Introduction
Gastric cancer (GC) is a common type of digestive
system tumor worldwide; specifically, it was the fourth most common
cancer worldwide in 2020. Although the mortality rate of GC has
declined in certain countries in recent years, it still ranks fifth
among cancers worldwide (1,2). At present, biomarkers for the
diagnosis and prognosis of GC are derived from serological and
tissue specimens, with serum markers such as carcinoembryonic
antigen (CEA), carbohydrate antigen (CA)19-9 and CA 72–4 have been
used for auxiliary diagnosis and monitoring of GC. However, the
optimal treatment window is often missed due to the low specificity
and sensitivity of these markers (3). Histological markers, such as Ki67, p27
and p53, also have problems of low sensitivity and specificity for
GC diagnosis. Therefore, as pathology is considered the gold
standard for cancer diagnosis (4),
the present study aimed to explore novel histological biomarkers
for the diagnosis and prognosis evaluation of GC.
Cluster-of-differentiation gene 44 (CD44), a cell
surface adhesion molecule, has been recognized as a cancer stem
cell marker for several types of cancer, including GC (5). CD44 variant isoform 9 (CD44v9), one of
the major protein splice variant isoforms, is highly expressed in
human GC tissues (6,7) and has been indicated to be associated
with a poor prognosis (8–10). Studies have reported that CD44v9
expression is associated with the pathological features of GC, lung
adenocarcinoma, urothelial cancer and bladder cancer (10–13);
however, certain studies which focused on the expression of CD44v9
in GC tissues reported inconsistencies in terms of the distribution
of patient sex, tumor size, tumor stage and tumor classification
strategies (9,10,14).
In addition, CD44 is expressed in lymphocytes and macrophages
surrounding tumor cells (15,16),
but little is known about CD44v9 expression in these cells.
T cell immunoglobulin and mucin domain-containing
protein 3 (TIM3), a crucial immune checkpoint protein, is expressed
mainly by immune cells, such as T helper cell 1 lymphocytes,
cytotoxic lymphocytes, monocytes, macrophages, natural killer (NK)
cells and dendritic cells, and is expressed at low levels in tumor
cells (17–19). Nevertheless, TIM3 in cancer cells
has been considered as an emerging target for GC treatment
(20). Recent studies have reached
different conclusions regarding the role of tumor-infiltrating
TIM3+ cells in predicting the overall survival (OS) of
patients with GC (20,21). Given that the CD44 and TIM3 proteins
are both expressed in lymphocytes and macrophages, we hypothesized
that synchronous expression of CD44v9 and TIM3 in tumor or stromal
cells could provide a promising marker for the diagnosis and
prognosis evaluation of GC.
In the present study, multiplex immunofluorescence
(mIF) was used to detect the expression of CD44v9 and TIM3 in GC
tissues. This method can be used to analyze the expression of
CD44v9 and TIM3 alone or combined, as well as their expression in
cytokeratin (CK)+ and CK− regions in GC
tissues. In addition, the present study assessed the potential
clinical value of these proteins in GC diagnosis and prognosis
evaluation.
Materials and methods
Study design
mIF was performed to detect the expression of CD44v9
and TIM3 in the CK+ and CK− regions of
tissues from patients with GC. A total of 96 patients who underwent
gastric resection without preoperative therapy between April and
November 2009 were included in the present study. The information
on the clinicopathological characteristics of the patients and the
corresponding tumor tissue microarray (TMA), which contained 180
cores, including 96 tumor tissue cores and 84 matched
tumor-adjacent normal tissues cores, was purchased solely from
Shanghai Outdo Biotech Co., Ltd. (catalog no. HStmA180Su13).
Matched tumor-adjacent normal tissues were unavailable for 12 of
the patients. The clinicopathological stage was based on the
seventh edition of the American Joint Committee on Cancer Staging
Manual (22). The present study was
performed with the approval of the Institutional Ethics Committee
of the Tangshan People's Hospital (Tangshan, China). Written
informed consent was obtained from all patients prior to the
study.
TMA preparation
Formalin-fixed, paraffin-embedded (FFPE) GC tissues
were evaluated by pathologists. Cores were cut from representative
tumor regions and arranged regularly in a blank paraffin block. The
tissue cores were then allowed to fuse in a thermostatic oven at
52°C. Subsequently, the tissue array block was continuously sliced
to make a TMA (gastric cancer TMA; cat. no. HStmA180Su13; Shanghai
Outdo Biotech Co., Ltd.), which was heated in an oven at 60°C for
16 h to fix the tissue on the slide. Each core was 1.5 mm in
diameter and 4 µm in thickness. Finally, pathologists checked the
quality of each core under the Nikon Eclipse E600 light microscope
and recorded the diagnosis based on hematoxylin and eosin (H&E)
staining. The H&E staining was performed using a fully
automatic dyeing machine (Leica ST5020; Leica Microsystems GmbH).
The H&E staining process was performed at room temperature. The
TMA was treated with xylene I and xylene II separately for 10 min,
followed by hydration in 100–70% ethanol for 5 min and rinsing in
running water for 1 min. The TMA was placed in hematoxylin staining
solution for 5 min, rinsed with running water for 2 min, washed
with 1% HCl and PBS buffer, and immersed in water for
counterstaining. The TMA was then placed in eosin staining solution
for 5 min. Subsequently, the TMA was dehydrated sequentially in
75–100% ethanol, followed by immersion in xylene I and xylene II to
complete the staining process.
mIF
The TMA was heated in an oven at 63°C for 1 h to
prepare for dewaxing. Dewaxing and hydration were then performed
using a fully automatic dyeing machine (Leica ST5020; Leica
Microsystems, Inc.) using xylene and gradient alcohol solution. The
TMA was then boiled in 1X repair solution (10X AR6 buffer; cat. no.
AR6001KT; Akoya Biosciences, Inc.) for 3 min, heated in a microwave
on low power for 15–20 min and cooled at room temperature to
complete antigen retrieval. The TMA was then treated with
H2O2 for 10 min to remove endogenous
peroxidase and washed with TBST (0.1% Tween20). The sections were
subsequently incubated with blocking buffer (1X Antibody
Diluent/Block; cat. no. ARD1001EA; Akoya Biosciences, Inc.) at room
temperature for 10 min, primary antibodies at room temperature for
1 h and HRP-conjugated secondary antibodies (EnVision™ FLEX+,
Mouse; catalog. no. K8002; and EnVision™ FLEX+, Rabbit; catalog.
no. K8009; both Agilent Technologies, Ltd.) at a 1:1 dilution at
room temperature for 10 min. After staining with Opal dye at a
1:100 dilution (Opal 7-colour Manual IHC Kit, catalog. no.
NEL801001KT; PerkinElmer, Inc.) and incubating at room temperature
for 10 min, the TMA was boiled in 1X repair solution (10X AR6
buffer; cat. no. AR6001KT; Akoya Biosciences, Inc.) for 3 min,
heated in a microwave on low power for 15–20 min and cooled at room
temperature to remove the primary and secondary antibodies (the
procedure was consistent with antigen retrieval). The TMA was then
washed with TBST (0.1% Tween20). Blocking, incubation with primary
antibody, incubation with secondary antibody, Opal staining, and
removal of primary and secondary antibodies was repeated until all
antibody staining was completed. Detailed information for the
primary antibodies is summarized in Table SI. The TMA was counterstained with
DAPI (D9542; Sigma-Aldrich; Merck KGaA) at room temperature for 5
min, washed with TBST (0.1% Tween20) and sealed with 1X
antifluorescence quenching agent (VECTASHIELD® HardSet™
Antifade Mounting Medium; catalog. no. H-1400; Vector Labs,
Inc.).
