Introduction
Lung cancer remains a formidable global health
challenge, claiming more lives than any other type of cancer.
According to the World Health Organization, lung cancer accounted
for ~1.8 million deaths in 2020. This represents a significant
burden on global health, particularly in developing countries.
Despite advancements in prevention and treatment, the incidence of
lung cancer continues to rise in many regions, underscoring the
need for urgent action (1). Lung
cancer bearing epidermal growth factor receptor (EGFR)
mutations was originally discovered in 2004, and precise
identification of the EGFR mutation type has laid the
foundation for subdividing the clinically relevant molecular
subtypes of non-small cell lung cancer (NSCLC) (2,3).
Notably, EGFR mutations have mostly been found in lung
adenocarcinoma (LUAD), which is the most classic histological form
of NSCLC, and the prevalence of EGFR-mutated lung tumor is
high in Asian populations, recorded at >40% (4). Clinically, patients with LUAD carrying
EGFR-positive mutations have markedly inconsistent responses
to EGFR tyrosine kinase inhibitors (TKIs) (5). Considering that clarifying whether a
patient carries a driver gene is a prerequisite for predicting the
clinical efficacy of molecularly targeted precision medicines,
certain clinical practice guidelines recommend performing
EGFR mutation testing in all lung cancer patients whose
tumors contain adenocarcinoma components (6).
In total >30 types of EGFR mutations are
scattered throughout the exon region encoding TK, of which deletion
mutations in exon 19 and point mutations in exon 21 (L858R) are the
most ‘classic’ (7,8). Patients with NSCLC harboring common
EGFR mutations represent >85% of all EGFR-positive
lung cancer cases. The remaining EGFR-positive cases
(<15%) have uncommon mutations gathered within exons 18–21 (rare
