Introduction
Hepatocellular carcinoma (HCC) is a common
malignancy of the liver with a poor prognosis. In 2020, there were
906,000 new cases and 830,000 deaths of HCC worldwide. In 2023, it
was estimated by the North American Association of Central Cancer
Registries that 41,210 cases of HCC were newly diagnosed in the
United States, with a morbidity rate that has tripled over the past
four decades (1). Currently, HCC is
the third leading cause of cancer-related deaths and the sixth most
common cancer worldwide (2).
Due to a lack of early symptoms of HCC, prompt
diagnosis and treatment fail to be provided, and most patients have
an advanced stage of the disease and have developed liver or
distant metastasis at the time of initial diagnosis. Furthermore,
only 20–30% of patients are suitable for surgical intervention, and
the postoperative recurrence rate is also as high as ~70% among
patients with early-stage HCC undergoing surgical resection
(3). Therefore, early
identification of patients with HCC is of great importance due the
poor responses to late treatment. In the past few decades,
alpha-fetoprotein (AFP) has been commonly used as a marker for
early screening of HCC, and has become the most widely used
biomarker for HCC (4). However, due
to its low sensitivity and specificity for detecting HCC, the
clinical effectiveness of AFP remains controversial. AFP serves as
the most common tumor marker for the diagnosis of liver cancer, but
its diagnostic efficacy still cannot meet the clinical needs. Many
patients with liver cancer have a low AFP level, whilst certain
people with a high AFP level do not develop liver cancer, thus
often leading to misdiagnosis or missed diagnosis to a certain
extent. Therefore, it is crucial to use new markers with
satisfactory levels of sensitivity and specificity to assist in
early detection (5).
Cancer stem cells (CSCs) are one of the chief
culprits of recurrence and metastasis of HCC. CSCs are a small
subpopulation of cancer cells with strong stemness within tumors
that serve a critical role in tumor heterogeneity, tumorigenesis,
tumor recurrence, metastasis and resistance to anticancer therapy
(6). Mounting evidence suggests
that tumor cells with stemness, namely liver CSCs (LCSCs), are
present in HCC, and liver stem cells are transformed into LCSCs
during the long-term inflammatory process induced by factors such
as chronic viral infection or alcohol (7–9). CSC
markers are the basis of the cellular and signaling functions of
LCSCs, which are essential for isolating CSCs, analyzing their
biological characteristics, and using them for targeted therapy.
Cluster of differentiation (CD)133, CD44, CD90 and epithelial cell
adhesion molecule (EpCAM) are common CSC markers (10). Recent studies have assessed the
potential of LCSC markers in HCC diagnosis, prognostic prediction
and new therapy development. For example, Tseeleesuren et al
(11) reported that CD133
positivity reduced the overall survival (OS) rate of patients with
HCC. Moreover, Sekar et al (12) reported that EpCAM-targeted therapy
enhanced chemotherapy sensitivity to cisplatin, EpCAM-targeted
therapy plus cisplatin delayed the progression of EpCAM-positive
HCC, and LCSC-targeted chemotherapy drugs improved the OS rate of
patients with HCC. In summary, certain potential prognostic
biomarkers and new therapeutic targets of HCC may be identified by
analyzing the aforementioned markers.
It remains unclear as to which CSC marker has the
greatest effect on clinical diagnosis and prognostic prediction.
Therefore, the present study performed a network meta-analysis
(NMA) to assess the diagnostic value of CSC markers for HCC and
their associations with prognosis. The analysis aimed to elucidate
one or a combination of CSC markers with the highest value for the
diagnosis and prognostic prediction of HCC, and to provide valuable
information for early diagnosis and prognostic prediction of
HCC.
Materials and methods
Design and registration
The reporting of the present NMA conformed to the
Preferred Reporting Items for Systematic Reviews and Meta-Analyses
statement (13). The present study
was registered in the International Prospective Register of
Systematic Reviews (identifier no. CRD42024504192).
Search methods
A total of two investigators (ZO and JY)
independently searched PubMed (https://pubmed.ncbi.nlm.nih.gov/), Embase (https://www.embase.com/), the Cochrane Central
Register of Controlled Trials (https://www.cochranelibrary.com/central) and Web of
Science (https://www.webofscience.com) from
January 1, 2013 to 17 November, 2023, without restrictions on the
type of literature, date/time and publication status. The keywords
from the Medical Subject Headings (https://libres.csu.edu.cn/vpn/249/https/P75YPLUPMNSGTLUPNSXT65UJNAYGP55X/mesh),
including ‘Hepatocellular Carcinoma’, ‘Liver Neoplasms’,
‘Neoplastic Stem Cells’, ‘AC133 Antigen’, ‘Thy-1 Antigens’,
‘Epithelial Cell Adhesion Molecule’ and ‘Hyaluronan Receptors’,
were used for literature retrieval. At the same time, the
references of the literature were searched to prevent the omission
of any relevant literature. The search strategies are provided in
Data S1.
Inclusion and exclusion criteria
The inclusion and exclusion criteria were defined in
strict accordance with the Population, Intervention, Comparison,
Outcomes and Study design principles (14). Eligible studies were those in which
patients with HCC had a cancer stem cell marker test and reported a
correlation analysis of survival prognosis or diagnostic value.
Only published cohort studies were considered. The study inclusion
criteria were as follows: i) All patients were diagnosed with HCC;
ii) all patients were tested for CSC markers; iii) diagnoses of HCC
were confirmed through liver histopathological examinations; iv) ≥1
of the following outcomes were reported: OS, recurrence-free
survival (RFS) and disease-free survival (DFS), as well as
diagnostic data on metastasis, differentiation and microvascular
invasion, including true positive, false positive, true negative
and false negative; and v) studies published in the English
language. The exclusion criteria were as follows: i) studies with
patients with cancers other than HCC; ii) studies that did not
involve the specific LCSC markers; iii) unclear criteria for
diagnosis and efficacy; iv) review articles, case reports, animal
experiments, conference summaries, guidelines, letters, opinion
articles and meeting abstracts; v) incomplete or erroneous data
that could not be extracted; and vi) studies published prior to
2013.
Study selection
According to the predefined inclusion and exclusion
criteria, two investigators (ZO and JY) independently selected the
studies. All potentially relevant studies were imported into
EndNote X9 (https://www.endnote.com/) to remove
duplicate publications. The titles and abstracts were then screened
to exclude ineligible ones. Preliminary eligible studies were
searched and further read in full to select the final eligible
studies. Disagreements were resolved through discussion or
consultation with a third investigator (JH).
Data extraction
The following data were extracted from the eligible
studies: First author, publication year, country, population
source, sample size, age, types of CSC markers, detection methods,
number of positive cases, number of negative cases, average
follow-up time, and outcome indicators including OS, RFS, DFS,
differentiation, vascular invasion, metastasis, sensitivity,
specificity and summary receiver operating characteristic (SROC)
curves. The data were extracted by two investigators (ZO and JY)
and further checked for accuracy by a third investigator (JH).