Multispectral imaging and cell
recognition
The TMA-stained images were captured using the
TissueFAXS Spectra Systems (TissueGnostics GmbH), which identifies
CK+ cells as tumor cells and CK− cells as
stromal cells in tumor tissue cores. The spectral characteristics
of each fluorophore were determined using single-antigen staining,
spectrograms were obtained using multispectral fluorescence
microscopy, and the signal was identified using StrataQuest
analysis software (version 7.1.129; TissueGnostics GmbH). Positive
cells were identified according to the intensity and area of the
nucleus and antibodies. The cells were divided into CK+
and CK− cells. In tumor-adjacent normal tissues cores,
CK+ cells are recognized as normal epithelial cells,
whilst CK− cells are identified as stromal cells
(21,23–26).
Protein expression was assessed based on the density of cells
(number of positive cells/mm2).
Statistical analysis
Non-normally distributed continuous variables are
expressed as the median (lower quartile, upper quartile). The
difference in protein expression in tissues was determined using
the Mann-Whitney U test and the comparison of protein expression
differences among tumor grades was performed using the
Kruskal-Wallis H test, followed by a post-hoc Steel-Dwass test. OS
rates were assessed using Kaplan-Meier analysis, and the log-rank
test was used to plot survival curves. The Cox proportional hazards
regression model was used to perform univariate and multivariate
survival analyses. Clinical associations were analyzed using the
χ2 test and correlations using Spearman's correlation
analysis. Receiver operating characteristic (ROC) curve analysis
was used to evaluate the diagnostic value of protein expression for
GC and to calculate the cut-off value. P<0.05 was considered to
indicate a statistically significant difference. The P-value is
indicative of a two-sided test. P<0.01 and P<0.001 were
considered to indicate strong significance. All the analyses were
performed using SPSS 22.0 (IBM Corp.) and JMP Pro 17.0 (SAS Corp.)
statistical software.
Results
Clinical specimens and clinical
characteristics
Human tissue samples were collected from 96 patients
with GC who had not received chemotherapy or radiotherapy prior to
radical resection. Among the 96 tumor tissues and 84 matched
tumor-adjacent normal tissues collected for use in the TMA
(Fig. S1), the normal tissue of
one patient was not included in the analysis due to the absence of
epithelial cells. The follow-up period ranged from 5.7 to 6.2
years. As of July 2015, 57/96 patients with GC had died. The mean
age of the patients was 64 years. Most of the tumors were
concentrated in the gastric antrum (n=50), stomach body (n=25), and
cardia (n=8). There was 13 cases of GC involving two or more than
two sites of the stomach. The pathological types of the patients
with GC included adenocarcinoma (n=76), signet ring cell carcinoma
(n=12), mucinous cell carcinoma (n=7) and undifferentiated
carcinoma (n=1). The clinical characteristics of the patients with
GC are presented in Table I.
 | Table I.Clinical characteristics of patients
with gastric cancer (n=96). |
Table I.
Clinical characteristics of patients
with gastric cancer (n=96).
| Characteristic | n (%) |
|---|
| Sex |
|
|
Male | 65 (67.7) |
|
Female | 31 (32.3) |
| Age, years |
|
|
<65 | 51 (53.1) |
|
≥65 | 45 (46.9) |
| Tumor size, cm |
|
| ≤5 | 49 (51.0) |
|
>5 | 47 (49.0) |
| T
classification |
|
| T1 | 2 (2.1) |
| T2 | 11 (11.5) |
| T3 | 50 (52.1) |
| T4 | 33 (34.4) |
| N
classification |
|
| N0 | 20 (20.8) |
| N1 | 19 (19.8) |
| N2 | 25 (26.0) |
| N3 | 32 (33.3) |
| M
classification |
|
| M0 | 94 (97.9) |
| M1 | 2 (2.1) |
| TNM stage |
|
| I | 8 (8.3) |
| II | 27 (28.1) |
|
III | 59 (61.5) |
| IV | 2 (2.1) |
| Pathological
grade |
|
| G2 | 16 (16.7) |
| G3 | 73 (76.0) |
| G4 | 7 (7.3) |
Expression of CD44v9 and TIM3 and
their co-expression in the CK+ and CK−
regions of tumor tissues and adjacent normal tissues from patients
with GC
The expression levels of CD44v9, TIM3 and CK were
assessed using mIF staining. CK was used as a marker to distinguish
tumor cells from stromal cells. Tumor cells were predominantly
present in the CK+ regions, and stromal cells were
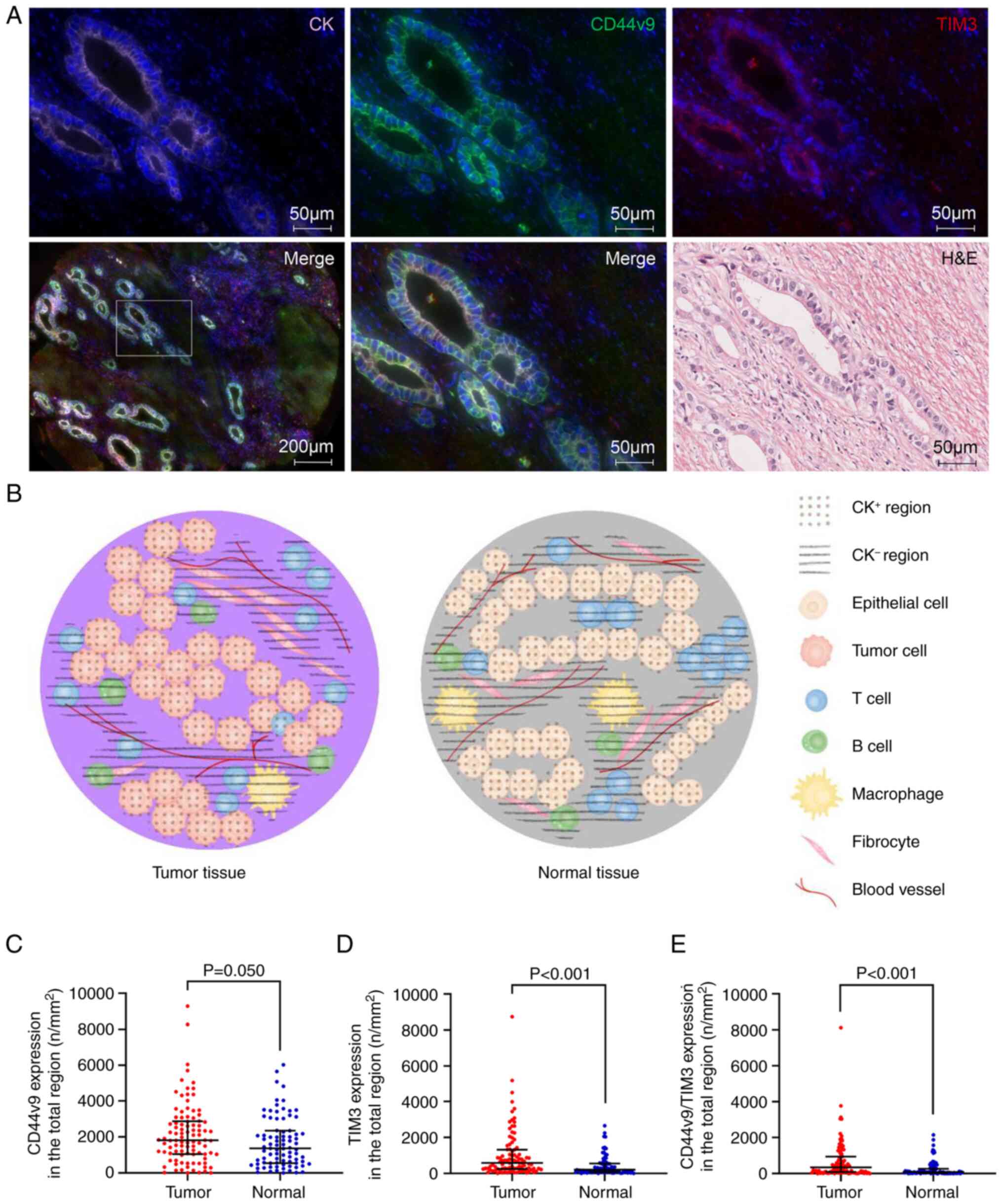
present in the CK− region (Fig. 1A). An ideograph of the
CK+ and CK− regions in GC tissues and normal
gastric tissues is presented in Fig.
