2C acted as a
protective factor in patients with KIRC (Fig. 3A). Next, the prognostic value of
KMT2C in KIRC, LUSC, OV and UVM was investigated using Kaplan-Meier
analysis. The OS results showed that elevated KMT2C expression was
related to a shorter OS time in LUSC and OV, and a longer OS time
in KIRC (Fig. 3B-E). The DSS
results showed that elevated KMT2C expression was positively
associated with a shorter DSS time in LUSC and UVM, but a longer
DSS time in KIRC and OV (Fig.
3F-I). The PFI results showed that elevated KMT2C was related
to a shorter PFI in LUSC and UVM, and a longer PFI in KIRC and OV
(Fig. 3J-M). The DFI results showed
that elevated KMT2C levels were positively associated with a
shorter DFI in LUSC, and a longer DFI in OV (Fig. 3N-P). The pan-cancer prognostic value
of KMT2C was also investigated using multivariate Cox regression
analysis. KMT2C acted as a risk factor for HNSC, KIRC and LUAD
(Table SII). The different
prognostic roles of KMT2C in different cancer types may be
regulated by the TME.
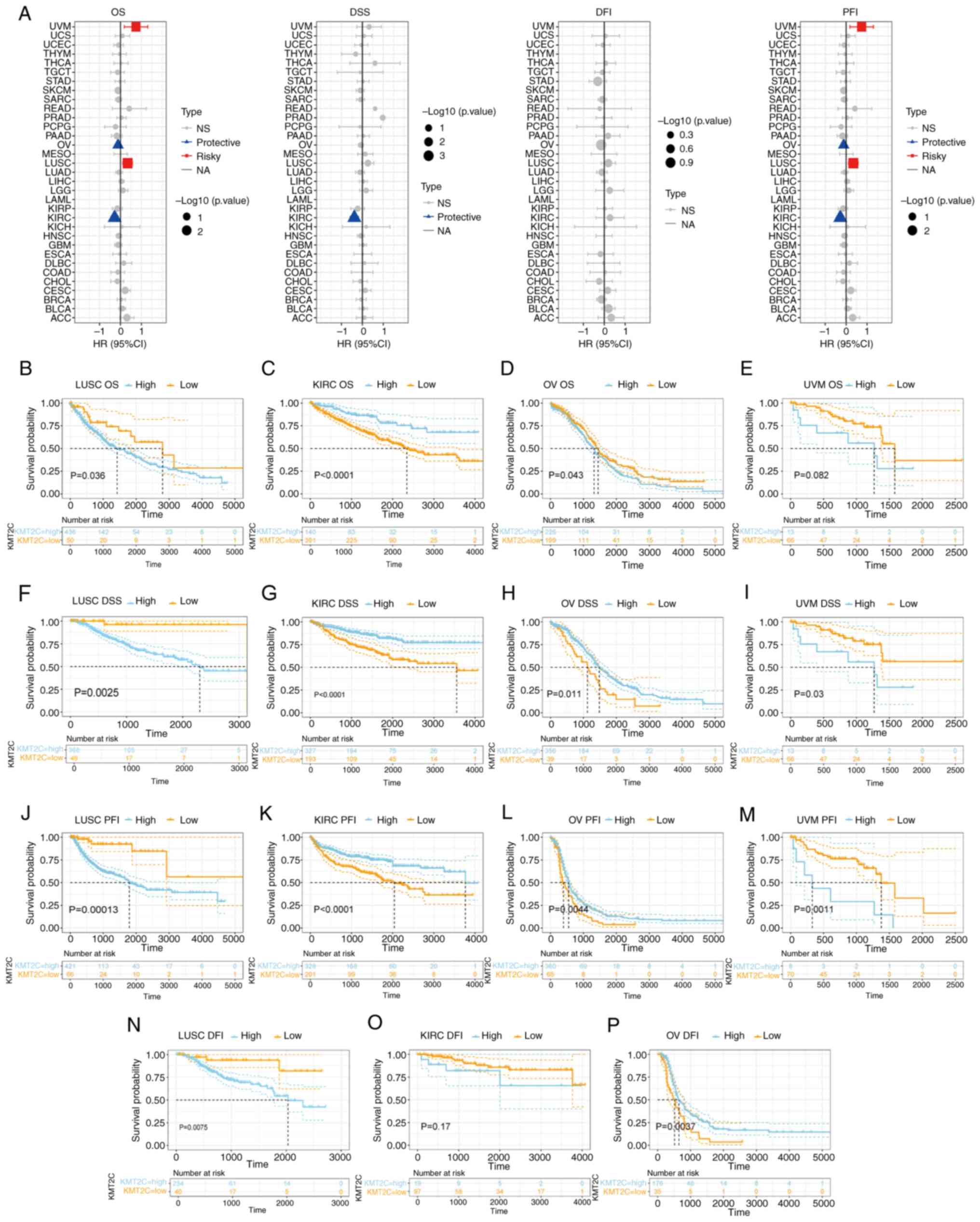 | Figure 3.Prognostic significance of KMT2C
expression. (A) Univariate Cox regression analysis of the effect of
KMT2C on OS, DSS, DFI and PFI in pan-cancer. (B) Kaplan-Meier
analysis of the effect of KMT2C on OS in LUSC. (C) Kaplan-Meier
analysis of the effect of KMT2C on OS in KIRC. (D) Kaplan-Meier
analysis of the effect of KMT2C on OS in OV. (E) Kaplan-Meier
analysis of the effect of KMT2C on OS in UVM. (F) Kaplan-Meier
analysis of the effect of KMT2C on DSS in LUSC. (G) Kaplan-Meier
analysis of the effect of KMT2C on DSS in KIRC. (H) Kaplan-Meier
analysis of the effect of KMT2C on DSS in OV. (I) Kaplan-Meier
analysis of the effect of KMT2C on DSS in UVM. (J) Kaplan-Meier
analysis of the effect of KMT2C on DFI in LUSC. (K) Kaplan-Meier
analysis of the effect of KMT2C on DFI in KIRC. (L) Kaplan-Meier
analysis of the effect of KMT2C on DFI in OV. (M) Kaplan-Meier
analysis of the effect of KMT2C on DFI in UVM. (N) Kaplan-Meier
analysis of the effect of KMT2C on PFI in LUSC. (O) Kaplan-Meier
analysis of the effect of KMT2C on PFI in KIRC. (P) Kaplan-Meier
analysis of the effect of KMT2C on PFI in OV. CI, confidence
interval; DFI, disease-free interval; DSS, disease specific
survival; HR, hazard ratio; KMT2C, histone lysine
N-methyltransferase 2C; NA, not applicable; NS, not significant;
OS, overall survival; PFI, progression free interval. KIRC, kidney
renal clear cell carcinoma; LUSC, lung squamous cell carcinoma; OV,
ovarian serous cystadenocarcinoma; UVM, uveal melanoma. |
Pan-cancer GSEA of KMT2C
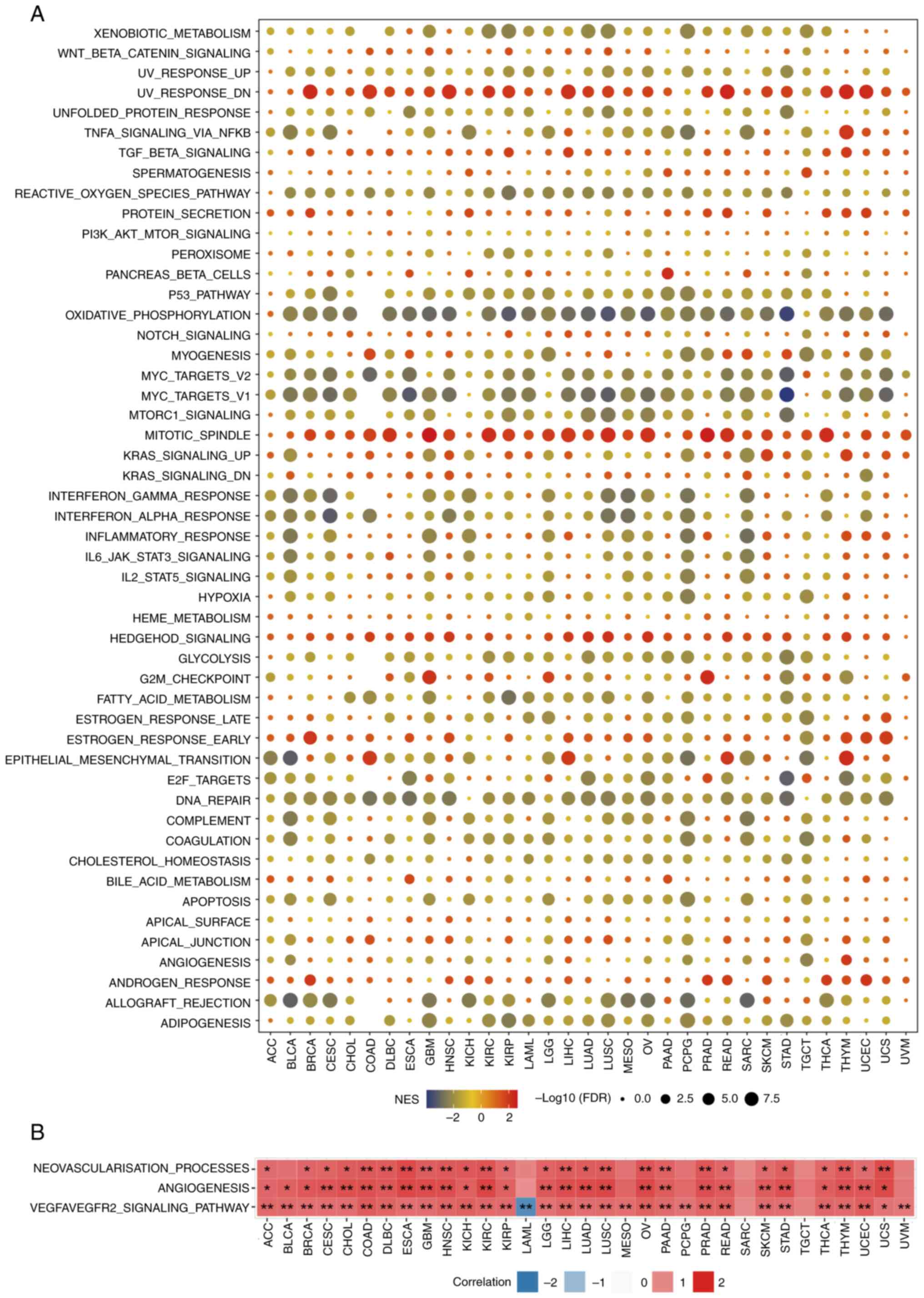
The signaling pathways through which KMT2C may be
involved in cancer was investigated using GSEA. The expression of