Quality assessment
The Newcastle-Ottawa Scale (NOS) (http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp)
was used for quality assessment of the studies included in the
prognostic analysis. The NOS mainly covers eight questions in three
domains, with a full score of 2 points for comparability and 1
point for the other seven questions. A study with a total score of
7–9 points was considered to be high-quality, and a total score of
4–7 points was defined as medium-quality. After the assessment was
completed, cross-checking was performed by two investigators. If
there was a disagreement, a third investigator assisted in the
adjudication process. Quality Assessment of Diagnostic Accuracy
Studies-2 (QUADAS-2; www.quadas.org), a revised tool for the quality
assessment of diagnostic accuracy studies, was used to
independently assess the risk of bias in the included studies and
the applicability of their results (15). In the case of disagreement, a
consensus was reached through discussion. Quality assessment was
performed on all aspects of the studies, including participant
profiles, index tests, target conditions, reference criteria,
process and timing. Missing participant data was considered a high
risk of bias in the study. If at least one domain of QUADAS-2 was
judged to be high risk, the study was judged to be at high risk of
bias.
Data synthesis and statistical
analysis
Statistical models based on the Bayesian framework
were established using the JAGS software (version 4.3.1, http://sourceforge.net/projects/mcmc-jags) in R
(version 4.3.1; RStudio; Posit Software, PBC). The
probabilistic-based Bayesian meta-analysis can reduce the
interference of confounding factors on the results and produce
stable results. Therefore, NMA on the prognosis was performed using
the Bayesian model (16). The
hazard ratio (HR) with a 95% confidence interval (CI) was
calculated for continuous data to assess their survival and
prognostic values. Random-effects models were used for all NMAs as
the studies included were clinically heterogeneous (namely,
different countries, clinical stages, histological grades, basic
physical conditions and anticancer treatments). The surface under
the cumulative ranking curve (SUCRA) was used to estimate the
relative ranking of different CSC markers in each outcome of
interest (17). The higher the