1B. First, the differences in CD44v9 and TIM3 expression
between tumor tissues and adjacent normal tissues of patients with
GC were evaluated. There was no significant difference in CD44v9
expression (P=0.05; Fig.
1C). However, the density of cells expressing TIM3 and
co-expressing CD44v9 and TIM3 (CD44v9/TIM3) was significantly
greater in tumor tissues than in normal tissues (P<0.001;
Fig. 1D and E). Second, differences
in the density of cells expressing CD44v9, TIM3 and CD44v9/TIM3
between the CK+ and CK− regions of the tumor
tissues were assessed. It was demonstrated that the density of
cells expressing CD44v9, TIM3 and CD44v9/TIM3 was significantly
higher in the CK+ region than in the CK−
region (P<0.001; Table II).
Finally, the differences in the density of cells expressing CD44v9,
TIM3 and CD44v9/TIM3 in the CK+ region or the
CK− region between the tumor tissues and normal tissues
was assessed. It was demonstrated that the density of cells with
these three expression patterns was significantly higher in the
CK+ and CK− regions of tumor tissues than in
normal tissues (P<0.05; Table III). Among the CD44v9+
cells, 16.67, 17.15 and 15.78% were dual-positive cells (the cells
co-expressing CD44v9 and TIM3) in the total, CK+ and
CK− regions, respectively, of the tumor tissues.
However, among the TIM3+ cells, 27.09, 92.57 and 37.28%
were dual-positive cells in the total, CK+ and
CK− regions, respectively, of the tumor tissues. These
data indicate that the expression levels of CD44v9, TIM3 and
CD44v9/TIM3 were higher in the CK+ regions compared with
in the CK− regions in tumor tissues, and overall, the
expression levels in tumor tissues were higher than in normal
tissues.
 | Table II.Comparison of the density of cells
expressing multiplex immunofluorescence markers in
cytokeratin-positive and -negative regions of gastric cancer
tissues. |
Table II.
Comparison of the density of cells
expressing multiplex immunofluorescence markers in
cytokeratin-positive and -negative regions of gastric cancer
tissues.
| mIF marker | CK+
region, n/mm2 | CK−
region, n/mm2 | P-value |
|---|
| CD44v9 | 5,413.08 (2,944.84,
7,726.14) | 952.08 (408.96,
1,617.90) |
<0.001a |
| TIM3 | 854.27 (267.89,
2861.04) | 401.74 (160.16,
958.02) |
<0.001a |
| CD44v9/TIM3 | 802.60 (200.58,
2423.22) | 156.93 (25.80,
431.16) |
<0.001a |
 | Table III.Comparison of the density of cells
expressing multiplex immunofluorescence markers in gastric cancer
and normal tissues of different regions. |
Table III.
Comparison of the density of cells
expressing multiplex immunofluorescence markers in gastric cancer
and normal tissues of different regions.
| A, CK+
region |
|---|
|
|---|
| mIF marker | Cancer tissue,
n/mm2 | Normal tissue,
n/mm2 | P-value |
|---|
| CD44v9 | 5,413.08 (2,944.84,
7726.14) | 3,162.35 (1,837.03,
4731.01) |
<0.001a |
| TIM3 | 854.27 (267.89,
2861.04) | 214.37 (57.40,
823.47) |
<0.001a |
| CD44v9/TIM3 | 802.60 (200.58,
2423.22) | 177.38 (37.03,
705.24) |
<0.001a |
|
| B,
CK− region |
|
| CD44v9 | 952.08 (408.96,
1617.90) | 502.40 (180.41,
1292.61) | 0.016a |
| TIM3 | 401.74 (160.16,
958.02) | 186.88 (76.22,
420.92) |
<0.001a |
| CD44v9/TIM3 | 156.93 (25.80,
431.16) | 33.03 (6.74,
143.50) |
<0.001a |
Correlations between clinical
characteristics and the expression of CD44v9 and TIM3, and their
co-expression in the CK+ and CK− regions of
tumor tissues from patients with GC
To assess the potential value of CD44v9 and TIM3 in
the diagnosis of GC, the present study analyzed the correlations
between the density of cells expressing CD44v9, TIM3 and
CD44v9/TIM3 in the CK+ and CK− regions of
tumor tissues and pathological features. It was revealed that the
density of cells positive for CD44v9, TIM3 and CD44v9/TIM3 in the
total, CK+ and CK− regions was significantly
positively correlated with patient age (P<0.05; Tables IV, SII and SIII). Additionally, the density of cells
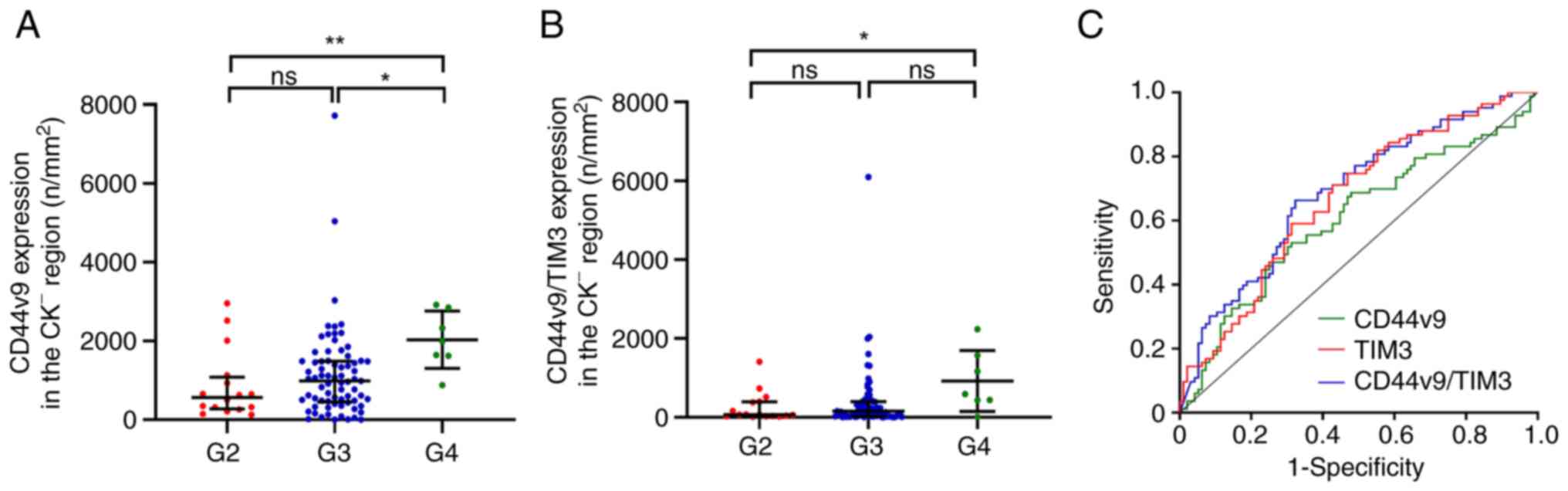
expressing CD44v9 in the CK− region was positively
correlated with tumor grade (P<0.01; Table IV), and the density of cells
expressing CD44v9 in the CK− region was significantly
higher in grade (G)4 GC samples than in G2 and G3 GC samples
(P<0.05; Fig. 2A). Similarly,
the density of cells expressing CD44v9/TIM3 in the CK−
region of tumor tissues was significantly positively correlated
with tumor grade and metastasis (P<0.05; Table IV), and the density of cells
expressing CD44v9/TIM3 in the CK− region was
significantly higher in G4 GC samples than in G3 samples
(P<0.05; Fig. 2B). The ROC
curves based on CD44v9, TIM3 and CD44v9/TIM3 expression patterns in
different regions are presented in Figs. 2C and S2. The expression of CD44v9 in the total
region of cancer tissues was not significant for diagnosis of GC
(P=0.050), whereas its expression was significant for GC
diagnosis when distinguishing CK+ (P<0.016)
and CK− (P=0.016) regions. TIM3 in the
CK+ region had the largest area under the curve (AUC)
and the highest diagnostic specificity (85.54%). Moreover, TIM3
also showed the highest diagnostic sensitivity (71.88%) in the
total region (Table V). The AUC of
CD44v9 and TIM3 and the CD44v9/TIM3 was higher in CK+
regions than in the total region and CK− region
(Table V; Figs. 2C and S2). These results indicate that the
expression of CD44v9 and TIM3 in accurate regions based on
CK− and CK+ could provide higher diagnostic
value for GC.