KMT2C was highly and negatively correlated with immune-activated
pathways, such as those associated with tumor necrosis factor
(TNF)-α, interferon (IFN)-γ and IFN-α signaling, the
pro-inflammatory response and allograft rejection, particularly in
BLCA, BRCA, ESCA, HNSC, LUAD, LUSC, PAAD and READ (Fig. 4A). These results suggested that high
expression of KMT2C may be a potential marker of the
immunosuppressive TME. It was further observed that KMT2C
expression was significantly positively correlated with activation
of signaling pathways implicated in the response to ultraviolet
radiation, reactive oxygen species, oxidative phosphorylation, MYC
signaling, mitotic spindle and DNA repair, consistent with the
function of KMT2C as a DNA damage regulator (Fig. 4A). Since tumor angiogenesis is an
important factor in tumor progression and has a key role in tumor
growth, metastasis and resistance to chemotherapy and
immunotherapy, it was also found that KMT2C expression was
significantly associated with the neovascularization process,
angiogenesis and the VEGFA/VEGFR2 signaling pathway (Fig. 4B). To verify the effect of KMT2C on
angiogenesis, a HUEVC angiogenesis experiment was performed. The
results showed that knockdown of KMT2C inhibited angiogenesis to a
certain extent (Fig. S3A and
B).
Single-cell pan-cancer analysis of
KMT2C expression
Next, the expression of KMT2C in tumor and stromal
cells from 58 single-cell cancer sample datasets was investigated
using the TISCH web tool. The results showed that KMT2C was
expressed in immune cells, endothelial cells (ECs), fibroblasts and
malignant cells (Fig. 5). In the
GSE146771 colon cancer dataset, which contained 54,285 single-cell
samples from 18 patients, KMT2C was widely expressed in a range of
immune cells, including B cells, conventional CD4+ T
cells (CD4Tconv), functional CD8+ T cells (CD8T),
exhausted CD8T cells (CD8Tex), monocytes, macrophages, natural
killer (NK) cells, proliferating T cells (Tprolif) and regulatory T
cells (Tregs). In the GSE131928 glioma cancer dataset, which
contained 7,930 single-cell samples from 35 patients, KMT2C was
widely expressed in astrocyte-like malignant cells, CD8Tex,
malignant cells, monocytes, macrophages, oligodendrocytes and
oligodendrocyte progenitor-like malignant cells. Finally, in the
GSE120575 melanoma dataset, which contained 16,291 single-cell
samples from 32 patients, KMT2C was widely expressed in B cells,
CD4Tconv, CD8T, CD8Tex, dendritic cells (DCs), monocytes,
macrophages, NK cells, Tprolif and Tregs. Together, these results
indicated that KMT2C was widely expressed in the TME (Fig. 5). The expression of KMT2C in
hepatocellular carcinoma was also investigated using the CDCP tool.
KMT2C was widely expressed in T, NK and myeloid cells (Fig. S4).