mutations: G719X, E709X or S768I) (9). With advances in precision detection
technology, it appears that EGFR compound mutations
(combinations of two or more unclassical mutations) are present in
>1% of patients with NSCLC (3,9). Known
mutation types help in selecting personalized effective treatment
decisions. Information from a previous case report suggested that
the combination of an EGFR TKI-sensitizing rare mutation
with another resistant uncommon mutation diminished the patient's
sensitivity to a TKI (10).
However, rare EGFR mutations are characterized by high
heterogeneity and low frequency, and combinations of duplex or even
triple rare mutations need to be further explored.
The present study reports the case of a patient with
LUAD carrying two rare EGFR mutations (a compound mutation
in exon 18 indel/p.G719C and exon 19 p.L747S) who received
osimertinib.
Case report
In October 2020, an 81-year-old man with a 68-year
history of smoking was admitted to The Tenth Affiliated Hospital of
Southern Medical University, Dongguan People's Hospital (Dongguan,
China) suffering from persistent shortness of breath for 1-month.
The patient developed a productive cough and worsened shortness of
breath within the week prior to admission to the hospital. The
chest radiograph presented a predominantly left-sided pleural
effusion and no significant right-sided pleural effusion. After
admission, the patient underwent thoracentesis catheter drainage
guided by B-ultrasound. The pleural fluid was bloody and turbid,
with lactate dehydrogenase (LDH) level of 1,028 U/l (normal range,
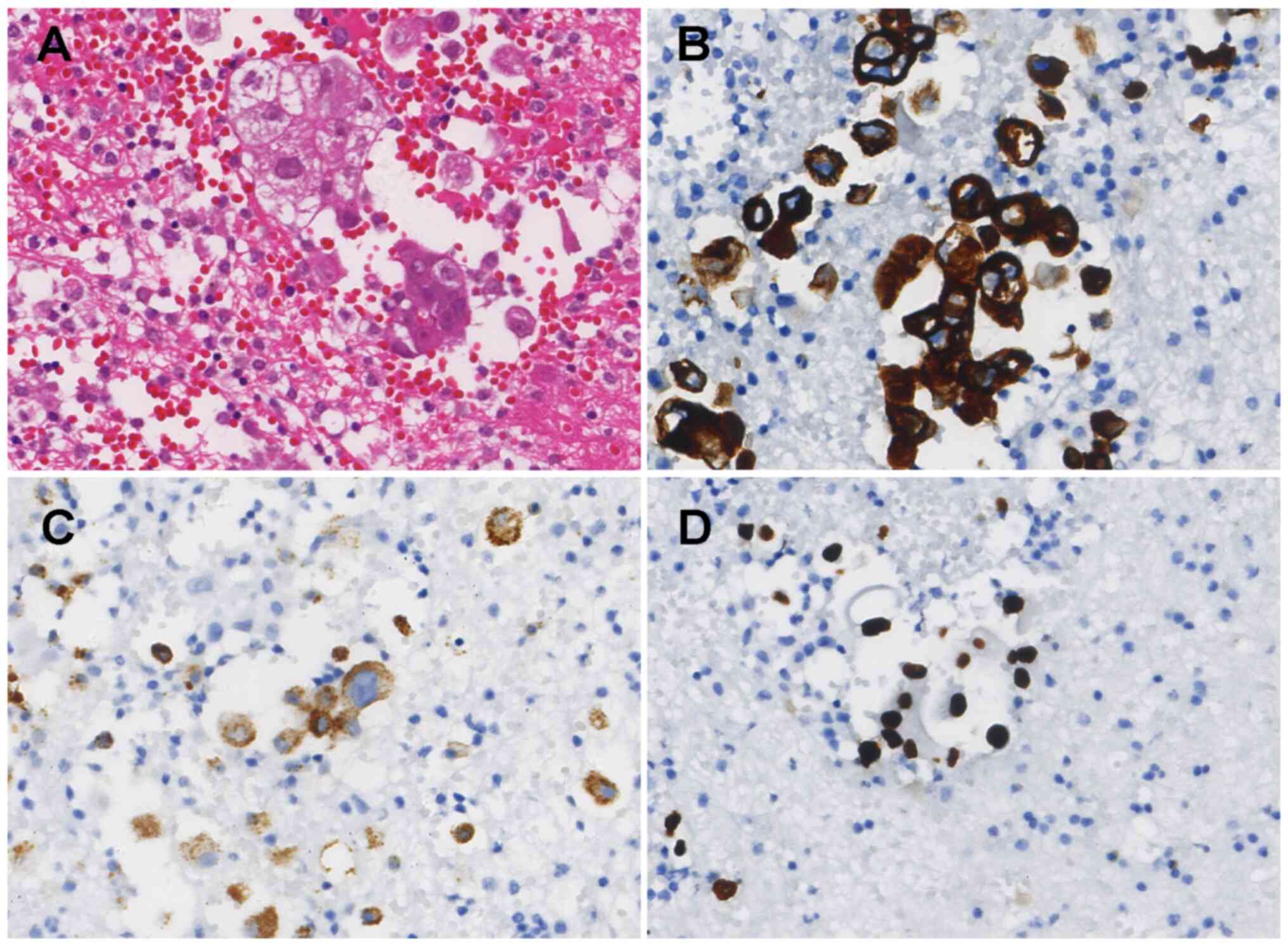
109-245 U/l). Hematoxylin and eosin staining identified a small
number of atypical cells in the pleural fluid (Fig. 1A; Data
S1). Immunohistochemical results also showed positive staining
for cytokeratin (CK)7, thyroid transcription factor-1 and napsin A,
while CK5/6 staining was negative (Fig.