 | Table IV.Correlation between clinical
characteristics and the expression of multiplex immunofluorescence
markers in the cytokeratin-negative region in patients with gastric
cancer (n=96). |
Table IV.
Correlation between clinical
characteristics and the expression of multiplex immunofluorescence
markers in the cytokeratin-negative region in patients with gastric
cancer (n=96).
|
| CD44v9 | TIM3 | CD44v9/TIM3 |
|---|
|
|
|
|
|
|---|
| Characteristic | rs | P-value | rs | P-value | rs | P-value |
|---|
| Sex | 0.180 | 0.080 | 0.152 | 0.141 | 0.127 | 0.216 |
| Age | 0.245 | 0.016a | 0.203 | 0.047a | 0.235 | 0.021a |
| Tumor size | 0.018 | 0.863 | 0.060 | 0.561 | 0.045 | 0.663 |
| T
classification | −0.096 | 0.350 | −0.057 | 0.582 | −0.065 | 0.526 |
| N
classification | 0.060 | 0.563 | 0.143 | 0.164 | 0.135 | 0.190 |
| M
classification | 0.176 | 0.086 | 0.192 | 0.061 | 0.208 | 0.042a |
| TNM stage | −0.023 | 0.826 | 0.055 | 0.596 | 0.041 | 0.690 |
| Pathological
grade | 0.272 | 0.007a | 0.197 | 0.054 | 0.202 | 0.049a |
 | Table V.Receiver operating characteristic
curve analysis of gastric cancer for the expression of multiplex
immunofluorescence markers in different regions. |
Table V.
Receiver operating characteristic
curve analysis of gastric cancer for the expression of multiplex
immunofluorescence markers in different regions.
| Index | AUC | P-value | Cut-off,
n/mm2 | Sensitivity, % | Specificity, % |
|---|
| Total |
|
|
|
|
|
|
CD44v9 | 0.585 | 0.050 | 1,035.81 | 77.08 | 40.96 |
|
TIM3 | 0.690 |
<0.001a | 261.08 | 71.88 | 60.24 |
|
CD44v9/TIM3 | 0.680 |
<0.001a | 613.59 | 62.50 | 69.88 |
| CK+ |
|
|
|
|
|
|
CD44v9 | 0.704 |
<0.001a | 3,922.86 | 67.71 | 69.88 |
|
TIM3 | 0.709 |
<0.001a | 1,225.51 | 45.83 | 85.54 |
|
CD44v9/TIM3 | 0.703 |
<0.001a | 750.57 | 52.08 | 81.93 |
| CK− |
|
|
|
|
|
|
CD44v9 | 0.604 | 0.016a | 512.57 | 68.75 | 53.01 |
|
TIM3 | 0.665 |
<0.001a | 320.76 | 57.29 | 71.08 |
|
CD44v9/TIM3 | 0.688 |
<0.001a | 63.65 | 67.71 | 66.27 |
Prognostic value of the expression of
CD44v9 and TIM3 and their co-expression in the CK+ and
CK− regions of tumor tissues from patients with GC
To analyze the prognostic value of CD44v9, TIM3 and
CD44v9/TIM3 expression in the CK+ and CK−
regions of tumor tissues, the patients with GC were divided into a
high- and low-expression group according to the median density of
cells expressing the indicator proteins. The differences between
the high- and low-expression groups were then compared. The results
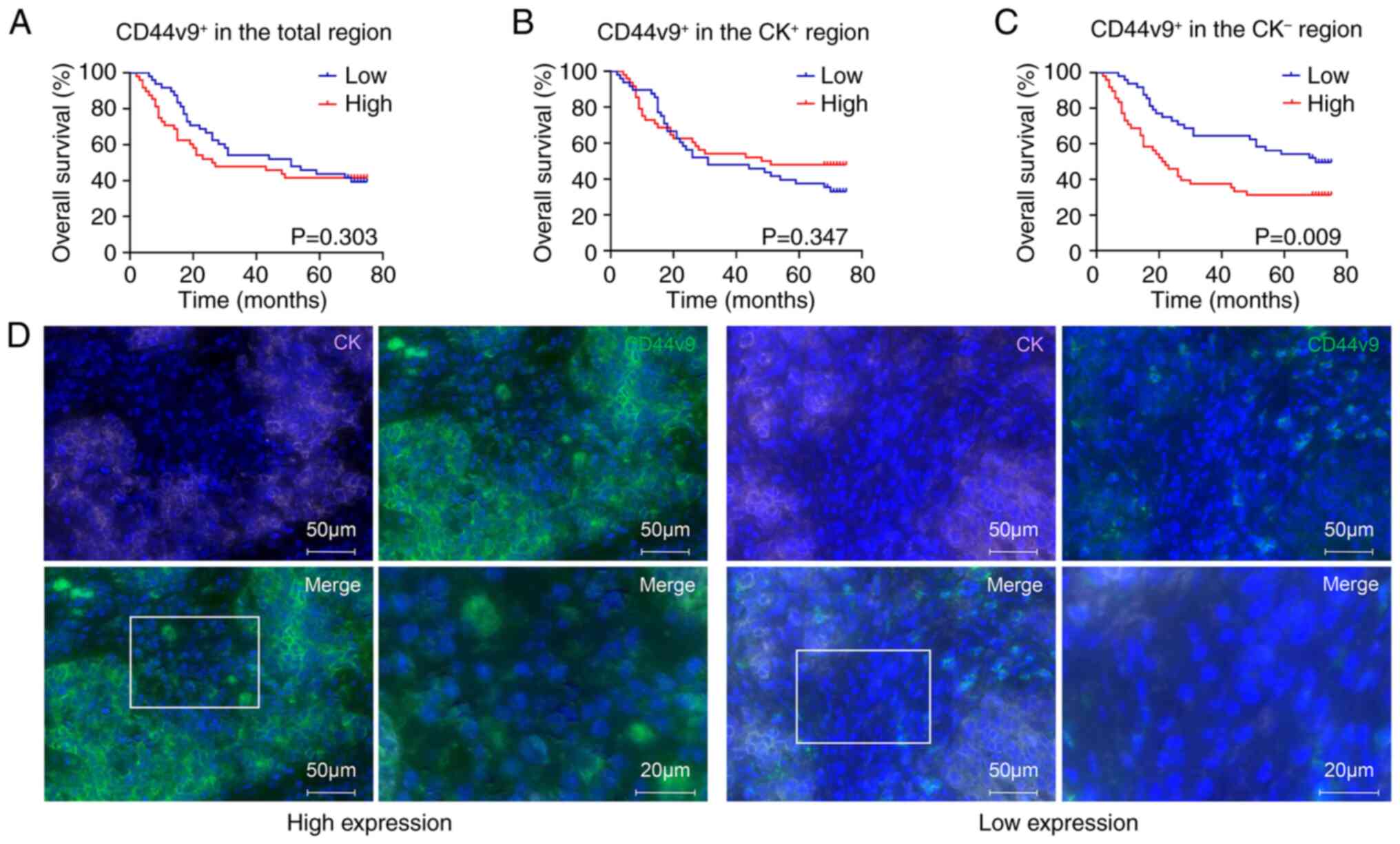
demonstrated that the 5-year survival rates of patients with high
and low CD44v9 expression in the CK− region were 31.25
and 68.75%, respectively. The OS of patients with GC with high
CD44v9 expression in the CK− region was significantly
shorter than that of patients with low CD44v9 expression
(P<0.01; Fig. 3C and D). This
indicates that a high expression of CD44v9 in the CK−
region predicts a poor prognosis in patients with GC. However, the
survival curves did not significantly differ between patients with
GC with high or low expression of other markers (P<0.05;
Figs. 3A and B, and S3). Furthermore, univariate analysis of
the markers and pathological features demonstrated that tumor (T)
classification, node (N) classification, metastasis (M)
classification, TNM stage, and the expression of CD44v9 in the
CK− region, were significant factors influencing OS in
patients with GC. Multivariate analysis indicated that high
expression of CD44v9 in the CK− region was a significant
independent risk factor predicting the OS of patients with GC
(P<0.01; hazard ratio, 2.387; 95% confidence interval,
1.384–4.118; Table VI). Taken
together, these findings indicate that the expression of CD44v9 in
the CK− region could be of great prognostic value for
patients with GC.
 | Table VI.Cox univariate multivariate
regression for overall survival in patients with gastric
cancer. |
Table VI.
Cox univariate multivariate
regression for overall survival in patients with gastric
cancer.
|
| Univariate Cox | Multivariate
Cox |
|---|
|
|
|
|
|---|
| Variable | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Sex (female vs.
male) | 1.109
(0.639–1.923) | 0.714 |
|
|
| Age (≥65 vs. <65
years) | 1.379
(0.820–2.318) | 0.226 |
|
|