Immune cell infiltration analysis
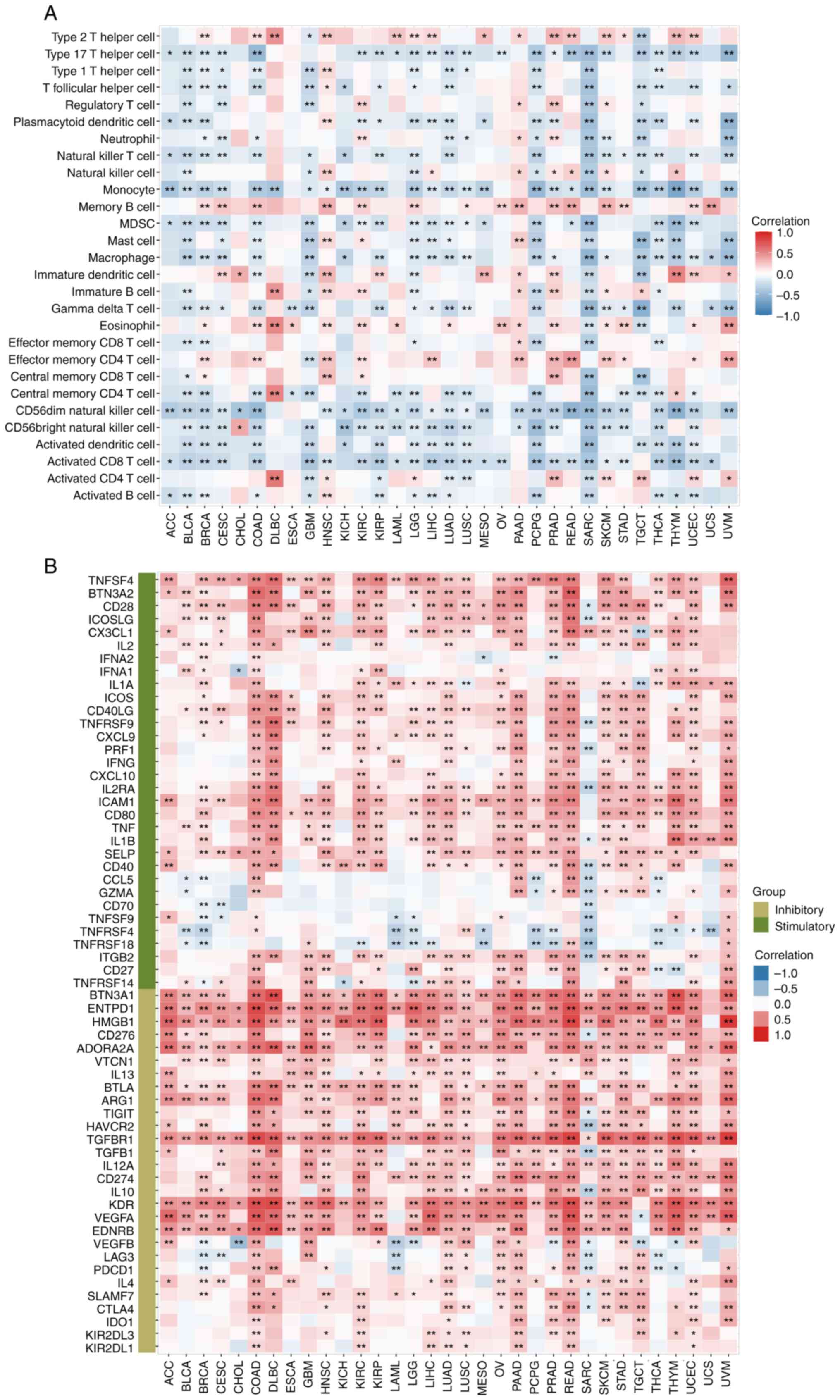
To investigate the relationship between KMT2C
expression and tumor immunity, the correlation between KMT2C levels
and tumor immune cell infiltration was investigated using ssGSEA.
The expression of KMT2C was positively correlated with the levels
of infiltrated type 2 T helper (Th2) cells, memory B cells,
immature DCs, eosinophils and effector memory CD4+ T
cells in most cancer types and negatively correlated with the
infiltration levels of Th17 cells, Th1 cells, plasmacytoid (p)DCs,
Natural killer T cells (NK-T), monocytes, myeloid-derived
suppressor cells (MDSCs), macrophages, γδ T cells, central memory
CD4+ T cells, CD56dim and
CD56bright NK cells, activated DCs and activated
CD8+ T cells (Fig. 6A).
In addition, the relationship between KMT2C expression and tumor
immune cell infiltration was investigated using data from the
TIMER2 database. The expression of KMT2C was positively correlated
with the infiltration levels of B cells, cancer-associated
fibroblasts (CAFs), DCs, ECs, M2 macrophages, mast cells,
monocytes, neutrophils, CD4+ T cells and Tregs in most
of the cancer types in TCGA, and negatively correlated with the
infiltration levels of pDCs, M1 macrophages, NK cells, γδ T cells,
NKT cells and Th1 cells in most of the cancer types (Fig. S5A). The potential correlation
between KMT2C expression and immune checkpoint genes, chemokines
and receptors were further investigated. KMT2C expression was
significantly positively correlated with the upregulation of immune
inhibitory genes, such as EDNRB, VEGFA, KDR, CD274, TGFBR1,
ADORA2A, CD276, HMGB1, ENTPD1 and BTN3A1, in most cancer
types, and was significantly negatively correlated with the
upregulation of immune stimulatory genes, such as TNFRSF4
and TNFRSF18, in multiple types of cancer (Fig. 6B). It was also found that KMT2C
expression was significantly positively correlated with the
expression of chemokines CXCL12, CXCL16, CX3CL1 and CCL28, and most
immune receptors (Fig. S5B and C).
Collectively, these results indicated that KMT2C expression was
significantly correlated with various components of the
immunosuppressive TME.
Predictive role of KMT2C in cancer
immunotherapy
To further investigate the relationship between
KMT2C expression and T cell function, the role of KMT2C in T cell
killing was reevaluated using published data (38). Knockout of KMT2C expression
improved T cell killing, implicating KMT2C as a potential
immunotherapeutic biomarker (Fig.
S6A). To verify the role of KMT2C in predicting the efficacy of
ICIs in patients, the correlation between KMT2C expression, TMB and
MSI were assessed. The results demonstrated that KMT2C was
significantly negatively correlated with the TMB in BRCA, COAD,
DLBC, KICH, KIRC, PAAD, STAD, TGCT and THCA, but positively
correlated with the TMB in HNSC, PRAD and THYM (Fig. 7A). Furthermore, KMT2C expression was
significantly negatively correlated with MSI in ACC, BRCA, COAD,
HNSC, KIRC, LIHC, PAAD, PCPG, PRAD, READ, SARC, SKCM, STAD, TGCT,
THCA and THYM (Fig. 7B). In
addition, TIDE was employed to investigate the correlation between
KMT2C levels and the response to immunotherapy and to compare the
predictive value of KMT2C with the values of other standardized
cancer biomarkers. The area under the receiver operator
characteristic curve for KMT2C was >0.5 in 12 immunotherapy
cohorts, suggesting that the KMT2C level exhibited a higher
predictive value than the TMB, T cell clonality (T.Clonality) and B
cell clonality (B.Clonality) biomarkers (Fig. 7C).