SUCRA value, the higher the probability that a marker was in the
top rank (17), and the
corresponding league tables were generated to compare the
differences in effects among markers. In addition, the consistency
and inconsistency models were compared using the deviation
information criterion (DIC). If there were <5 points of
difference in the DIC, the consistency was considered to be good
and the consistency model was used (18). Publication bias was detected using
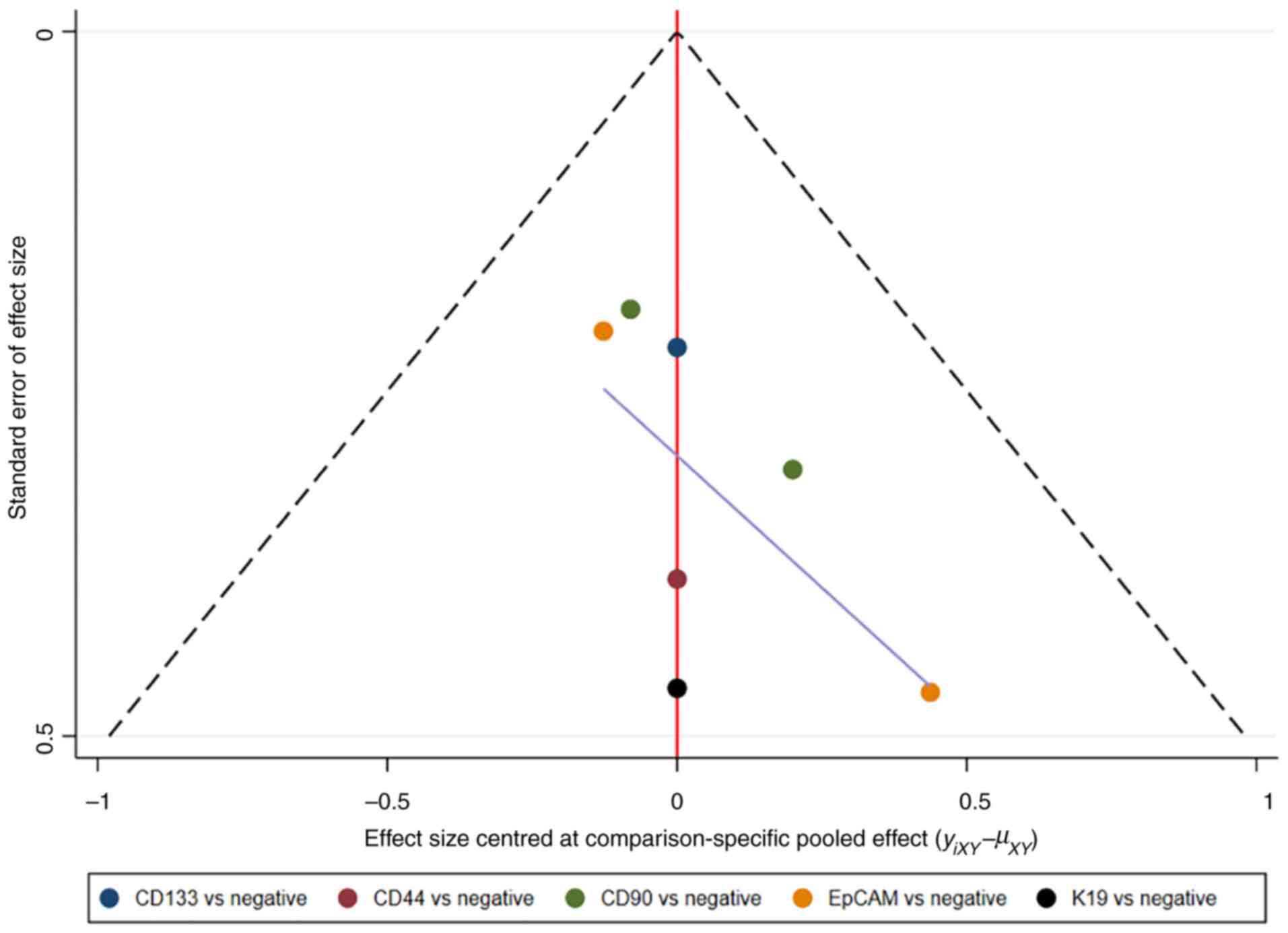
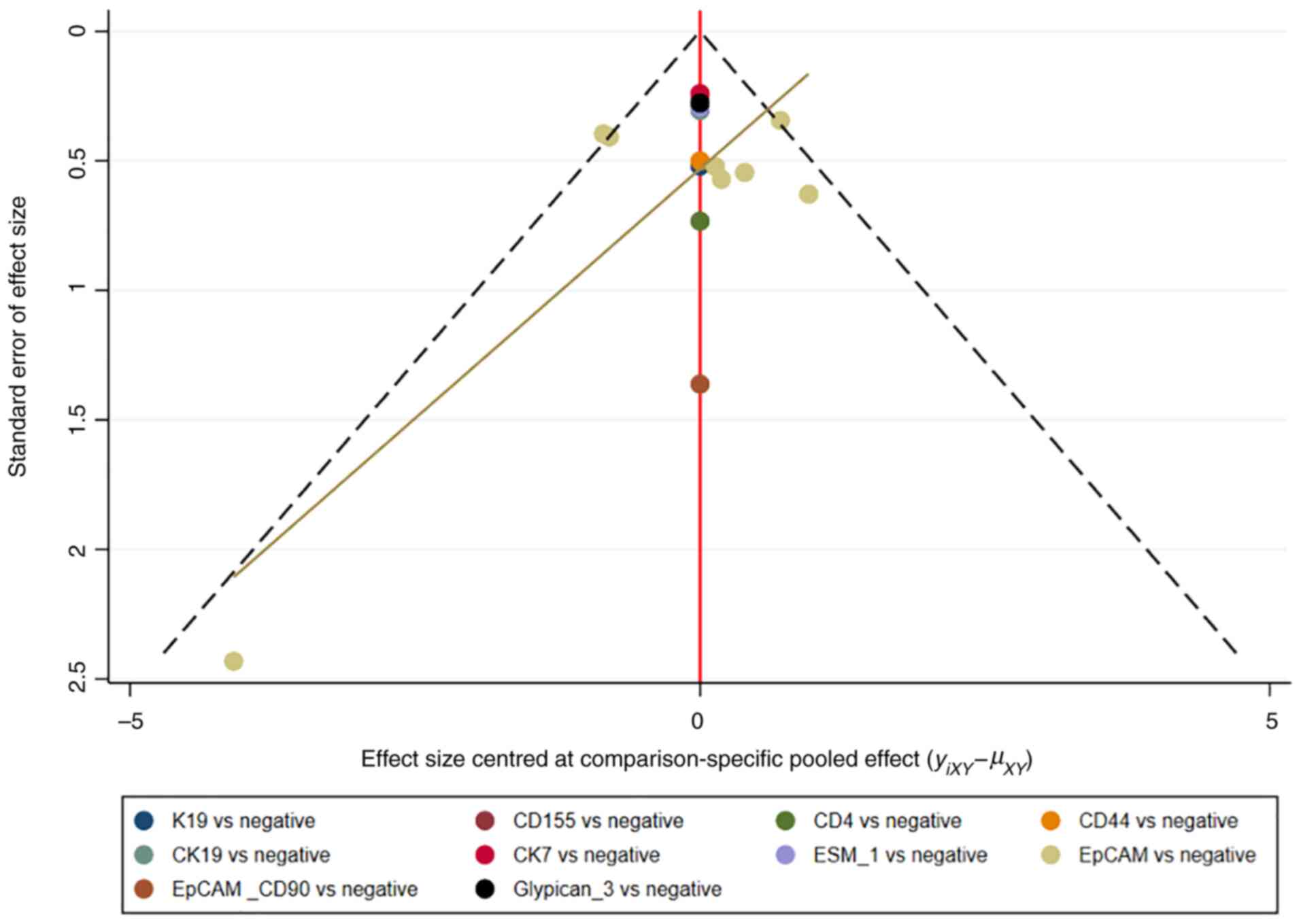
comparison-adjusted funnel plots. Network plots and
comparison-adjusted funnel plots of NMA were plotted by Stata
(version 15.0; StataCorp LP). In addition, the CSC marker that had
the most accurate predictive effect was assessed using an
NMA-diagnostic test (NMA-DT) based on the ANOVA model. As more
stable results can be obtained using Bayesian ANOVA models, the
diagnostic performance of different CSCs was compared using the
ANOVA model (19). The diagnostic
value of markers was evaluated using their sensitivity, specificity
and 95% CI. The diagnostic value of several markers was compared
with the reference standard simultaneously at different thresholds
by NMA-DT using R software (version 4.3.1; http://www.R-project.org; The R Foundation). The
relative sensitivity and specificity of CSC markers were evaluated
against standard diagnostic methods, and these CSC markers were
ranked based on the diagnostic odds ratio (DOR) and superiority
index. The higher the DOR and superiority index, the higher the
accuracy of CSC markers. Review Manager (version 5.3; http://training.cochrane.org/online-learning/core-software/revman)
was used to calculate the area under the SROC curve. Literature
heterogeneity was considered during Bayesian modeling, and thus the
impact of heterogeneity was reduced by the random-effects model in
the analysis (20). The funnel plot
has the advantages of intuitiveness, detection of publication bias,
identification of heterogeneity, and enhancement of conclusion
reliability and credibility (21).
Therefore, publication bias was presented using the funnel
plot.
Results
Search results
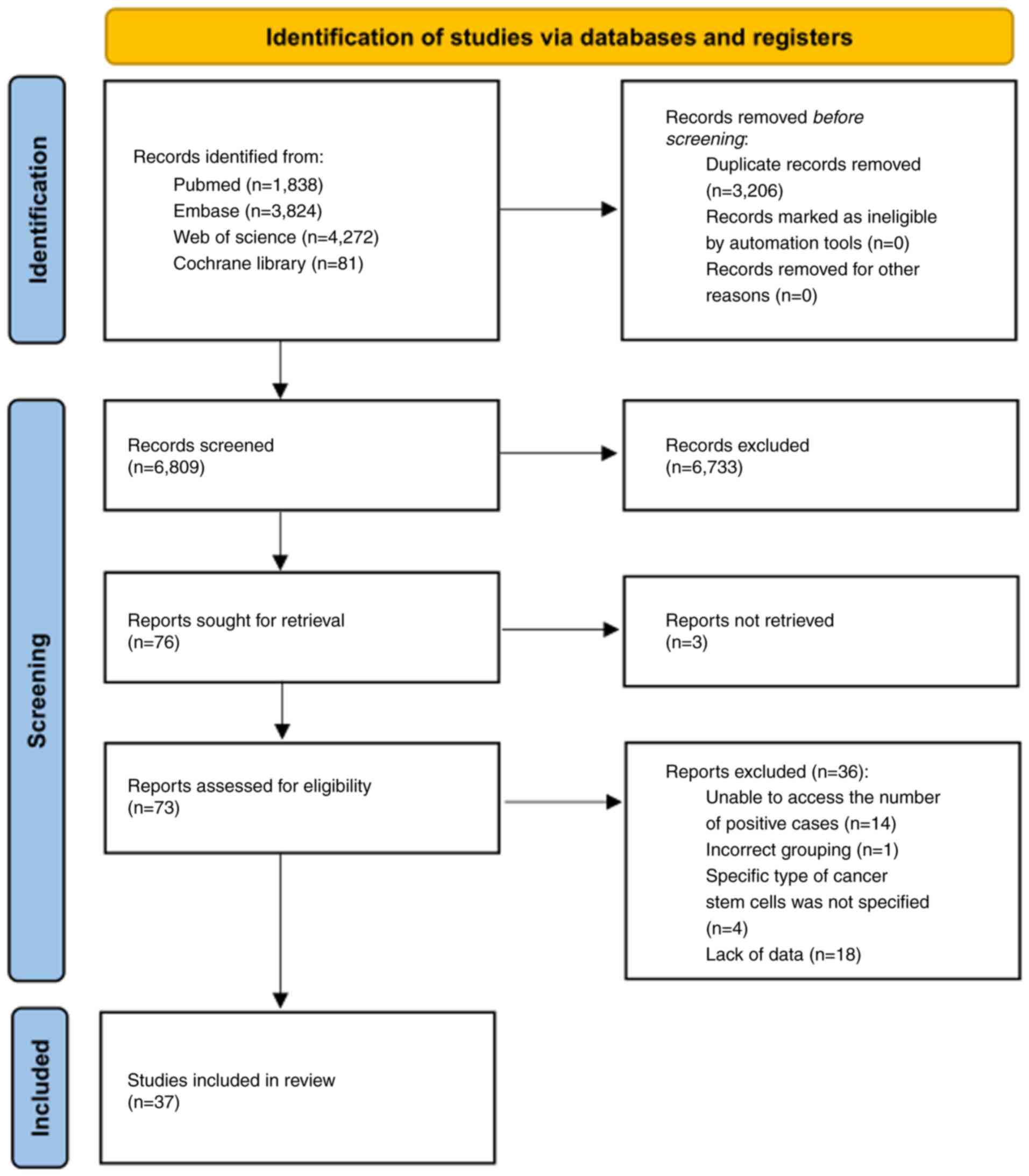
Initially, a total of 10,015 potentially relevant
studies were identified from the four aforementioned electronic