| Tumor size (>5
vs. ≤5 cm) | 1.648
(0.976–2.785) | 0.062 |
|
|
| Tumor grade (G4 vs.
G2 and G3) | 2.025
(0.807–5.085) | 0.133 |
|
|
| TNM stage (III and
IV vs. I and II) | 3.016
(1.592–5.715) | 0.001a | 1.655
(0.747–3.668) | 0.215 |
| T classification
(T3 and T4 vs. T1 and T2) | 4.066
(1.270–13.020) | 0.018a | 2.196
(0.911–10.235) | 0.222 |
| N classification
(N1, N2 and N3 vs. N0) | 5.315
(1.920–14.713) | 0.001a | 3.054
(0.747–3.668) | 0.070 |
| M classification
(M1 vs. M0) | 6.678
(1.576–28.290) | 0.010a | 3.073
(0.708–13.345) | 0.134 |
| CD44v9
expression |
|
|
|
|
| In
total region (High vs. low) | 1.156
(0.687–1.943) | 0.585 |
|
|
| In
CK+ region (High vs. low) | 0.780
(0.462–1.317) | 0.352 |
|
|
| In
CK− region (High vs. low) | 1.989
(1.173–3.373) | 0.011a | 2.387
(1.384–4.118) | 0.002a |
| TIM3
expression |
|
|
|
|
| In
total region (High vs. low) | 1.374
(0.816–2.314) | 0.231 |
|
|
| In
CK+ region (High vs. low) | 1.352
(0.803–2.275) | 0.257 |
|
|
| In
CK− region (High vs. low) | 1.595
(0.943–2.696) | 0.082 |
|
|
| CD44v9/TIM3
expression |
|
|
|
|
| In
total region (High vs. low) | 1.272
(0.756–2.139) | 0.365 |
|
|
| In
CK+ region (High vs. low) | 1.423
(0.845–2.397) | 0.184 |
|
|
| In
CK− region (High vs. low) | 1.539
(0.913–2.594) | 0.106 |
|
|
Discussion
The tumor microenvironment (TME) has been reported
to serve crucial roles in the development, differentiation,
survival and proliferation of cancer cells (27,28).
In addition to tumor cells, fibroblasts, immune cells (such as T
lymphocytes, B lymphocytes, NK T cells and tumor-associated
macrophages) and stromal cells (such as pericytes and certain
adipocytes) are involved in the TME (28). In the past, studies on biomarkers in
cancer tissues have focused on tumor cells in tumor tissues,
ignoring stromal cells. However, certain biomarkers, such as TIM3,
are expressed in both tumor cells and stromal cells (18). Therefore, the present study aimed to
assess the expression of CD44v9 and TIM3 and the co-expression of
CD44v9 and TIM3 in tumor cells and stromal cells using mIF to
evaluate the potential clinical value of these biomarkers for the
evaluation if the diagnosis and prognosis of GC.
In the present study, the assessment of CD44v9 and
TIM3 expression in tissues differs from traditional qualitative and
semi-quantitative immunohistochemistry (IHC) methods (29–32).
H&E staining and mIF assays were performed, which used an
automated pathological imaging system to identify and count the
number of positive cells/mm2. This is a quantitative
approach for assessing protein expression levels in tissues and
qualified for histopathology of cancer study (21,23–24).
Although the scoring system of traditional IHC has been used in
many studies to measure protein expression, inconsistent results
are common (8,10,14).
For example, tissues with low densities of positive cells are
recorded as zero (8). Thus, the
present study directly adopted the method of measuring the density
of positive cells instead of immunofluorescence scores.
Furthermore, the cancer cells could be distinguished from stromal
cells according to CK expression, not the normal cells. Therefore,
the diagnosis of GC was according to the density of cells
expressing CD44v9 or TIM3 in the CK+ region. It was
demonstrated that the expression of CD44v9 in the CK+
region of tumor tissues was greater than that in normal tissues,
which indicates that CD44v9 function as an oncogene in GC
development. Considering the disruption of the intracellular
reactive oxygen species (ROS)/glutathione (GSH) balance (such as
excessive ROS production) leading to energy homeostasis imbalance
and tumor cell death (33), and the
decrease in intracellular ROS casused by CD44v9 via upregulation of
cystine uptake and promotion of GSH synthesis in GC cells (34,35),
we hypothesize that CD44v9 may promote GC cell proliferation by
intracellular decrease of ROS. Moreover, the present study
demonstrated that CD44v9+ epithelial cells could be
scattered or clustered in normal tissues. Similar studies reported
that the CD44v9 expression could occur in the regenerated gastric
epithelium cells or gastric epithelium cells infected with H.
pylori (36–38), as well as the expansion of stem
cells of gastric tissues (37).