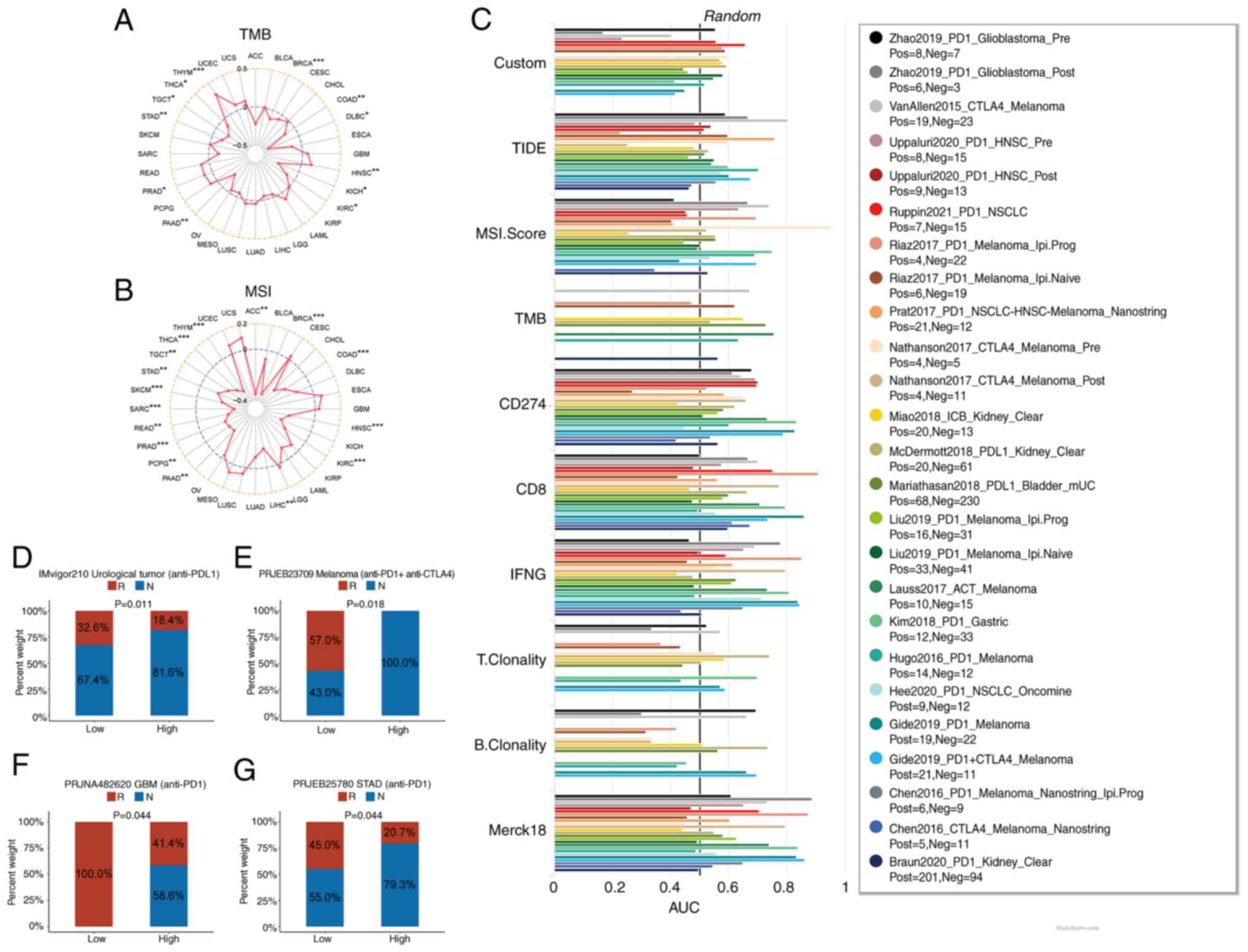 | Figure 7.Predictive role of KMT2C in cancer
immunotherapy. (A) The correlation between KMT2C expression and TMB
in pan-cancer. (B) The correlation between KMT2C expression and MSI
in pan-cancer. (C) The predictive value of KMT2C as a biomarker in
immunotherapy cohorts. The objective response rates of low and high
KMT2C expression subgroups of patients in the (D) IMvigor210, (E)
PRJEB23709, (F) PRJNA482620 and (G) PRJEB25780 immunotherapy
cohorts. *P<0.05, **P<0.01, ***P<0.001. AUC, area under
the curve; R, response; N, no response; KMT2C, histone lysine
N-methyltransferase 2C; TMB, tumor mutational burden; MSI,
microsatellite instability; PD-1, programmed cell death protein
1. |
Several clinical databases were searched to
investigate the role of KMT2C in immunotherapy. The IMvigor210
cohort included the transcriptomic and immunophenotypic profiles of
348 patients with urological tumors who had been treated with
anti-PD-L1 immunotherapy. The PRJEB23709 cohort included 91
patients with metastatic melanoma who had been treated with
anti-programmed cell death protein 1 (PD-1) monotherapy or
anti-PD-1 and anti-CTLA-4 in a combination immunotherapy. The
PRJNA482620 cohort included the transcriptomic and clinical
profiles of 34 patients with GBM who had been treated with
anti-PD-1 immunotherapy. The PRJEB25780 cohort included data from
78 patients with metastatic gastric tumors, who had been treated
with anti-PD-1 immunotherapy. These immunotherapy cohort datasets
were used to evaluate the ability of KMT2C to predict the ORR.
Patients with low KMT2C expression had higher ORRs following
immunotherapy in all 4 cohorts (Fig.
7D-G). The ORR for patients with high KMT2C expression in the
IMvigor210 cohort was 18.4% and the ORR for patients with low KMT2C
expression was 32.6% (Fig. 7D). The
association between KMT2C and the clinical response rates to
immunotherapy drugs was also examined. In the PRJNA482620 dataset,
the patients were treated with pembrolizumab or nivolumab. The
response rate of patients with low KMT2C expression (100.0%) was
significantly higher than that of patients with high KMT2C
expression (41.4%) (Fig. 7F). In
the GSE78220 dataset, patients were treated with pembrolizumab or
nivolumab. The response rate of patients with low KMT2C expression
(80.0%) was higher than that of patients with high KMT2C expression
(45.5%) (Fig. S6B). In the
PRJEB25780 dataset, patients were treated with pembrolizumab. The
response rate of patients with low KMT2C expression (45.0%) was
higher than that of patients with high KMT2C expression (20.7%)
(Fig. 7G). These results implicated
KMT2C expression as a potential biomarker of the clinical response
to pembrolizumab or nivolumab. Tissue samples from 29 patients with
kidney cancer were also analyzed and a significant positive
correlation between KMT2C expression and PD-L1 expression was
observed (Figs. S6C, S7 and S8). This was consistent with the results
of the pan-cancer analysis. However, as there is no available KMT2C
antibody with good specificity, there was a problem with high