databases. After 3,206 duplicate studies were excluded, the title
and/or abstract of each study was screened based on the inclusion
and exclusion criteria, following which 6,733 studies were
excluded. The remaining 76 studies were further assessed for
eligibility by reading their full texts; however, the full text of
three studies could not be found so these were subsequently
excluded. Furthermore, 36 studies were excluded as they did not
report the number of positive cases (n=14), did not use the correct
grouping method (n=1), did not specify the type of CSCs (n=4), and
lacked data (n=18). Ultimately, 37 studies were deemed eligible and
incorporated into the NMA. Fig. 1
details the study selection procedure.
Characteristics of included
studies
Among the 37 eligible studies published between 2013
and 2023, there were six multicenter studies (22–27)
and 31 single-center studies (11,28–57).
Of the 3,980 patients involved, men and women accounted for 79.31
and 20.69%, respectively. A total of 30 studies were performed in
Asia (11,22,23,27–30,33–35,37–49,51–57),
five in Europe (24–26,31,50),
one in America (36), and one in
Africa (32). A total of 15 CSC
markers were involved. EpCAM was reported in 25 studies (11,22–24,26–32,34–37,39,42–44,46,48–50,53,57),
CD133 in 11 studies (11,30,38,40,41,43,44,48,51,54,56),
CD44 in two studies (41,52), CD90 in four studies (29,45,47,51),
CD56 in three studies (22,44,48),
CK19 in five studies (23,27,35,44,53)
and keratin 16 (K19) in three studies (48–50).
The methods used to test CSC markers involved immunohistochemistry,
reverse transcription-quantitative PCR, fluorescence-activated cell
sorting and peripheral blood testing. Furthermore, the mean age and
mean follow-up time were statistically analyzed. Table SI presents the characteristics and
details of each study included in the NMA.
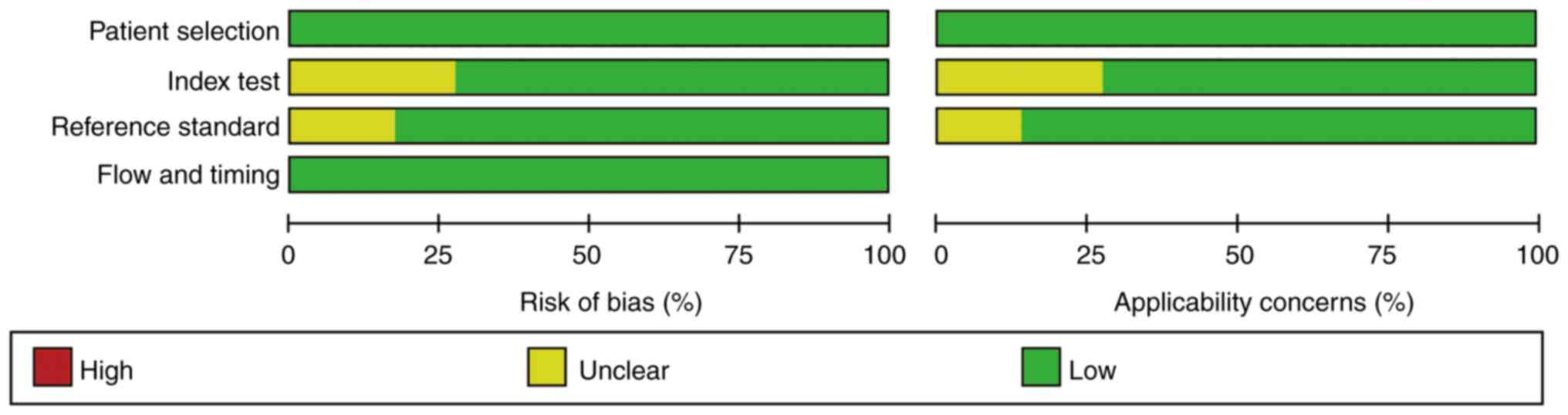
Quality assessment
There was no sufficient information used to judge
the index tests in several studies (20,21,33,35,36,40,44,51),
which were assessed as at unclear risk of bias. In several studies
(21,30,37,39,41),
the risk assessment of bias was not clear, so there was
insufficient information to judge whether blind interpretation of
the reference criteria results was used. The overall quality of the
included studies was favorable (Figs.
2 and 3).