1B-D; Data S1). The laboratory
tests also revealed that the serum NSCLC-associated antigen (CK19
fragment antigen 21-1) level was 4.32 µg/ml (normal level, 3.3
µg/ml), which was considered a more likely cause of LUAD. Further
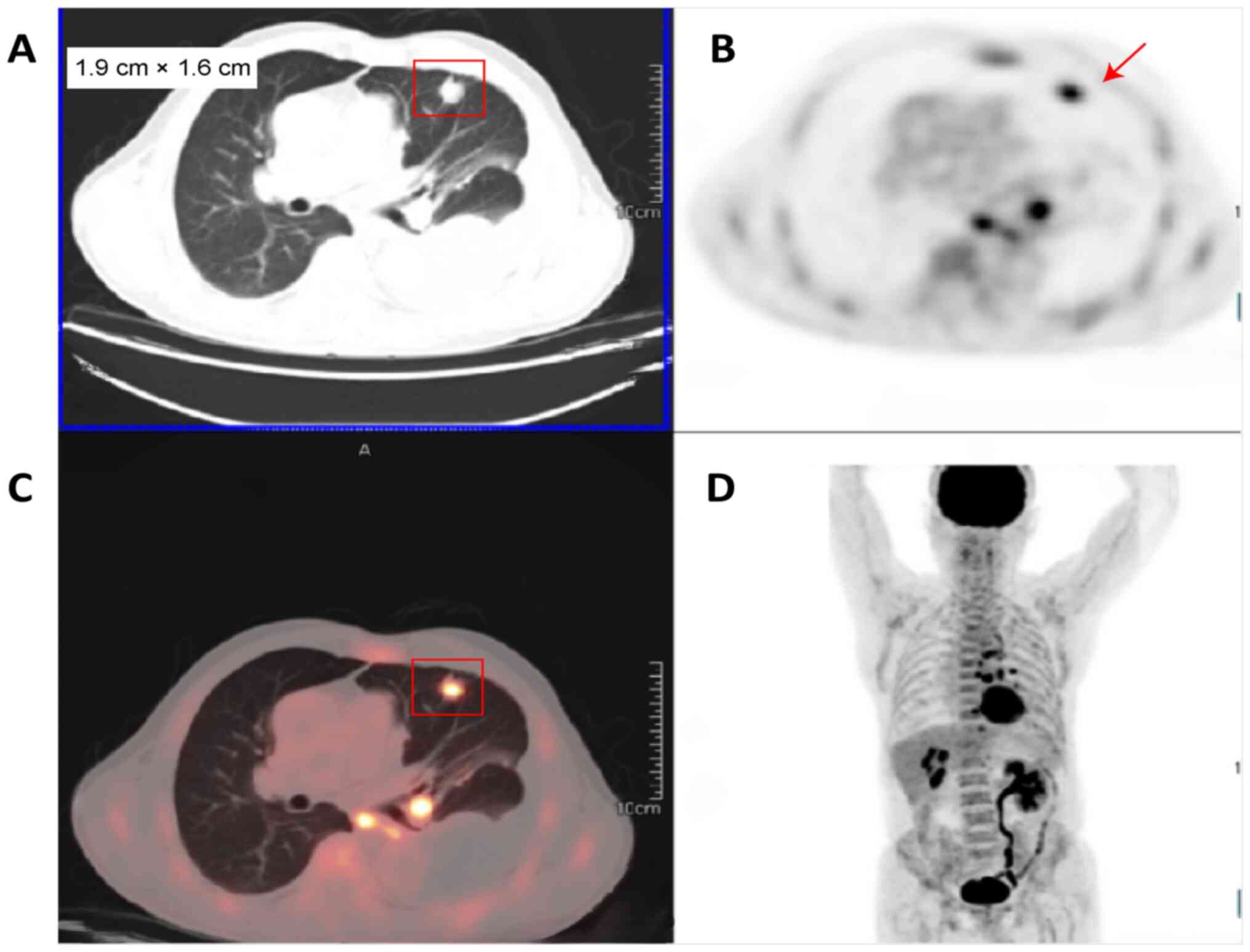
whole-body positron emission tomography/computed tomography (CT)
examination revealed a lingual nodule in the upper lobe of the left
lung, measuring 1.9×1.6 cm, with increased fluorodeoxyglucose
metabolism, which was considered peripheral lung cancer (Fig. 2). In combination with the
pathological findings, this confirmed that the patient had LUAD.
Another whole-body scan imaging revealed multiple metastatic
lesions involving the pleura, lymph nodes and bones. In order to
find a reliable treatment strategy, in October 2020, the patient's
chest fluid sediment biopsy tissue and plasma were submitted for
next-generation sequencing (NGS) separately. DNA sequencing was
performed by Geneseeq Technology, Inc., using a panel of 139 lung
cancer-related genes. Briefly, the genomic DNA from FFPE sections,
biopsy samples and whole blood control samples was extracted using
the QIAamp DNA FFPE Tissue Kit (Qiagen, Inc.) and the DNeasy Blood
and Tissue Kit (Qiagen, Inc.), respectively. Circulating cell-free
DNA (cfDNA) from plasma was extracted using the QIAamp Circulating
Nucleic Acid Kit (Qiagen, Inc.). Sequencing libraries were prepared
using the KAPA Hyper Prep Kit (Kapa Biosystems, Inc.) following the
manufacturer's instructions. Customized xGen Lockdown probes
(Integrated DNA Technologies, Inc.) targeting 139 cancer-relevant
genes were used for hybridization enrichment. The capture reaction
was performed with Dynabeads M-270 (Thermo Fisher Scientific, Inc.)