background signal in the immunohistochemistry results, which
affected the conclusions to a certain extent.
KMT2C-mediated prediction of
pan-cancer drug sensitivity
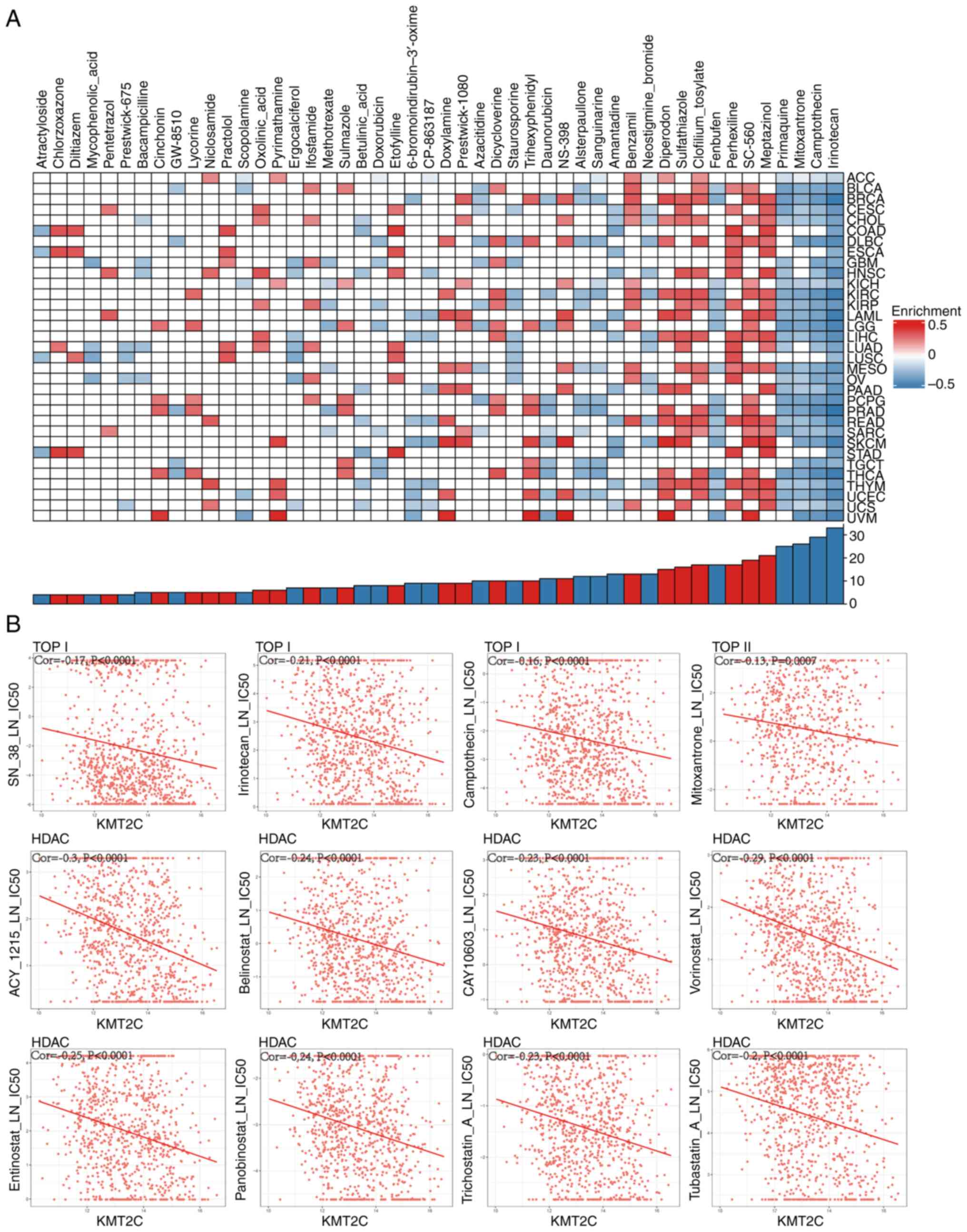
To date and to the best of our knowledge, no small
molecule inhibitors that target KMT2C have been reported. Thus,
potential inhibitors of the KMT2C-regulated signaling pathway were
identified using the CMap dataset. High KMT2C expression was
negatively correlated with the topoisomerase (TOP) I inhibitors,
irinotecan and camptothecin, in 33 and 29 cancer types,
respectively. KMT2C expression was negatively correlated with the
TOP II inhibitor, mitoxantrone, in 26 cancer types. In addition,
primaquine, a potent antimalarial agent, was also enriched in 25
cancer types (Fig. 8A). An analysis
of a GDSC dataset (https://www.cancerrxgene.org/) found that KMT2C
expression was significantly negatively correlated with several TOP
I inhibitors (SN38, irinotecan and camptothecin), a TOP II
inhibitor (mitoxantrone), several histone deacetylase (HDAC)
inhibitors (ACY1215, belinostat, CAY10603, vorinostat, entinostat,
panobinostat, trichostatin A and tubastatin A), DOT1-like histone
H3K79 methyltransferase (DOT1L) inhibitors (EPZ004777 and EPZ5676)
and a G9A nuclear histone lysine methyltransferase inhibitor
(UNC0638) (Figs. 8B and S9A). These results indicated that these
small molecule inhibitors may be suitable for treating tumors with
high levels of KMT2C expression. In addition, it was found that
KMT2C expression was significantly correlated with TOP I, HDAC 1–9,
DOT1L and G9A expression, indicating that KMT2C interacted with
these proteins via direct or indirect mechanisms (Fig. S9B). The antiproliferative effect of
TOP I, HDAC, DOT1L and G9A inhibitors in different cancer cells
in vitro was also verified. The result indicated that SN38
(TOP I inhibitor) and vorinostat (HDAC inhibitor) inhibited the
proliferation of A549 and H1975 cells (Fig. S9C and D).
Cellular in vitro assays
The role of KMT2C in a variety of tumor cell lines
has been previously studied, including breast, prostate and ovarian
cancer cell lines (39–41). In addition, KMT2C mutations have
been significantly associated with the metastasis of small-cell
lung cancer (21), but the role of
KMT2C in non-small cell lung cancer (NSCLC) metastasis has not, to
the best of our knowledge, been studied. Therefore, the role of
KMT2C in NSCLC was explored by knocking down KMT2C expression in
A549 and H1975 cells using shRNA. The RT-qPCR and dot blot results
showed that the expression of KMT2C in A549 and H1975 cells was
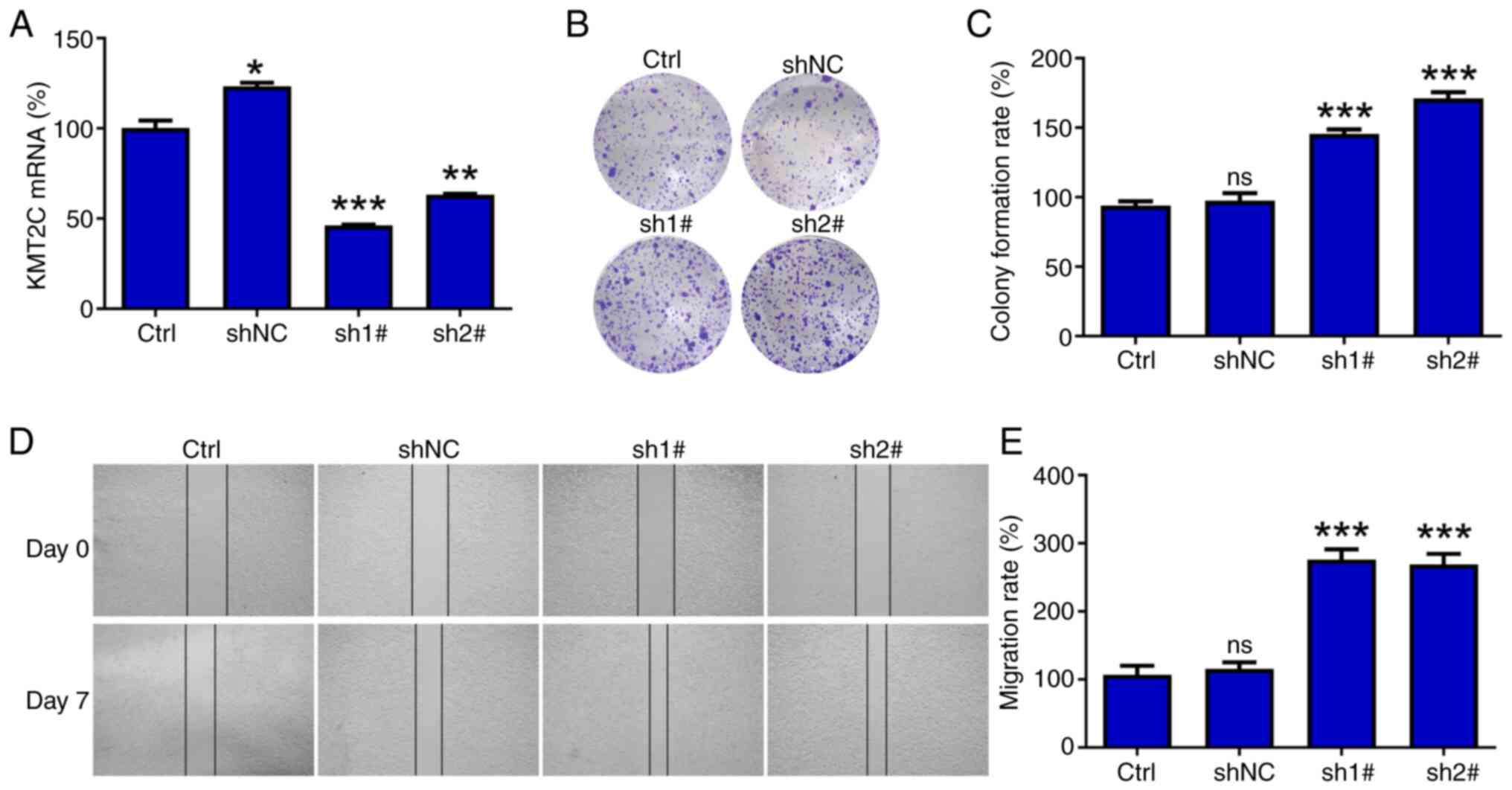
significantly reduced following shRNA treatment (Figs. 9A, S10A and S10B). The reduction in KMT2C expression
significantly improved the proliferation and migration of A549
cells in colony formation and wound healing assays, respectively
(Fig. 9B-E). However, knocking down
KMT2C expression reduced the proliferation of H1975 cells (Fig. S10C). These differing results may be
related to the different expression levels of KMT2C in these cells
(Fig. S10D).