NMA
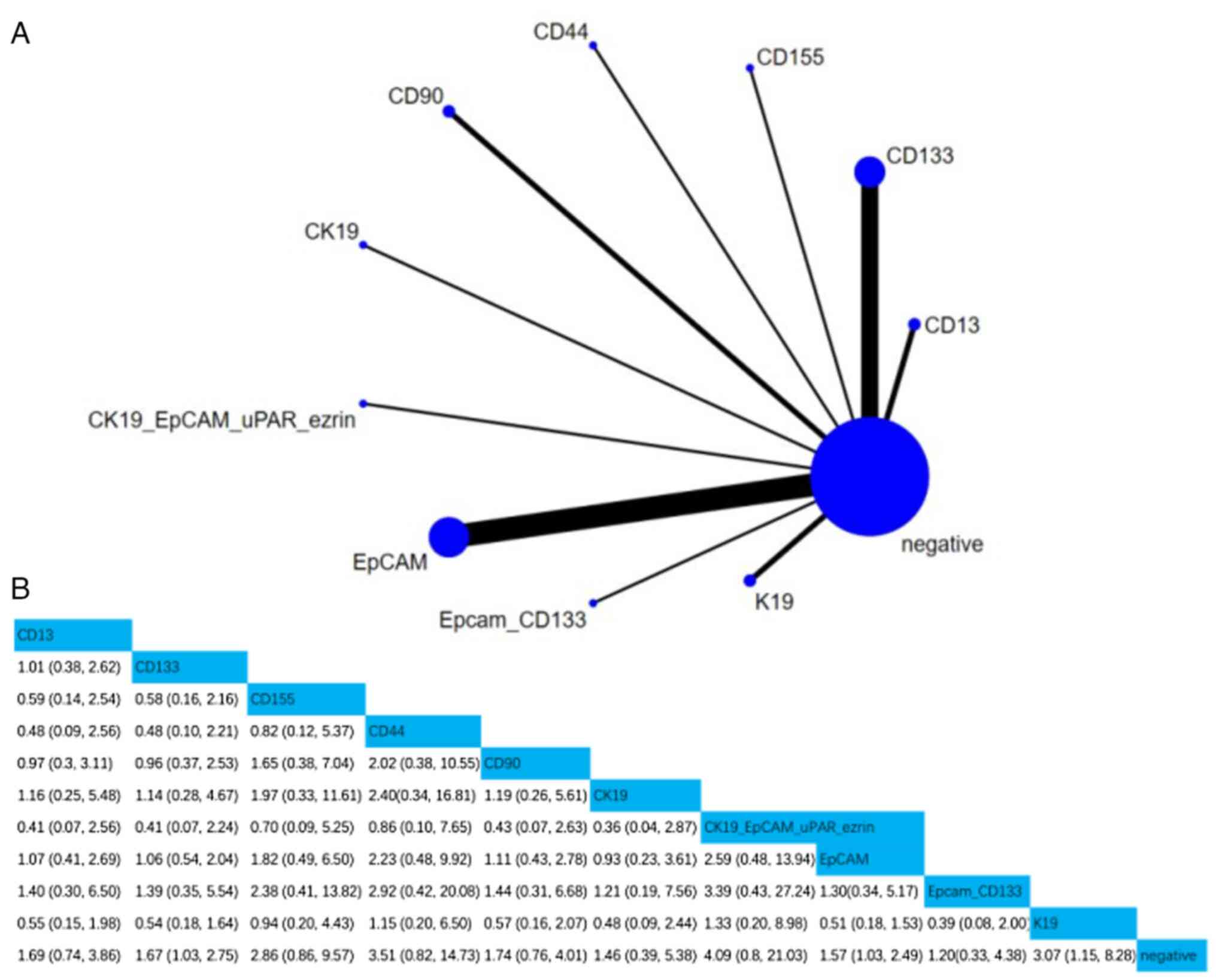
Survival prediction
i) OS. A total of 20 studies involving 2,400
patients assessed the associations between 10 CSC markers and OS.
Compared with negative CSC markers, the HR (95% CI) of CD133, EpCAM
and K19 were 1.67 (1.03, 2.75), 1.57 (1.03, 2.49) and 3.07 (1.15,
8.28), respectively, indicating that CD133, EpCAM and K19
positivity were notably associated with OS. Furthermore, according
to the SUCRA value, simultaneous positivity of CK19 and EpCAM was
the strongest predictor of OS (SUCRA, 78.65%; Figs. 4 and S1).
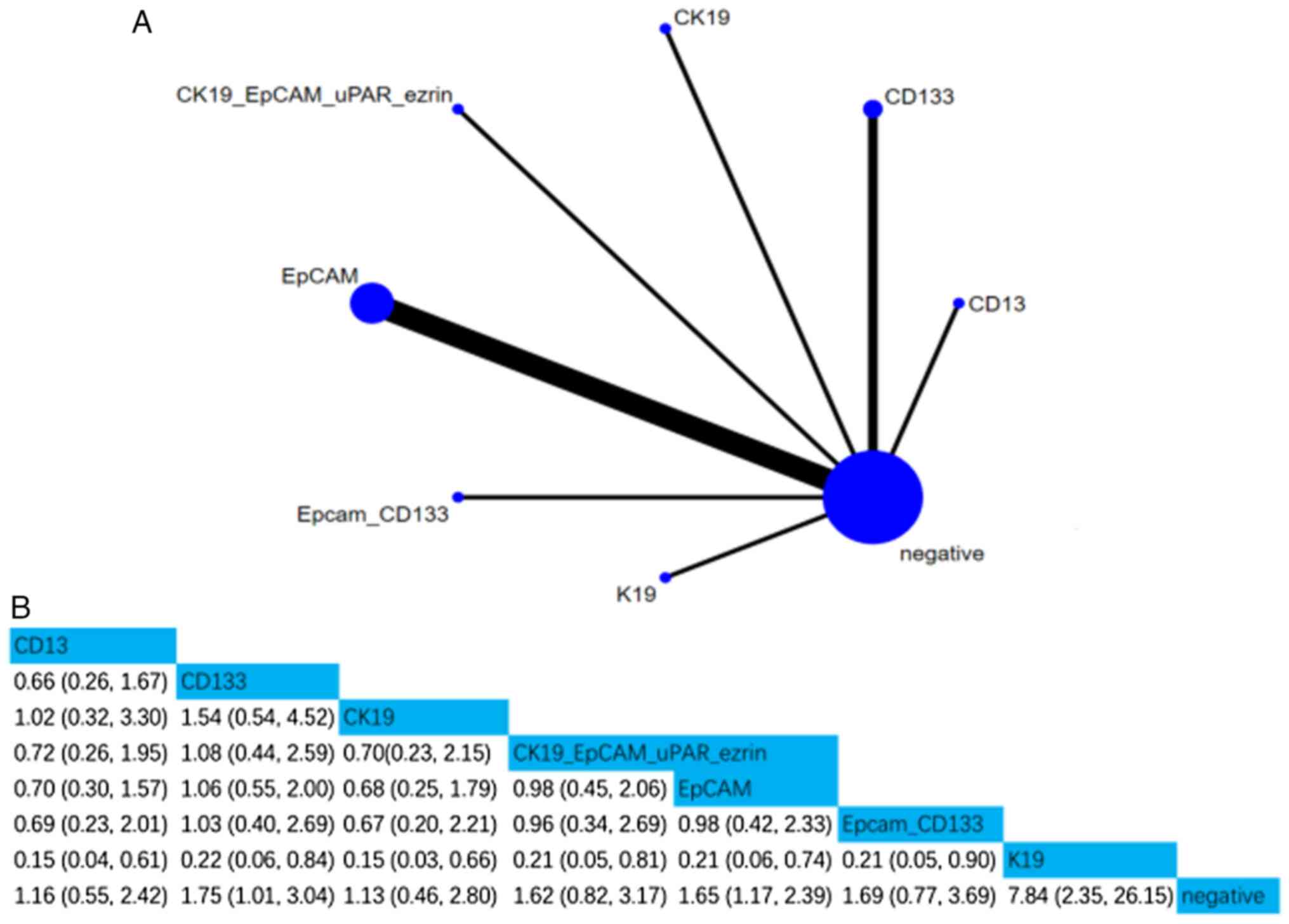
ii) RFS. A total of 10 studies involving 1,258
patients assessed the associations between seven CSC markers and
RFS. Compared with negative CSC markers, the HR (95% CI) of CD133,
EpCAM and K19 were 1.75 (1.01, 3.04), 1.65 (1.17, 2.39), and 7.84
(2.35, 26.15), respectively, indicating that CD133, EpCAM and K19
positivity were notably associated with RFS. Moreover, according to
the SUCRA value, K19 was the strongest predictor of RFS (SUCRA,
98.93%; Figs. 5 and S2).