Nevertheless, a previous study ignored the
distinction between the parenchyma and stroma in GC tissues
(10). CD44 was previously reported
to be expressed in T cells, B cells and macrophages (16,39–41)
and it has been reported to be upregulated in M2 macrophages, and
the deletion of CD44 hinders the polarization of macrophages,
suppressed the migration of tumor cells (41). As a variant subtype of CD44, CD44v9
has been reported to be overexpressed in myeloma plasma cells,
which could promote the adhesion of stromal cells and the secretion
of IL-6, which promotes cell proliferation, anti-apoptosis,
invasion and metastasis through the activation of the JAK/STAT3
signaling pathway (42,43). In addition, CD44v9 could be also
expressed in T cells (44). A
previous review revealed that ROS can modulate the TME by affecting
several stromal cells that provide metabolic support and blood
supply, and facilitate immune responses to tumors (45). The T cell-dependent antitumor
response is also dependent on ROS (45). Overexpression of CD44v9 in stromal
cells may disrupt the balance of ROS in T cells and affect the
antitumor function of T cells. Based on this evidence, we
hypothesize that high expression of CD44v9 in stromal cells may
promote tumor growth. In the present study, it was demonstrated
that the density of CD44v9+ cells in the CK−
region of tumor tissues was greater than that in normal tissues,
but lower than that in the CK+ region. The expression of
CD44v9 in the CK− region of tumor tissues was positively
correlated with tumor grade and age. In addition, patients with GC
with high CD44v9 expression in the CK− region
experienced a worse OS rate than those with low CD44v9 expression,
and CD44v9 was identified as an independent risk factor for GC. The
univariate Cox analysis revealed a difference between stages I–II
and stages III–IV (P<0.05). However, the multivariate Cox
analysis did not demonstrate a statistical difference between the
two groups. TNM staging was considered to be a potential
confounding factor in the statistical analysis, which highlighted
the importance of the expression level of CD44v9 in the
CK− region for prognosis of GC. The TNM staging may be a
confounding variable that indirectly affected the survival rates,
exhibiting a ‘false association’ with the outcome. Adjusting for
confounders in regression allows for the identification of
independent contributors to the outcomes (46,47).
Additionally, similar studies have also indicated that certain
prognostic factors, such as TNM stage and tumor size (21,48,49),
do not always hold statistical significance in Cox regression
analyses.
TIM3 is an immune checkpoint, and its activation can
increase T-cell dysfunction and exhaustion (50). TIM3 has been also recognized to
inhibit NK cell activity by binding to phosphatidyl serine
(51). The two aforementioned
studies reported that overexpression of TIM3 in immune cells could
promote tumor cell growth. However, previous studies have reported
that TIM3 is mainly expressed in immune cells, and rarely expressed
in epithelial-origin tumor cells, such as GC cells and lung
adenocarcinoma cells (18,19,52).
The present study demonstrated that the expression of TIM3 in tumor
tissues was higher than that in normal tissues, which is consistent
with the findings of Wang et al (18) and Chen et al (20). Furthermore, the present study
revealed that the expression of TIM3 in the CK+ region
was higher than that in the CK− region in tumor tissues,
contrary to the findings of Wu et al (21). Thus, we hypothesize that TIM3
originated from epithelial cells, and the different results based
on the same method may be caused by the histological heterogeneity
of GC tissue cores or the limited number of cases. The histological
heterogeneity includes individual characteristics, GC tissue
collection site and the quantity of T cell infiltration (53). Although the CK+ and
CK− regions were distinguished based on rigorous
examination, certain CK− immune cells were inevitably
incorporated into the CK+ region at the edge of the
region, so this analysis method needs to be improved. Moreover, in
the present study, the sensitivity of TIM3 in the total region was
71.88%, and the specificity of TIM3 in CK+ region for
diagnosing GC was 85.54%. Previous studies have reported that the
sensitivity of Ki67 and p27 for diagnosis in different histological
types of GC ranges from 46.3–62.4% (54,55),
which are all lower than the sensitivity of the biomarkers in the
present study. A previous study also reported that the sensitivity
of p53 in intestinal type adenocarcinoma was 83%, and sensitivity
for diagnosis of signet ring cell carcinoma was only 69% (56). A previous study showed that soluble
TIM3 in the serum of patients with GC could be detected by ELISA,
with diagnostic sensitivity (73.98%) and specificity (95.89%)
higher than TIM3 in the tissues of the present study (57). However, the sensitivity of TIM3 in
tissue specimens in the present study for GC diagnosis is higher
than the combined sensitivity of CA 72–4, CA 19-9 and CEA in serum
(58). The role of TIM3 in serum
and tissue for GC diagnosis should be further researched through
comparative studies in future.
In the present study, the AUC for CD44v9/TIM3 in the
CK− region was 0.688, which was higher the AUCs for
CD44v9 and TIM3 in the CK− region alone. Moreover, the
density of cells expressing CD44v9/TIM3 was positively correlated
with age, M classification and tumor grade. The diagnostic value of
CD44v9 combined with TIM3 in the CK− region was greater
than that of each factor individually. This indicates that the
combination of CD44v9 and TIM3 provides better clinical
significance than either factor alone. However, considering the
time-consuming and high cost of mIF, the expression of CK, TIM3 and
CD44v9 could be detected by individual or combined staining of the
traditional IHC method in clinical application. This traditional
IHC requires thorough training for pathologists, which is
time-consuming, labor-intensive and requires more tissues for
multiple biomarkers. For the TMA used in the present study,
multiple biomarkers could be detected in one experiment by using a
few tissue samples, and the StrataQuest analysis software could
quantitatively analyze different fluorophores, not depending on the
subjectivity of pathologists. Although the present study did not
use IHC, it is recommended that appropriate testing methods are
selected according to the sample size and cost in clinical
applications.
The present study has certain limitations: i) The
FFPE tissue specimens of patients with GC were preserved for a
relatively long time. Although several studies used samples that
were preserved for similar amount of time to those in the present
study (20,21), the length of time inevitably
affected the detection effect of CK, TIM3 and CD44v9; ii) the size
of the tissue samples were small, increasing the heterogeneity. It
is therefore necessary to recruit recent patients and increase the
sample size to reduce limitations in future studies; and iii) the
role of CD44v9 in GC diagnosis determined in the present study is
based on clinical samples, not in vitro and in vivo
experiments. Thus, we can only hypothesize that CD44v9 acted as an
oncogene through ROS on immune responses. Further study on the
mechanism of CD44v9 in GC development needs to be researched in the
future.
In summary, dividing tissue regions based on CK
expression is important for the diagnosis of GC. Moreover, TIM3 in
the CK+ region demonstrated diagnostic potential for GC.
High expression of CD44v9 in the CK− region was an
independent risk factor predicting the prognosis of GC. Although
CD44v9/TIM3 expression in the CK− region exhibited lower
diagnostic value than TIM3 in the CK+ region,
CD44v9/TIM3 expression in the CK− region was associated
with the degree of GC metastasis and differentiation, highlighting
the need for future research.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present work was supported by the Natural Science Foundation
of Hebei Province (grant no. H2021105019).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
YL and JL conceived and designed the project. LL
and RY collected and sorted the data. XW and LL performed the
statistical analysis of the data. RY, ZW and QL analyzed the
pathological images. The first draft of the manuscript was written
by XW and all authors commented on previous versions of the
manuscript. XW and YL confirm the authenticity of all the raw data.