Discussion
Immunotherapies using ICIs or adoptively transferred
immune cells have revolutionized cancer treatment, especially for
metastatic cancer (42,43), and patients previously considered as
incurable can now achieve long-term remission and survival
(44). Although immunotherapy can
produce a lasting response, ICI monotherapy typically has an ORR of
only ~20% for solid tumors, and numerous patients eventually
developed drug resistance. A number of mechanisms, including
intrinsic cancer cell factors and immunosuppressive cells, create a
TME that is hostile to tumor-targeting immune cells (45,46).
Consequently, researchers have begun to explore biomarkers to
achieve precise immunotherapy. Biomarkers can screen patients who
may benefit from immunotherapy and avoid unnecessary medical costs,
hyper-progression and potentially severe toxicity in individuals
who are unlikely to respond to treatment (47–49).
However, the identification of effective and reliable biomarkers
remains a significant challenge in immunotherapy. In the present
study, the value of KMT2C in predicting response to ICI
immunotherapy was investigated and it was discovered that KMT2C was
a robust pan-cancer prognostic biomarker.
The pan-cancer expression of KMT2C was first
evaluated and it was found that KMT2C was highly expressed in BRCA,
CHOL, ESCA, GBM, KICH, LAML, LGG, PAAD, PRAD and STAD. In contrast,
KMT2C expression was low in ACC, BLCA, CESC, COAD, LUAD, LUSC, OV,
SKCM, TGCT, THCA, THYM, UCEC and UCS. Paired expression analyses
revealed that KMT2C expression was high in CHOL, COAD, LIHC, KICH
and THCA, suggesting that KMT2C plays an important role in the
development and progression of these cancer types. The pan-cancer
prognostic value of KMT2C was also evaluated using univariate Cox
regression and Kaplan-Meier analyses. These two methods yielded
consistent results. KMT2C was a protective factor for patients with
KIRC and OV, but a risk factor for patients with LUSC and UVM,
indicating that KMT2C might have different roles in different
cancer types.
Tumor-infiltrating immune cells have critical roles
in the eradication of tumors, and cancer cells can inhibit immune
cell infiltration by reshaping the TME (50,51).
In the present study, the GSEA results suggested that KMT2C
expression was significantly negatively correlated with immune
activation, such as the TNF-α, IFN-γ and IFN-α pro-inflammatory
responses and allograft rejection, indicating that tumors with high
levels of KMT2C were poorly immunogenic. Angiogenesis is another
cancer hallmark that is necessary for tumor cell survival and plays
an important role in tumor growth, invasion and metastasis
(52). In the present study, it was
demonstrated that KMT2C expression was significantly positively
correlated with the neovascularization process, angiogenesis and
the VEGFA/VEGFR2 signaling pathway. Tumor angiogenesis is a marker
of cancer progression, and there is growing evidence that it also
causes immunosuppression and evasion of antitumor immunity.
Angiogenesis factors are known to directly or indirectly inhibit T
cell development and function, promote T cell exhaustion by
upregulating immune checkpoints, inhibit DC maturation, regulate
macrophage polarization and increase the number of intratumoral
regulatory T cells and MDSCs (53–55).
In addition, tumor vascular dysfunction leads to insufficient blood
perfusion and oxygenation, in turn leading to tumor hypoxia, which
produces various immunosuppressive effects (56). The results of the present study
suggested that KMT2C may promote tumor vascularization by
regulating the expression of angiogenesis factors such as VEGF and
FGF. Thus, targeting KMT2C may be a good way to inhibit
vascularization.
In the present study, the association between KMT2C
expression and the intratumoral infiltration of various immune
cells that play critical and diverse roles in tumor suppression was
investigated. Cytotoxic CD8+ T cells recognize and kill
tumor cells (57). Monocytes play
key roles in the maintenance of homeostasis, pathogen recognition,
clearance and inflammation (58).
NK cells have antitumor functions and are involved in immune
regulation (59). Macrophages are
important autologous immune cells that participate in the clearance
of infected and tumor-transformed cells and in immunomodulation
(60). CAFs are an important
component of the TME and have multiple functions such as
extracellular matrix remodeling, regulation of metabolism and
angiogenesis, and interaction with cancer cells and infiltrating
immune cells. CAFs promote tumorigenesis, tumor development, and
resistance to a various therapeutic strategies, including
chemotherapy, radiotherapy, targeted therapy, antiangiogenic
therapy, immunotherapy and endocrine therapy (61). ECs also have an important role in
tumor angiogenesis. Tumor-associated neutrophils enhance the
proliferation of tumor cells by releasing neutrophil extracellular
traps (62). The results of the
present study indicated that KMT2C expression was negatively
correlated with enrichment of infiltrated CD8+ T cells,
monocytes, NK cells and macrophages in most cancer types, and
positively correlated with the presence of CAFs, ECs, neutrophils
and Tregs in the TME. In addition, it was found that KMT2C levels
were positively correlated with the expression of inhibitory immune
genes, such as CD274, KDR and IDO1. Collectively, these results
suggested that KMT2C expression could maintain the
immunosuppressive TME by regulating immune cell infiltration.
The predictive value of KMT2C was then evaluated in
25 immunotherapy cohorts and it was found that KMT2C exhibited a
higher predictive value than TMB, T.Clonality and B.Clonality. By
evaluating whether KMT2C could predict the ORR in patients with
cancer receiving immunotherapy, it was found that patients with low
KMT2C expression had a higher ORR following immunotherapy than
those with high KMT2C expression. ORRs of patients in the
IMvigor210, PRJEB23709, PRJNA482620 and PRJEB25780 cohorts were
22.8, 53.8, 50.0 and 26.9%, respectively. The respective ORRs of
patients with low KMT2C expression were 32.6, 57.0, 100.0 and
45.0%, respectively. These results indicated that KMT2C may
effectively predict the response to immunotherapy. The correlation
between KMT2C and PD-L1 expression in kidney tumors was also
verified, and it was found that KMT2C expression was significantly
and positively correlated with PD-L1 expression. As one of the most
important immune checkpoints, PD-L1 can be induced by inflammatory
cytokines, including IFN, TNF-α and VEGF, in addition to
constitutively low expression in Antigen-presenting cells (APCs)
and a variety of non-hematopoietic cells (63,64).
Tumor cells and tumor-associated antigen presenting cells highly
express PD-L1 in the TME, whereas tumor-infiltrating lymphocytes
express PD-1 in response to long-term tumor antigen stimulation.
The combination of PD-L1 and PD-1 can induce apoptosis,
incapacitation and depletion of T cells, and then inhibit the
activation, proliferation and antitumor function of tumor
antigen-specific CD8+ T cells, leading to tumor immune
escape (65). KMT2C is a
methyltransferase that regulates gene expression via epigenetic
modifications (66). Therefore,
tumor cells may regulate the expression of PD-L1 through KMT2C,
thereby facilitating immune escape. This may also explain why
patients with low KMT2C expression have better treatment outcomes
when undergoing immunotherapy.
Since KMT2C is a DNA damage repair regulator,
targeting KMT2C could enhance the antitumor effects of
chemotherapeutic drugs, particularly those that induce DNA damage.
However, to the best of our knowledge, no KMT2C inhibitors have
been developed to date. In the present study, CMap and GDSC
datasets were used to analyze the association between drug
sensitivity and KMT2C expression. TOP I, HDAC, DOT1L and G9A
inhibitor sensitivities were significantly correlated with KMT2C
expression. The drug sensitivity test results also showed that TOP
I (SN38) and HDAC (vorinostat) inhibitors potently inhibited the
proliferation of A549 and H1975 cells.
In conclusion, in the present study, a pan-cancer
data analysis revealed that KMT2C was a protective factor for
patients with KIRC and OV, but a risk factor for patients with LUSC
and UVM. High KMT2C expression was negatively correlated with
immune cell infiltration, immune stimulatory regulators, TMB and
MSI in various cancer types. In addition, patients with low KMT2C
levels showed higher ORRs following immunotherapy. Taken together,
these findings demonstrated that KMT2C may be a biomarker for
predicting response to immunotherapy. However, the present study
had certain limitations, such as the relatively small sample size
in the immunotherapy cohort, which may have led to inevitable
systematic bias and affected the resulting conclusions. Therefore,
the role of KMT2C in the context of cancer immunotherapy requires
further validation using larger datasets. Furthermore, the use of
only lung cancer cell lines was another limitation of the present
study and thus, more studies including additional cell lines are
required in the future.