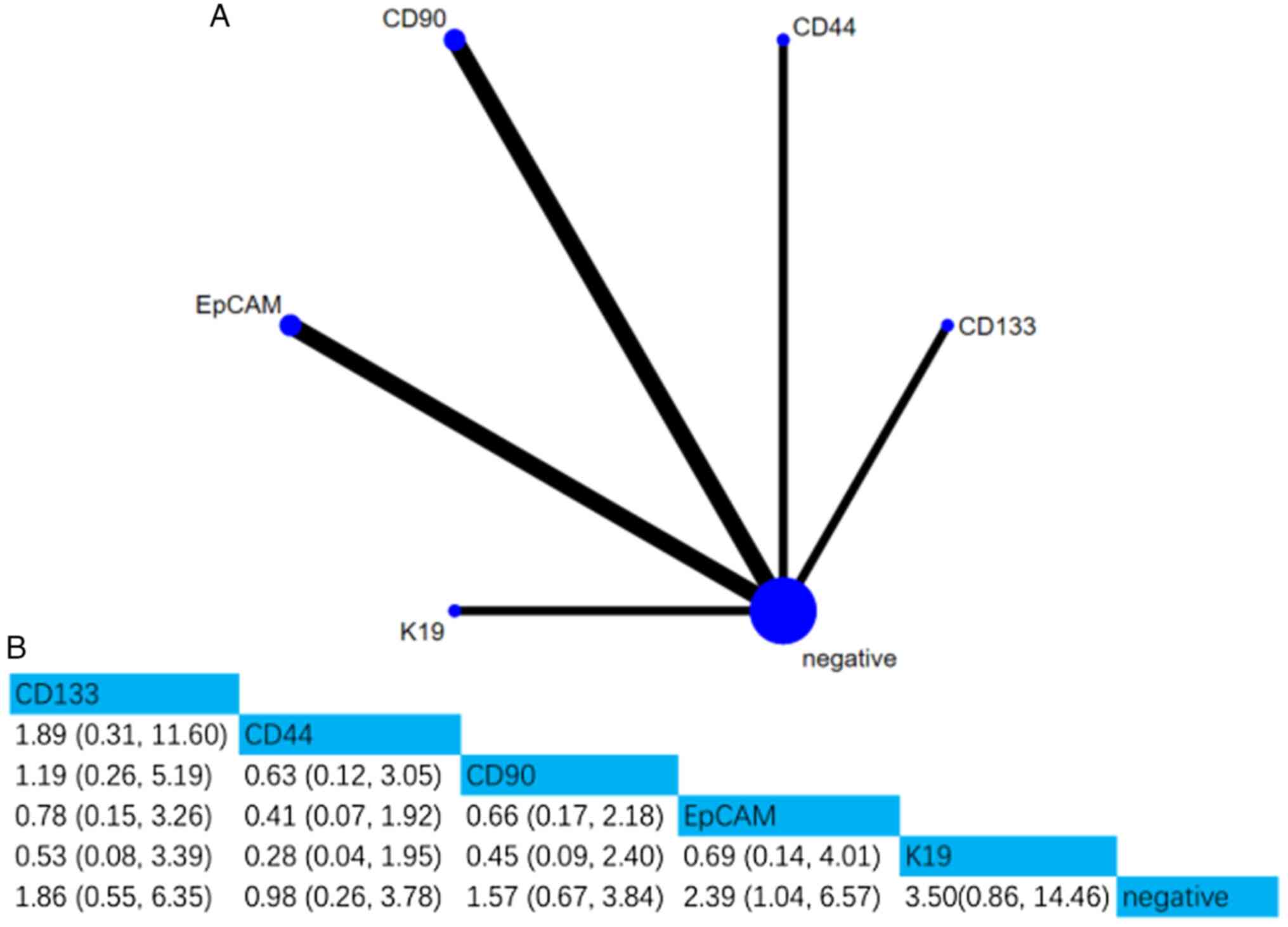
iii) DFS. A total of five studies involving 608
patients assessed the associations between five CSC markers and
DFS. Compared with negative CSC markers, the HR (95% CI) of EpCAM
was 2.39 (1.04, 6.57), indicating that EpCAM positivity was notably
associated with DFS. Furthermore, according to the SUCRA value, K19
was the strongest predictor of DFS (SUCRA, 84.95%; Figs. 6 and S3).