All authors have read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was performed in line with the
principles of the Declaration of Helsinki. Approval was granted by
the Ethics Committee of the Tangshan People's Hospital (Tangshan,
China; approval no. RMYY-LLKS-2023203). Written informed consent
was obtained from all participants included in the present
study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sedeta E, Sung H, Laversanne M, Bray F and
Jemal A: Recent mortality patterns and time trends for the major
cancers in 47 countries worldwide. Cancer Epidemiol Biomarkers
Prev. 32:894–905. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
Globocan estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Matsuzaki J, Tsugawa H and Suzuki H:
Precision medicine approaches to prevent gastric cancer. Gut and
Liver. 15:3–12. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang FH, Zhang XT, Tang L, Wu Q, Cai MY,
Li YF, Qu XJ, Qiu H, Zhang YJ, Ying JE, et al: The Chinese society
of clinical oncology (CSCO): Clinical guidelines for the diagnosis
and treatment of gastric cancer. Cancer Commun (Lond). 44:127–172.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Morath I, Hartmann TN and Orian-Rousseau
V: CD44: More than a mere stem cell marker. Int J Biochem Cell
Biol. 81:166–173. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Choi ES, Kim H, Kim HP, Choi Y and Goh SH:
CD44v8-10 as a potential theranostic biomarker for targeting
disseminated cancer cells in advanced gastric cancer. Sci Rep.
7:49302017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Harn HJ, Ho LI, Chang JY, Wu CW, Jiang SY,
Lee HS and Lee WH: Differential expression of the human metastasis
adhesion molecule CD44V in normal and carcinomatous stomach mucosa
of Chinese subjects. Cancer. 75:1065–71. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hirata K, Suzuki H, Imaeda H, Matsuzaki J,
Tsugawa H, Nagano O, Asakura K, Saya H and Hibi T: CD44 variant 9
expression in primary early gastric cancer as a predictive marker
for recurrence. Br J Cancer. 109:379–386. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Go SI, Ko GH, Lee WS, Kim RB, Lee JH,
Jeong SH, Lee YJ, Hong SC and Ha WS: CD44 variant 9 serves as a
poor prognostic marker in early gastric cancer, but not in advanced
gastric cancer. Cancer Res Treat. 48:142–152. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yamakawa Y, Kusuhara M, Terashima M,
Kinugasa Y, Sugino T, Abe M, Mochizuki T, Hatakeyama K, Kami K and
Yamaguchi K: CD44 variant 9 expression as a predictor for gastric
cancer recurrence: Immunohistochemical and metabolomic analysis of
surgically resected tissues. Biomed Res. 38:41–52. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Akamine T, Tagawa T, Ijichi K, Toyokawa G,
Takamori S, Hirai F, Okamoto T, Oda Y and Maehara Y: The
significance of CD44 variant 9 in resected lung adenocarcinoma:
Correlation with pathological early-stage and FGFR mutation. Ann
Surg Oncol. 26:1544–1551. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hagiwara M, Kikuchi E, Tanaka N, Kosaka T,
Mikami S, Saya H and Oya M: Variant isoforms of CD44 involves
acquisition of chemoresistance to cisplatin and has potential as a
novel indicator for identifying a cisplatin-resistant population in
urothelial cancer. BMC Cancer. 18:1132018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ogihara K, Kikuchi E, Okazaki S, Hagiwara
M, Takeda T, Matsumoto K, Kosaka T, Mikami S, Saya H and Oya M:
Sulfasalazine could modulate the CD44v9-xCT system and enhance
cisplatin-induced cytotoxic effects in metastatic bladder cancer.
Cancer Sci. 110:1431–141. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jogo T, Oki E, Nakanishi R, Ando K,
Nakashima Y, Kimura Y, Saeki H, Oda Y, Maehara Y and Mori M:
Expression of CD44 variant 9 induces chemoresistance of gastric
cancer by controlling intracellular reactive oxygen spices
accumulation. Gastric Cancer. 24:1089–1099. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang Y, Gao J, He Y, Qi Z, Qian L, Chen
W, Xu H, Yue Y, Mao X, Guo S, et al: Vascular normalization was
associated with colorectal tumor regression upon anti-PD-L1
combinational therapy. J Immunology Res. 2023:58670472023.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Solier S, Müller S, Cañeque T, Versini A,
Mansart A, Sindikubwabo F, Baron L, Emam L, Gestraud P, Pantoș GD,
et al: A druggable copper-signalling pathway that drives
inflammation. Nature. 617:386–394. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
He Y, Cao J, Zhao C, Li X, Zhou C and
Hirsch FR: TIM-3, a promising target for cancer immunotherapy. Onco
Targets Ther. 11:7005–7009. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang Y, Zhao E, Zhang Z, Zhao G and Cao H:
Association between Tim-3 and Gal-9 expression and gastric cancer
prognosis. Oncol Rep. 40:2115–2126. 2018.PubMed/NCBI
|
|
19
|
Qin S, Dong B, Yi M, Chu Q and Wu K:
Prognostic values of TIM-3 expression in patients with solid
tumors: A meta-analysis and database evaluation. Front Oncol.
10:12882020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen K, Gu Y, Cao Y, Fang H, Lv K, Liu X,
He X, Wang J, Lin C, Liu H, et al: TIM3+ cells in gastric cancer:
Clinical correlates and association with immune context. Br J
Cancer. 126:100–108. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wu Y, Chen YQ, Shi TG, Tan NJ and Chen WC:
Study of immunophenotypic characteristics, clinicopathological
parameters and prognosis in gastric cancer microenvironment.
Zhonghua Yi Xue Za Zhi. 103:2786–2794. 2023.(In Chinese).
PubMed/NCBI
|
|
22
|
Edge SB, Byrd DR, Compton CC, Fritz AG,
Greene FL and Trotti A: AJCC cancer staging manual. Seventh
edition. Springer; New York: pp. 117–126. 2010
|
|
23
|
Lin L, Li H, Wang X, Wang Z, Su G, Zhou J,
Sun S, Ma X, Chen Y, You C, et al: Components of the tumor immune
microenvironment based on m-IHC correlate with prognosis and
subtype of triple-negative breast cancer. Cancer Med.
12:21639–21650. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zuo WW, Zhao CF, Li Y, Sun HY, Ma GM, Liu
YP and Kang S: High expression of PARP1 in tumor and stroma cells
predicts different prognosis and platinum resistance in patients
with advanced epithelial ovarian cancer. Front Oncol.