Supplementary Material
Supporting Data
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
This work was supported by The National Natural Science
Foundation of China (grant no. 81972040), The Anhui Provincial
Natural Science Foundation (grant nos. 2108085MH321 and
2108085QH379), The Research Fund of Anhui Medical University (grant
no. 2020×kj013) and the Foundation of Science and Technology
Department of Anhui Province of China (grant no. KJ2021A0313).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
WLW, XSL and RL designed the research. WC, LC, YWX,
MQW, ZYC, ZTW, YQW, JJX and YW performed the experiments. WC and
WLW wrote the manuscript. MQW, ZYC, ZTW, YQW and JJX provided
experiment materials. WLW revised the manuscript. All authors have
read and approved the final manuscript. WLW and XSL confirm the
authenticity of all the raw data.
Ethics approval and consent to
participate
Paraffin-embedded kidney tumor tissue samples were
obtained from patients treated at the Second Affiliated Hospital of
Anhui Medical University (Hefei, China), with written informed
consent. All experiments were approved by The Medical Ethics
Committee of the Second Affiliated Hospital of Anhui Medical
University (approval no. 81220282).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kubli SP, Berger T, Araujo DV and Mak L:
Beyond immune checkpoint blockade: Emerging immunological
strategies. Nat Rev Drug Discov. 20:899–819. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
He X and Xu C: Immune checkpoint signaling
and cancer immunotherapy. Cell Res. 30:660–669. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Huang Q, Lei Y, Li X, Guo F and Liu M: A
Highlight of the mechanisms of immune checkpoint blocker
resistance. Front Cell Dev Biol. 8:5801402020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Waldman AD and Lenardo J: A guide to
cancer immunotherapy: From T cell basic science to clinical
practice. Nat Rev Immunol. 20:651–668. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kruger S, Ilmer M, Kobold S, Cadilha B,
Endres S, Ormanns S, Schuebbe G, Renz BW, D'Haese JG, Schloesser H,
et al: Advances in cancer immunotherapy 2019-latest trends. J Exp
Clin Cancer Res. 38:2682019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ribas A and Wolchok JD: Cancer
immunotherapy using checkpoint blockade. Science. 359:1350–1355.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Darvin P, Toor S and Elkord V: Immune
checkpoint inhibitors: Recent progress and potential biomarkers.
Exp Mol Med. 50:1–11. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Acevedo J, Kimbrough E and Lou Y: Next
generation of immune checkpoint inhibitors and beyond. J Hematol
Oncol. 14:452021. View Article : Google Scholar
|
|
9
|
Johnson D, Nebhan C, Moslehi J and Balko
J: Immune-checkpoint inhibitors: Long-term implications of
toxicity. Nat Rev Clin Oncol. 19:254–267. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Franzin R, Netti G, Spadaccino F, Porta C,
Gesualdo L, Stallone G, Castellano G and Ranieri E: The use of
immune checkpoint inhibitors in oncology and the occurrence of AKI:
Where do we stand? Front Immunol. 11:5742712020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wright J, Powers A and Johnson D:
Endocrine toxicities of immune checkpoint inhibitors. Nat Rev
Endocrinol. 17:389–399. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rao R and Dou Y: Hijacked in cancer: The
KMT2 (MLL) family of methyltransferases. Nat Rev Cancer.
15:334–346. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Park K and Kim J: Transcriptional
regulation by the KMT2 histone H3K4 methyltransferases. Biochim
Biophys Acta Gene Regul Mech. 1863:1945452020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhai X and Brownell J: Biochemical
perspectives on targeting KMT2 methyltransferases in cancer. Trends
Pharmacol Sci. 42:688–699. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhu J, Liu Z, Liang X, Wang L, Wu D, Mao W
and Shen D: A Pan-Cancer study of KMT2 family as therapeutic
targets in cancer. J Oncol. 2022:39822262022. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shen E, Shulha H, Weng Z and Akbarian S:
Regulation of histone H3K4 methylation in brain development and
disease. Philos Trans R Soc Lond B Biol Sci. 369:201305142014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bochyńska A, Firzlaff J and Lüscher B:
Modes of Interaction of KMT2 Histone H3 Lysine 4
Methyltransferase/COMPASS complexes with chromatin. Cells.
7:172018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rampias T, Karagiannis D, Avgeris M,
Polyzos A, Kokkalis A, Kanaki Z, Kousidou E, Tzetis M, Kanavakis E,
Stravodimos K, et al: The lysine-specific methyltransferase
KMT2C/MLL3 regulates DNA repair components in cancer. EMBO Rep.
20:e468212019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chang A, Liu L, Ashby J, Wu D, Chen Y,
O'Neill SS, Huang S, Wang J, Wang G, Cheng D, et al: Recruitment of
KMT2C/MLL3 to DNA damage sites mediates DNA damage responses and
regulates PARP inhibitor sensitivity in cancer. Cancer Res.
81:3358–3373. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mendiratta G, Ke E, Aziz M, Liarakos D,
Tong M and Stites E: Cancer gene mutation frequencies for the U.S.
population. Nat Commun. 12:59612021. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Na F, Pan X, Chen J, Chen X, Wang M, Chi
P, You L, Zhang L, Zhong A, Zhao L, et al: KMT2C deficiency
promotes small cell lung cancer metastasis through DNMT3A-mediated
epigenetic reprogramming. Nat Cancer. 3:753–767. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen X, Zhang G, Chen B, Wang Y, Guo L,
Cao L, Ren C, Wen L and Liao N: Association between histone lysine
methyltransferase KMT2C mutation and clinicopathological factors in
breast cancer. Biomed Pharmacother. 116:1089972019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang P and Huang Y: Genomic alterations
in KMT2 family predict outcome of immune checkpoint therapy in
multiple cancers. J Hematol Oncol. 14:392021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang R, Wu H, Xu M and Xie X: KMT2A/C
mutations function as a potential predictive biomarker for
immunotherapy in solid tumors. Biomark Res. 8:712020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tu Z, Ouyang Q, Long X, Wu L, Li J, Zhu X
and Huang K: Protein Disulfide-Isomerase A3 is a robust prognostic
biomarker for cancers and predicts the immunotherapy response
effectively. Front Immunol. 13:8375122022. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sun D, Wang J, Han Y, Dong X, Ge J, Zheng
R, Shi X, Wang B, Li Z, Ren P, et al: TISCH: A comprehensive web
resource enabling interactive single-cell transcriptome
visualization of tumor microenvironment. Nucleic Acids Res.
49:D1420–D1430. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lamb J: The connectivity map: A new tool
for biomedical research. Nat Rev Cancer. 7:54–60. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Barretina J, Caponigro G, Stransky N,
Venkatesan K, Margolin A, Kim S, Wilson CJ, Lehár J, Kryukov GV,
Sonkin D, et al: The cancer cell line encyclopedia enables
predictive modelling of anticancer drug sensitivity. Nature.
483:603–607. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jin Y, Wang Z, He D, Zhu Y, Chen X and Cao
K: Identification of novel subtypes based on ssGSEA in
immune-related prognostic signature for tongue squamous cell
carcinoma. Cancer Med. 10:8693–8707. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu J, Liu Q, Shen H, Liu Y, Wang Y, Wang
G and Du J: Identification and validation of a three
Pyroptosis-Related lncRNA signature for prognosis prediction in
lung adenocarcinoma. Front Genet. 13:8386242022. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Mariathasan S, Turley S, Nickles D,
Castiglioni A, Yuen K, Wang Y, Kadel EE III, Koeppen H, Astarita
JL, Cubas R, et al: TGFβ attenuates tumour response to PD-L1
blockade by contributing to exclusion of T cells. Nature.