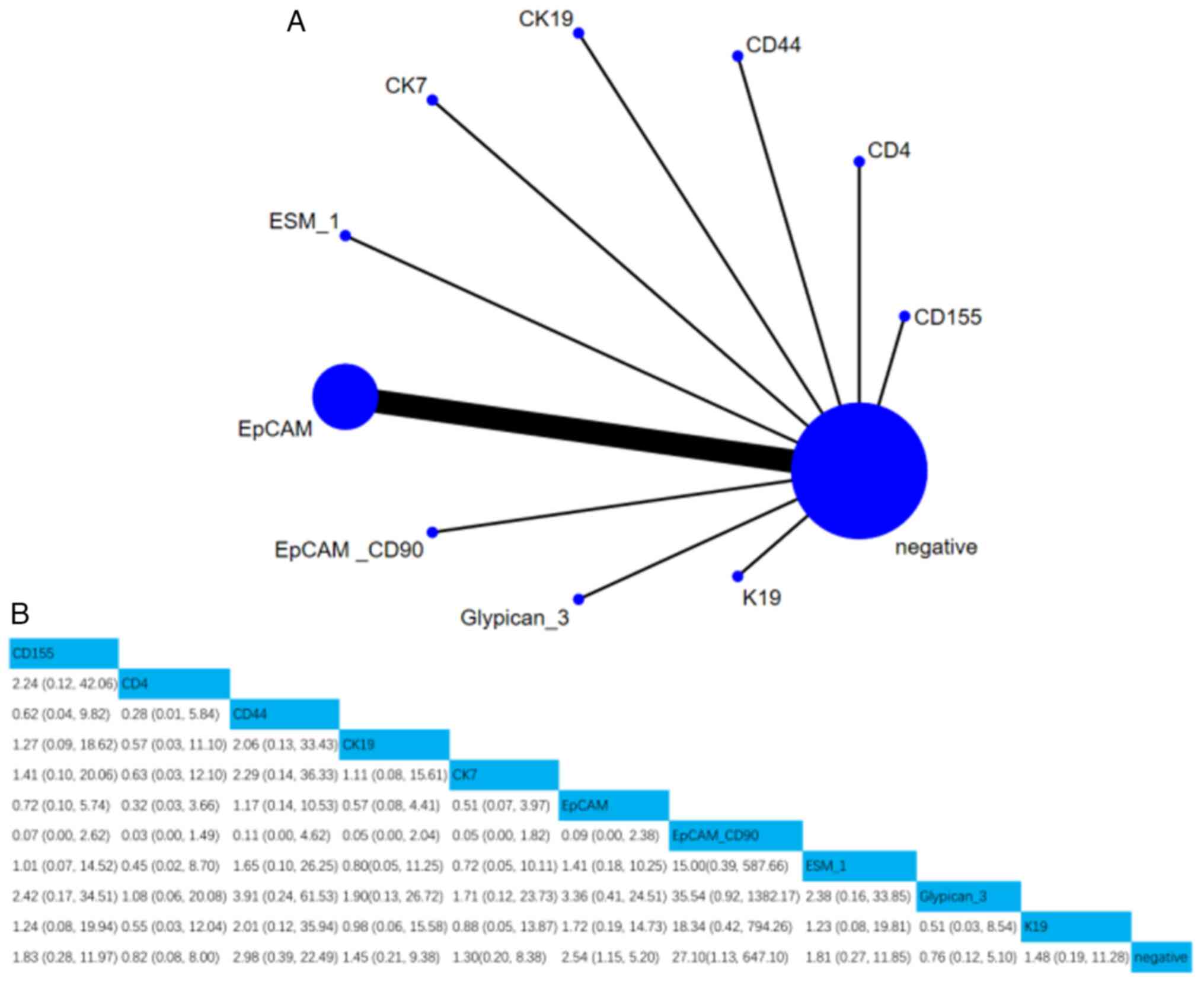
iv) Recurrence. A total of 11 studies involving
1,258 patients assessed the association between 10 CSC markers and
recurrence. Compared with negative CSC markers, the HR (95% CI) of
EpCAM and CD90 were 2.54 (1.15, 5.2) and 27.1 (1.13, 647.1),
respectively, indicating that EpCAM and CD90 positivity were
notably associated with recurrence. Moreover, according to the
SUCRA value, simultaneous positivity of EpCAM and CD90 was the
strongest predictor of recurrence (SUCRA, 5.61%; Figs. 7 and S4).
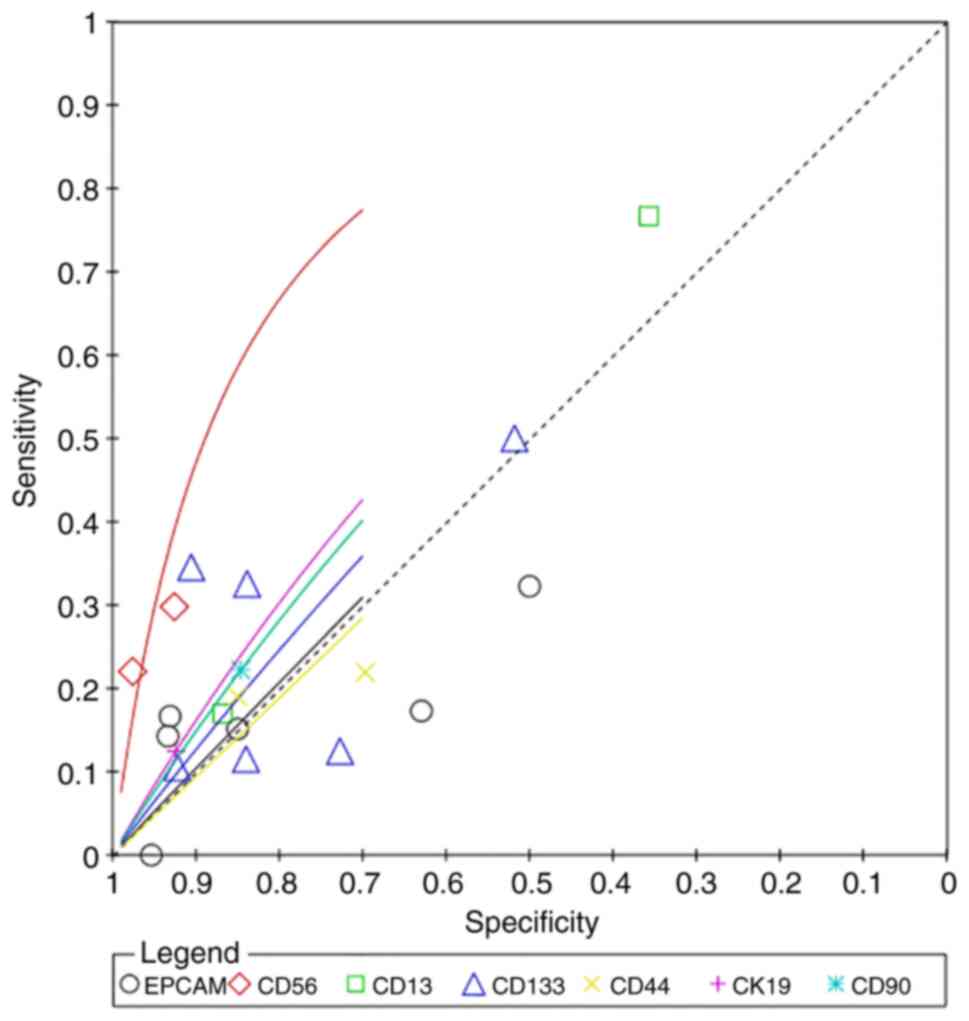
Diagnostic prediction
CD56 had the highest superiority index for
predicting poor differentiation (7.4498; 95% CI: 0.3333, 13.0000),
and its corresponding sensitivity, specificity and DOR values were