12:9314452022. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Moll R, Divo M and Langbein L: The human
keratins: Biology and pathology. Histochem Cell Biol. 129:705–733.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chu PG and Weiss LM: Keratin expression in
human tissues and neoplasms. Histopathology. 40:403–439. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Antonio MJ, Zhang C and Le A: Different
tumor microenvironments lead to different metabolic phenotypes. Adv
Exp Med Biol. 1311:137–147. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hui L and Chen Y: Tumor microenvironment:
Sanctuary of the devil. Cancer Lett. 368:7–13. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Taube JM, Akturk G, Angelo M, Engle EL,
Gnjatic S, Greenbaum S, Greenwald NF, Hedvat CV, Hollmann TJ, Juco
J, et al: The society for immunotherapy of cancer statement on best
practices for multiplex immunohistochemistry (IHC) and
immunofluorescence (IF) staining and validation. J Immunother
Cancer. 8:e0001552020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Harms PW, Frankel TL, Moutafi M, Rao A,
Rimm DL, Taube JM, Thomas D, Chan MP and Pantanowitz L: Multiplex
immunohistochemistry and immunofluorescence: A practical update for
pathologists. Mod Pathol. 36:1001972023. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Stack EC, Wang C, Roman KA and Hoyt CC:
Multiplexed immunohistochemistry, imaging, and quantitation: A
review, with an assessment of Tyramide signal amplification,
multispectral imaging and multiplex analysis. Methods. 70:46–58.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Granier C, Vinatier E, Colin E, Mandavit
M, Dariane C, Verkarre V, Biard L, El Zein R, Lesaffre C,
Galy-Fauroux I, et al: Multiplexed immunofluorescence analysis and
quantification of intratumoral PD-1+ Tim-3+ CD8+ T Cells. J Vis
Exp. 132:566062018.
|
|
33
|
Liu T, Sun L, Zhang Y, Wang Y and Zheng J:
Imbalanced GSH/ROS and sequential cell death. J Biochemical Mol
Toxicol. 36:e229422022. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hayashi H, Yasufuku I, Higashi T,
Chikaishi W, Yokoi R, Fukada M, Sato Y, Asai R, Tajima JY, Saigo C,
et al: Late recurrent gastric carcinoma 12 years after surgery with
attenuation of CD44 variant 9 expression. Surg Case Rep. 9:872023.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ishimoto T, Nagano O, Yae T, Tamada M,
Motohara T, Oshima H, Oshima M, Ikeda T, Asaba R, Yagi H, et al:
CD44 variant regulates redox status in cancer cells by stabilizing
the xCT subunit of system xc(−) and thereby promotes tumor growth.
Cancer Cell. 19:387–400. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Bertaux-Skeirik N, Wunderlich M, Teal E,
Chakrabarti J, Biesiada J, Mahe M, Sundaram N, Gabre J, Hawkins J,
Jian G, et al: CD44 variant isoform 9 emerges in response to injury
and contributes to the regeneration of the gastric epithelium. J
Pathol. 242:463–475. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Jang BI, Li Y, Graham DY and Cen P: The
role of CD44 in the pathogenesis, diagnosis, and therapy of gastric
cancer. Gut and Liver. 5:397–405. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Engevik AC, Feng R, Choi E, White S,
Bertaux-Skeirik N, Li J, Mahe MM, Aihara E, Yang L, DiPasquale B,
et al: The development of spasmolytic polypeptide/TFF2-expressing
metaplasia (SPEM) during gastric repair is absent in the aged
stomach. Cell Mol Gastroenterol Hepatol. 2:605–624. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Topham DJ and Reilly EC: Tissue-resident
memory CD8+ T cells: From phenotype to function. Front Immunol.
9:5152018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yi P, Cao P, Yang M, Xiong F, Jiang J, Mei
Y, Xin Y, Zhao M, Wu H and Lu Q: Overexpressed CD44 is associated
with B-cell activation via the HA-CD44-AIM2 pathway in lupus B
cells. Clin Immunol. 255:1097102023. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang Q, Wang X, Liu Y, Xu H and Ye C:
Pan-cancer and single-cell analyses identify CD44 as an
immunotherapy response predictor and regulating macrophage
polarization and tumor progression in colorectal cancer. Front
Oncol. 14:13808212024. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Van Driel M, Günthert U, van Kessel AC,
Joling P, Stauder R, Lokhorst HM and Bloem AC: CD44 variant
isoforms are involved in plasma cell adhesion to bone marrow
stromal cells. Leukemia. 16:135–143. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Johnson DE, O'Keefe RA and Grandis JR:
Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat Rev
Clin Oncol. 15:234–248. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Fricke H, Hartmann J, Sitter T, Steldinger
R, Rieber P and Schiffl H: Continuous ambulatory peritoneal
dialysis impairs T lymphocyte selection in the peritoneum. Kidney
Int. 49:1386–1395. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Cheung EC and Vousden KH: The role of ROS
in tumour development and progression. Nat Rev Cancer. 22:280–297.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Pieters M, Kruger IM, Kruger HS, Breet Y,
Moss SJ, van Oort A, Bester P and Ricci C: Strategies of modelling
incident outcomes using cox regression to estimate the population
attributable risk. Int J Environ Res Public Health. 20:64172023.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Sauerbrei W, Perperoglou A, Schmid M,
Abrahamowicz M, Becher H, Binder H, Dunkler D, Harrell FE Jr,
Royston P and Heinze G: State of the art in selection of variables
and functional forms in multivariable analysis-outstanding issues.
Diagn Progn Res. 4:32020. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhu N, Zhao Y, Yan W, Wei L, Sang Q, Li J,
Liu B and Yu B: Characterization of alternative splicing events and
prognostic signatures in gastric cancer. Cancer Cell Int.
24:1672024. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Cao M, Hu C, Pan S, Zhang Y, Yu P, Zhang
R, Cheng X and Xu Z: Development and validation of nomogram for
predicting early recurrence after radical gastrectomy of gastric
cancer. World J Surg Oncol. 22:212024. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Cai H, Li M, Deng R, Wang M and Shi Y:
Advances in molecular biomarkers research and clinical application
progress for gastric cancer immunotherapy. Biomark Res. 10:672022.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yang X and Li M, Qin X, Tan S, Du L, Ma C
and Li M: Photophosphatidylserine guides natural killer cell
photoimmunotherapy via TIM-3. J Am Chem Soc. 144:3863–3874. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Jiang J, Jin MS, Kong F, Cao D, Ma HX, Jia
Z, Wang YP, Suo J and Cao X: Decreased galectin-9 and increased
Tim-3 expression are related to poor prognosis in gastric cancer.
PLoS One. 8:e817992013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zhou Y, Li S, Hu Y, Xu X, Cui J, Li S, Li
Z, Ji J and Xing R: Multi-regional sequencing reveals the genetic
and immune heterogeneity of non-cancerous tissues in gastric
cancer. J Pathol. 263:454–465. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Wang YK, Lv XX, Wang ZQ, Zhou YM, Jiang B,
Wang SN and Chen XD: The significance of the
microlymphangiogenesis, microangiogenesis, and combined detection
of programmed cell death-1 protein (PD-1)/ki67 in gastric cancer
tissues. J Cancer Res Clin Oncol. 149:9129–9137. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Li L, Wu W, Zheng W, Hui Q and Zhao C:
Analysis of the correlation between p27 expression and helicobacter
pylori infection in gastric cancer. APMIS. 130:21–25. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Awadh M and Darwish A, Alqatari H, Buzaid
FM and Darwish A: A descriptive analysis of gastric cancer with an
immunohistochemical study of ki67 and p53 as prognostic factors.:
Bahrain experience. Saudi Med J. 44:1300–1309. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Chen L, Hong J, Hu R, Yu X, Chen X, Zheng
S, Qin Y, Zhou X, Wang Y, Zheng L, et al: Clinical value of
combined detection of serum sTim-3 and pepsinogen for gastric
cancer diagnosis. Cancer Manag Res. 13:7759–7769. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Wang H, Jin W, Wan C and Zhu C: Diagnostic
value of combined detection of CA72-4, CA19-9, and carcinoembryonic
antigen comparing to CA72-4 alone in gastric cancer: A systematic
review and meta-analysis. Transl Cancer Res. 11:848–856. 2022.
View Article : Google Scholar : PubMed/NCBI
|