and xGen Lockdown Hybridization and Wash Kit (Integrated DNA
Technologies, Inc.). Captured libraries were PCR amplified on beads
using Illumina p5 and p7 primers in KAPA HiFi HotStart ReadyMix
(Kapa Biosystems, Inc.) and purified with Agencourt AMPure XP
beads. Libraries were quantified by qPCR using the KAPA Library
Quantification Kit (Kapa Biosystems, Inc.), with a final loading
concentration of 2.5 nM. The library fragment size was determined
using a Bioanalyzer 2100 (Agilent Technologies, Inc.). The
target-enriched library was sequenced on the HiSeq4000 NGS platform
(Illumina, Inc.) using a paired-end 150 bp sequencing strategy.
Data analysis involved quality control with Trimmomatic (version
0.39; http://github.com/usadellab/Trimmomatic), alignment
using BWA (version 0.7.17; http://github.com/lh3/bwa), PCR deduplication with
Picard (version 2.23.8; http://github.com/broadinstitute/picard), and variant
calling using Mutect [version 1.1.7 (older version) or part of GATK
4.x series (current); https://gatk.broadinstitute.org/hc/en-us/articles/360037593851-Mutect2)
and Scalpel (version 0.5.3; http://github.com/lezonlab/scalpel), followed by
annotation with vcf2maf (version 1.6.19; http://github.com/mskcc/vcf2maf). The sequencing depth
was 150X for whole blood control samples, 1500X for tumor tissues,
and 5000X for cfDNA samples.
Two uncommon EGFR mutations [exon 18
c.2154_2155delinsTT (p.G719C) missense mutation and exon 19
c.2240T>C (p.L747S) missense mutation] were present (Table I). In addition, the tumor protein
p53 (TP53) exon 4 c.228del (p.P77Qfs*46) frameshift mutation
and the cyclin-dependent kinase 6 (CDK6) exon 2 c.112C>G
(p.R38G) missense mutation were also identified as common
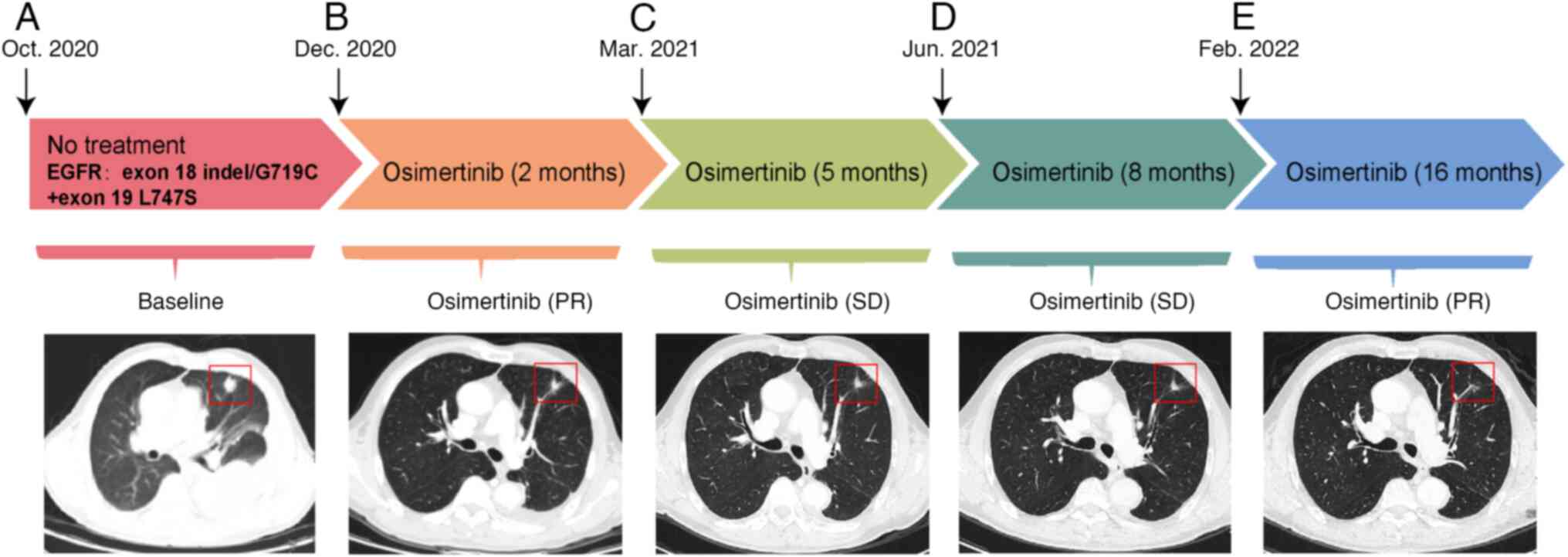
mutations. In response to the EGFR mutation, the patient was
administered osimertinib treatment orally (80 mg per day) from 1
week after admission (Fig. 3A), and
evaluation of the tumor by CT scans displayed a persistent partial
response to the disease for the next 2 months.
 | Table I.Patient tumor and plasma
next-generation sequencing results. |
Table I.
Patient tumor and plasma
next-generation sequencing results.
| Genes | Alterations | Nucleotide
change | Plasma | Tissue |
|---|
| EGFR | p.G719C |
c.2154_2155delinsTT | 0.1% | 31.4% |
|
| p.L747S | c.2240T>C | 0.2% | 28.6% |
| TP53 | p.P77Qfs*46 | c.228del | - | 18.1% |