554:544–548. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xiong D, Wang Y and You M: A gene
expression signature of TREM2hi macrophages and γδ T cells predicts
immunotherapy response. Nat Commun. 11:50842020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhao J, Chen A, Gartrell R, Silverman A,
Aparicio A, Chu T, Bordbar D, Shan D, Samanamud J, Mahajan A, et
al: Immune and genomic correlates of response to anti-PD-1
immunotherapy in glioblastoma. Nat Med. 25:462–469. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kim S, Cristescu R, Bass A, Kim M,
Odegaard J, Kim K, Liu XQ, Sher X, Jung H, Lee M, et al:
Comprehensive molecular characterization of clinical responses to
PD-1 inhibition in metastatic gastric cancer. Nat Med.
24:1449–1458. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Amin MB, Greene FL, Edge SB, Compton CC,
Gershenwald JE and Brookland R: The Eighth Edition AJCC Cancer
Staging Manual: Continuing to build a bridge from a
population-based to a more ‘personalized’ approach to cancer
staging. CA Cancer J Clin. 67:93–99. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang WL, Jiang ZR, Hu C, Chen C, Hu ZQ,
Wang AL, Wang L, Liu J, Wang WC and Liu QS: Pharmacologically
inhibiting phosphoglycerate kinase 1 for glioma with NG52. Acta
Pharmacol Sin. 42:633–640. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Pan D, Kobayashi A, Jiang P, Ferrari de
Andrade L, Tay RE, Luoma AM, Tsoucas D, Qiu X, Lim K, Rao P, et al:
A major chromatin regulator determines resistance of tumor cells to
T cell-mediated killing. Science. 359:770–775. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Gala K, Li Q, Sinha A, Razavi P, Dorso M,
Sanchez-Vega F, Chung YR, Hendrickson R, Hsieh JJ, Berger M, et al:
KMT2C mediates the estrogen dependence of breast cancer through
regulation of ERα enhancer function. Oncogene. 37:4692–4710. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Xiong W, Deng H, Huang C, Zen C, Jian C,
Ye K, Zhong Z, Zhao X and Zhu L: MLL3 enhances the transcription of
PD-L1 and regulates anti-tumor immunity. Biochim Biophys Acta Mol
Basis Dis. 1865:454–463. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zheng J, Wang C, Gao C, Xiao Q, Huang C,
Wu M and Li LY: MLL3 suppresses tumorigenesis through regulating
TNS3 enhancer activity. Cell Death Dis. 12:3642021. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Sugie T: Immunotherapy for metastatic
breast cancer. Chin Clin Oncol. 7:282018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wrobel P and Ahmed S: Current status of
immunotherapy in metastatic colorectal cancer. Int J Colorectal
Dis. 34:13–25. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Sharma P and Allison J: The future of
immune checkpoint therapy. Science. 348:56–61. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Vesely M, Zhang T and Chen L: Resistance
mechanisms to Anti-PD cancer immunotherapy. Annu Rev Immunol.
40:45–74. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Schoenfeld A and Hellmann M: Acquired
resistance to immune checkpoint inhibitors. Cancer Cell.
37:443–455. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Chan T, Yarchoan M, Jaffee E, Swanton C,
Quezada S, Stenzinger A and Peter S: Development of tumor mutation
burden as an immunotherapy biomarker: Utility for the oncology
clinic. Ann Oncol. 30:44–56. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Walk E, Yohe S, Beckman A, Schade A,
Zutter M, Pfeifer J and Berry AB; College of American Pathologists
Personalized Health Care Committee, : The cancer immunotherapy
biomarker testing landscape. Arch Pathol Lab Med. 144:706–724.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Chang L, Chang M, Chang H and Chang F:
Microsatellite instability: A predictive biomarker for cancer
immunotherapy. Appl Immunohistochem Mol Morphol. 26:e15–e21. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Chen Y, Zhao B and Wang X: Tumor
infiltrating immune cells (TIICs) as a biomarker for prognosis
benefits in patients with osteosarcoma. BMC Cancer. 20:10222020.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Jiang B, Mason J, Jewett A, Liu M, Chen W,
Qian J, Ding Y, Ding S, Ni M, Zhang X and Man YG:
Tumor-infiltrating immune cells: Triggers for tumor capsule
disruption and tumor progression? Int J Med Sci. 10:475–497. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Li T, Kang G, Wang T and Huang H: Tumor
angiogenesis and anti-angiogenic gene therapy for cancer. Oncol
Lett. 16:687–702. 2018.PubMed/NCBI
|
|
53
|
Liu ZL, Chen HH, Zheng LL, Sun LP and Shi
L: Angiogenic signaling pathways and anti-angiogenic therapy for
cancer. Signal Transduct Target Ther. 8:1982023. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Fang JW, Lu Y, Zheng JY, Jiang XC, Shen
HX, Shang X, Lu Y and Fu P: Exploring the crosstalk between
endothelial cells, immune cells, and immune checkpoints in the
tumor microenvironment: New insights and therapeutic implications.
Cell Death Dis. 14:5862023. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zhu L, Yu X, Wang L, Liu J, Qu Z, Zhang H,
Li L, Chen J and Zhou Q: Angiogenesis and immune checkpoint dual
blockade in combination with radiotherapy for treatment of solid
cancers: Opportunities and challenges. Oncogenesis. 10:472021.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Guo FF and Cui JW: Anti-angiogenesis:
Opening a new window for immunotherapy. Life Sci. 258:1181632020.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Iwahori K: Cytotoxic CD8+ lymphocytes in
the tumor microenvironment. Adv Exp Med Biol. 1224:53–62. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Ugel S, Canè S, Sanctis F and Bronte V:
Monocytes in the tumor microenvironment. Annu Rev Pathol.
16:93–122. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Wu S, Fu T, Jiang Y and Shao Z: Natural
killer cells in cancer biology and therapy. Mol Cancer. 19:1202020.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Li X, Liu R, Su X, Pan Y, Han X, Shao C
and Shi Y: Harnessing tumor-associated macrophages as aids for
cancer immunotherapy. Mol Cancer. 18:1772020. View Article : Google Scholar
|
|
61
|
Liao Z, Tan Z, Zhu P and Tan N:
Cancer-associated fibroblasts in tumor microenvironment-Accomplices
in tumor malignancy. Cell Immunol. 343:1037292021. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Kim J and Bae J: Tumor-Associated
macrophages and neutrophils in tumor microenvironment. Mediators
Inflamm. 2016:60581472016. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Dirix V, Corbière V, Thomas CW, Selis E,
Allard S, Hites M, Aerts L, Giese T and Mascart F: Blood
tolerogenic monocytes and low proportions of dendritic cell
subpopulations are hallmarks of human tuberculosis. J Leukoc Biol.
103:945–954. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Safi M, Ahmed H, Al-Azab M, Xia YL, Shan
X, Al-Radhi M, Al-Danakh A, Shopit A and Liu J: PD-1/PDL-1
inhibitors and cardiotoxicity; molecular, etiological and
management outlines. J Adv Res. 29:45–54. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Hashimoto M, Kamphorst AO, Im SJ, Kissick
HT, Pillai RN, Ramalingam SS, Araki K and Ahmed R: CD8 T cell
exhaustion in chronic infection and cancer: Opportunities for
interventions. Annu Rev Med. 69:301–318. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Yang HS, Cui W and Wang LH: Epigenetic
synthetic lethality approaches in cancer therapy. Clin Epigenetics.
11:1362019. View Article : Google Scholar : PubMed/NCBI
|