| CDK6 | p.R38G | c.112C>G | 2.5% | - |
In December 2020, CT of the chest and upper abdomen
showed that the focal nodule in the upper lobe of the left lung had
regressed (Fig. 3B), with a size of
1.3×1.2 cm, that the number and size of pleural nodules were
reduced compared with before, and that a small amount of effusion
had appeared in the pericardial cavity and the left pleural cavity
(Fig. 3C-E). The left-sided lung
disease was in partial remission, which suggested that the
osimeratinib treatment strategy was effective. However, CT also
revealed new lesions in the liver (S4) and thyroid gland. By
February 2022, a repeat CT examination indicated that the patient's
left lung lesion remained stable after 16 months of treatment with
osimertinib. During the whole case period, the patient underwent
monthly physical examinations and quarterly imaging studies.
Discussion
In recent years, NSCLC treatment has moved from
histology-based therapy to molecularly targeted precision therapy
for driver genes. In this therapeutic context, EGFR
genotyping based on NGS technology has also become the preferred
approach for assessing NSCLC (11,12).
As an event usually excluded from clinical trials, the discovery of
non-classical or complex EGFR mutations has opened up new
opportunities for patients with NSCLC, especially for those with
LUAD. However, there is a paucity of data obtained on rare
EGFR mutations in the clinical setting. Given that
EGFR mutations are highly heterogeneous genetic alterations
that occur within tumor lesions, and that rare EGFR
mutations occur at a low frequency in LUAD tumors, there is a need
to perform a more refined analysis of these patients. The present
study reports the rare case of a patient in whom two non-classical
EGFR mutations, indel/G719C (exon 18) and L747S (exon 19),
were observed in the tumor tissue. Furthermore, a significantly
better outcome in contrast to the typical outcome of patients with
NSCLC, was observed in this patient, who responded well to
osimertinib. Notably, the presence of CDK6 and TP53
mutations may be a key oncogenic event common to advanced
EGFR-mutated LUAD. CDK6 mutations have been
associated with tumorigenesis and progression in certain cancer
types, such as breast (13), lung
(14) and prostate (15) cancer. However, specific data on the
prognostic impact in LUAD, particularly in the context of TP53
mutations, may be limited (16).
Numerous studies have, however, established a strong correlation
between TP53 mutations and a poor prognosis in various
cancer types such as breast (17),
lung (18) and colorectal (19) cancer, as well as LUAD. Patients with
TP53 mutations often exhibit advanced tumor stages,
increased metastasis and reduced overall survival times (20).
EGFR exon 18 mutations have a low frequency,
being observed in <4% of lung cancer patients with EGFR
mutations (7). Among them, the
G719X (X can be replaced by any base) point mutation is the
predominant subtype among EGFR exon 18 mutations.. To the best of
our knowledge, the present case may be the first reported case on
the application of the third-generation TKI osimertinib in a
patient with LUAD carrying the EGFR exon 18 indel/G719C
mutation. An encouraging efficacy of osimertinib has been reported
in several case reports on LUAD with uncommon EGFR mutations. Fang
and Liu (21) illustrated that
patients with LUAD harboring EGFR T751_I759delinsS sustained
a marked response to osimertinib for 16 months (21). In addition, a 2021 case report by
Shan et al (22) also
founded that high-dose osimertinib achieved great clinical benefit
in a patient with metastatic lung cancer carrying two rare
EGFR mutations (G719S and L861Q). Notably, the use of
osimertinib was effective in extending the median overall survival
time of the patients by >12 months. In a multicenter phase II
study that included 36 patients with lung cancer, the objective
response rate of osimertinib demonstrated in patients carrying the
G719X mutation was ~50%, which was comparable to the response rate
of a first-generation TKI (23).
According to the present report, the patient was able to achieve a
durable objective response with no adverse effects to osimertinib,
suggesting a better efficacy of osimertinib against exon 18 indel
compared with other treatment options.
Referring to previous datasets, more than
three-quarters of G719X mutations are complex, that is,
combinations of G719X with other rare or classical mutations. In
one study, G719X and other mutations were present in 8 of 15
patients with lung cancer (24). In
the present report, the patient carried the complex mutations of
exon 18 indel/G719C + exon 19 L747S, which is a de novo
compound mutation, before receiving TKI therapy. Considering that
EGFR L747S is a crucial factor in the poor response to
first-generation TKIs, the use of osimertinib may be feasible for
patients with the aforementioned uncommon mutations. The G719C +
L747S mutation was previously found in a female patient with LUAD
whose symptoms of dyspnea and coughing disappeared within 1 month
of osimertinib treatment, and a significant and sustained reduction
in the lesion area was observed thereafter (25). The current case study is the first
reported instance of a patient with EGFR L747S mutations
responding positively to osimertinib treatment. The same appeared
to be true for the exon 18 indel/G719C + exon 19 L747S mutations
described in the present report and provided new clinical data for
the treatment of patients with metastatic lung cancer harboring the
G719C and L747S mutations.
The current study presents a detailed case report on
a patient with LUAD carrying rare EGFR mutations and the response
to osimertinib treatment. However, there are several limitations
and areas that could be improved. It is important to note that the
study was restricted to a single patient, limiting the
generalizability of the findings. Additionally, the lack of
long-term follow-up data prevents a comprehensive understanding of
the patient's overall survival and potential resistance
development. The article also lacks detailed mechanistic insights
into the efficacy of osimertinib for the specific EGFR mutations
observed, and a more extensive review of comparative treatments
would provide a broader context for evaluating the effectiveness of
osimertinib.
In conclusion, the present study describes the case
of a patient with LUAD carrying two rare EGFR mutations
(exon 18 indel/G719C + exon 19 L747S) who responded well to
osimertinib. This newly emerged complex mutation may be associated
with a good progression-free survival time when using a
third-generation TKI, but more convincing evidence is required to
support this. To determine a more appropriate and comprehensive
treatment strategy, a more extensive molecular analysis (e.g. NGS)
is indispensable when dealing with lung cancer carrying EGFR
mutations to maximize the identification of the complex genetic
features of the cancer.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was supported and funded by the Dongguan Social
Development Science and Technology Project Mission Statement (Key
Project in 2021; grant no. 20211800905042).
Availability of data and materials
Due to the restrictions imposed by China's national
legislation on patient privacy, the genome sequencing data
generated and analyzed during the current study cannot be publicly
shared. According to the Personal Information Protection Law and
the Regulation on the Administration of Human Genetic Resources,
the sharing of genetic data is strictly regulated to protect
individual privacy and sensitive information. Researchers seeking
access to this data for specific purposes may contact the
corresponding author, subject to approval from relevant regulatory
bodies.
Authors' contributions
All authors contributed to the manuscript. YL, LL,
ZL and KL were responsible for the conception and design of the
study. Data were collected and analyzed by YL, LL, CS and CL. ZL
and KL confirm the authenticity of all the raw data. The manuscript
was drafted by YL and LL, and was critically revised by ZL and KL.
The whole study was supervised by ZL and KL. All authors have read
and approved the final manuscript.
Ethics approval and consent to
participate
This study was performed according to institutional
guidelines and was approved by Ethical Committee of The Tenth
Affiliated Hospital of Southern Medical University, Dongguan
People's Hospital (Dongguan, China; approval no. 2023KZPJ063),
following the Declaration of Helsinki guidelines. Written informed
consent was obtained from the patient for participation in this
study.
Patient consent for publication
The patient provided written consent for the
publication of the present case report.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Dong J, Li B, Lin D, Zhou Q and Huang D:
Advances in targeted therapy and immunotherapy for non-small cell
lung cancer based on accurate molecular typing. Front Pharmacol.
10:2302019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bi X, Song P, Wang C, Zhang X and Liu C:
Genomic profiling reveals non-small cell lung cancer with common
mutations of EGFR exon 20 and exon 21: A case report. Transl Cancer
Res. 11:1423–1428. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shi Y, Au JS, Thongprasert S, Srinivasan
S, Tsai CM, Khoa MT, Heeroma K, Itoh Y, Cornelio G and Yang PC: A
prospective, molecular epidemiology study of EGFR mutations in
Asian patients with advanced non-small-cell lung cancer of
adenocarcinoma histology (PIONEER). J Thorac Oncol. 9:154–162.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wu JY, Yu CJ, Chang YC, Yang CH, Shih JY
and Yang PC: Effectiveness of tyrosine kinase inhibitors on
‘uncommon’ epidermal growth factor receptor mutations of unknown
clinical significance in non-small cell lung cancer. Clin Cancer
Res. 17:3812–3821. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Azzoli CG, Baker S Jr, Temin S, Pao W,
Aliff T, Brahmer J, Johnson DH, Laskin JL, Masters G, Milton D, et
al: American Society of Clinical Oncology clinical practice
guideline update on chemotherapy for stage IV non-small-cell lung
cancer. J Clin Oncol. 27:6251–6266. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Beau-Faller M, Prim N, Ruppert AM,
Nanni-Metéllus I, Lacave R, Lacroix L, Escande F, Lizard S, Pretet
JL, Rouquette I, et al: Rare EGFR exon 18 and exon 20 mutations in
non-small-cell lung cancer on 10 117 patients: A multicentre
observational study by the French ERMETIC-IFCT network. Ann Oncol.
25:126–131. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Won YW, Han JY, Lee G, Park SY, Lim KY,
Yoon KA, Yun T, Kim HT and Lee JS: Comparison of clinical outcome
of patients with non-small-cell lung cancer harbouring epidermal
growth factor receptor exon 19 or exon 21 mutations. J Clin Pathol.
64:947–952. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Qin BD, Jiao XD, Yuan LY, Liu K, Wang Z,
Qin WX and Zang YS: The effectiveness of afatinib and osimertinib
in a Chinese patient with advanced lung adenocarcinoma harboring a
rare triple EGFR mutation (R670W/H835L/L833V): A case report and
literature review. Onco Targets Ther. 10:4739–4745. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li K, Yang M, Liang N and Li S:
Determining EGFR-TKI sensitivity of G719X and other uncommon EGFR
mutations in non-small cell lung cancer: Perplexity and solution
(Review). Oncol Rep. 37:1347–1358. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang T, Wan B, Zhao Y, Li C, Liu H, Lv T,
Zhan P and Song Y: Treatment of uncommon EGFR mutations in
non-small cell lung cancer: New evidence and treatment. Transl Lung
Cancer Res. 8:302–316. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chang YS, Tu SJ, Chen YC, Liu TY, Lee YT,
Yen JC, Fang HY and Chang JG: Mutation profile of non-small cell
lung cancer revealed by next generation sequencing. Respir Res.
22:32021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Goel S, Bergholz JS and Zhao JJ: Targeting
CDK4 and CDK6 in cancer. Nat Rev Cancer. 22:356–372. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gong W, Wang L, Zheng Z, Chen W, Du P and
Zhao H: Cyclin-dependent kinase 6 (CDK6) is a candidate diagnostic
biomarker for early non-small cell lung cancer. Transl Cancer Res.
9:95–103. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen X, Wu Y, Wang X, Xu C, Wang L, Jian
J, Wu D and Wu G: CDK6 is upregulated and may be a potential
therapeutic target in enzalutamide-resistant castration-resistant
prostate cancer. Eur J Med Res. 27:1052022. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sitthideatphaiboon P, Teerapakpinyo C,
Korphaisarn K, Leelayuwatanakul N, Pornpatrananrak N, Poungvarin N,
Chantranuwat P, Shuangshoti S, Aporntewan C, Chintanapakdee W, et
al: Co-occurrence CDK4/6 amplification serves as biomarkers of de
novo EGFR TKI resistance in sensitizing EGFR mutation non-small
cell lung cancer. Sci Rep. 12:21672022. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Børresen AL, Andersen TI, Eyfjörd JE,
Cornelis RS, Thorlacius S, Borg A, Johansson U, Theillet C,
Scherneck S, Hartman S, et al: TP53 mutations and breast cancer
prognosis: particularly poor survival rates for cases with
mutations in the zinc-binding domains. Genes Chromosomes Cancer.
14:71–75. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gu J, Zhou Y, Huang L, Ou W, Wu J, Li S,
Xu J, Feng J and Liu B: TP53 mutation is associated with a poor
clinical outcome for non-small cell lung cancer: Evidence from a
meta-analysis. Mol Clin Oncol. 5:705–713. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
González-Aguilera JJ, Oliart S, Azcoita MM
and Fernández-Peralta AM: Simultaneous mutations in K-ras and TP53
are indicative of poor prognosis in sporadic colorectal cancer. Am
J Clin Oncol. 27:39–45. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Canale M, Petracci E, Delmonte A, Bronte
G, Chiadini E, Ludovini V, Dubini A, Papi M, Baglivo S, De Luigi N,
et al: Concomitant TP53 mutation confers worse prognosis in
EGFR-mutated non-small cell lung cancer patients treated with TKIs.
J Clin Med. 9:10472020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fang YF and Liu PC: Afatinib and
osimertinib in lung adenocarcinoma harbored EGFR T751_I759delinsS
mutation: A case report. Thorac Cancer. 12:3429–3432. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shan CG, Wang H, Lin T, Liu D, Wen L, Chen
ZJ, Zhen JJ, Lai MY, Zhang L, Zou X, et al: A non-small cell lung
cancer (NSCLC) patient with leptomeningeal metastasis harboring
rare epidermal growth factor receptor (EGFR) mutations G719S and
L861Q benefited from doubling dosage of osimertinib: A case report.
Ann Palliat Med. 10:5897–5901. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cho JH, Lim SH, An HJ, Kim KH, Park KU,
Kang EJ, Choi YH, Ahn MS, Lee MH, Sun JM, et al: Osimertinib for
patients with non-small-cell lung cancer harboring uncommon EGFR
mutations: A multicenter, open-label, phase II trial
(KCSG-LU15-09). J Clin Oncol. 38:488–495. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kobayashi S, Canepa HM, Bailey AS,
Nakayama S, Yamaguchi N, Goldstein MA, Huberman MS and Costa DB:
Compound EGFR mutations and response to EGFR tyrosine kinase
inhibitors. J Thorac Oncol. 8:45–51. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Grolleau E, Haddad V, Boissière L,
Falchero L and Arpin D: Clinical efficacy of osimertinib in a
patient presenting a double EGFR L747S and G719C mutation. J Thorac
Oncol. 14:e151–e153. 2019. View Article : Google Scholar : PubMed/NCBI
|