41.84% (95% CI: 14.01–71.10), 80.61% (95% CI: 60.57, 96.17) and
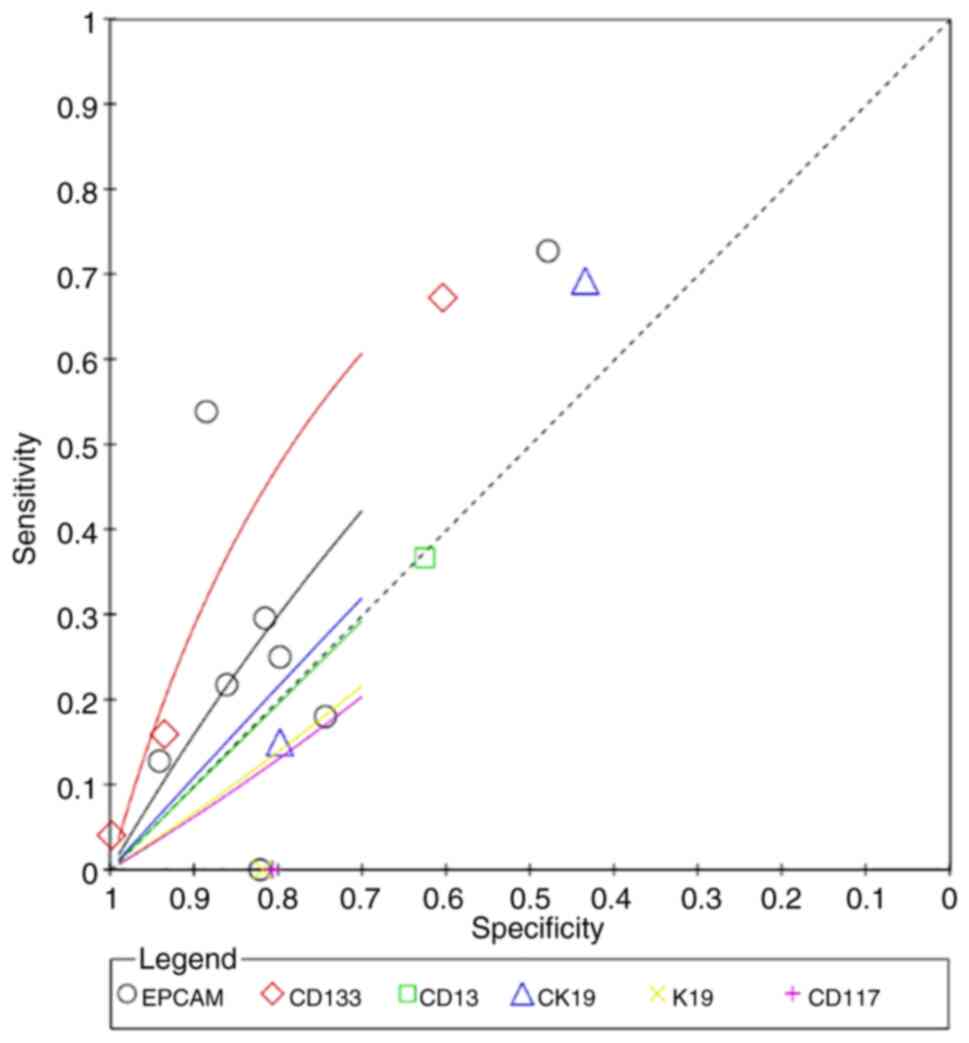
4.9086 (95% CI: 0.5793, 21.2615), respectively (Fig. 8; Table
I). K19 had the highest superiority index for predicting
microvascular invasion (8.4777; 95% CI: 0.2308, 17.0000), and its
corresponding sensitivity, specificity and DOR values were 79.60%
(95% CI: 44.80, 98.64), 61.19% (95% CI: 42.11, 77.00) and 22.6900
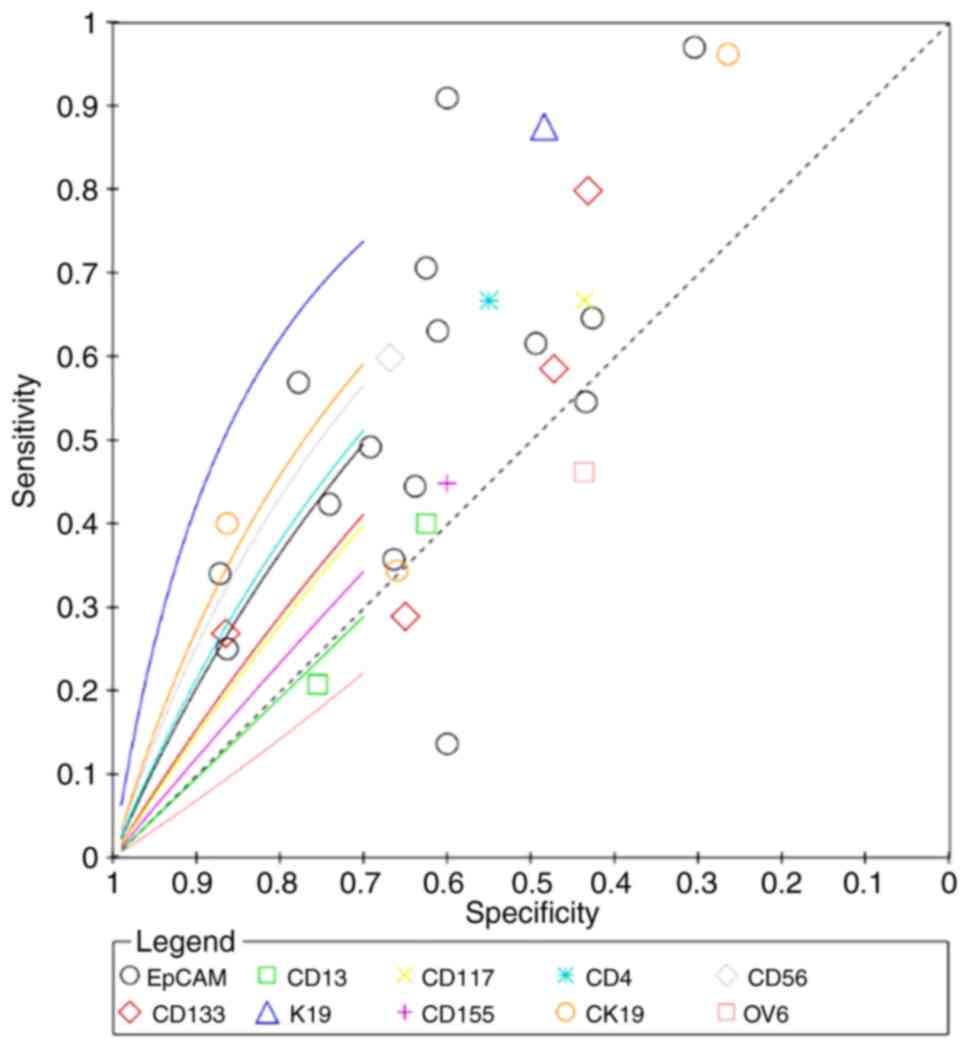
(95% CI: 1.1803, 122.8203), respectively (Fig. 9; Table
II. The superiority index of CD133 was the highest for
predicting metastasis (5.6097; 95% CI: 0.3333, 11.0000), with
sensitivity, specificity and DOR values of 35.95% (95% CI: 15.48,
59.44), 81.19% (95% CI: 61.60, 93.76) and 3.2171 (95% CI: 0.7013,
10.0291), respectively (Fig. 10;
Table III). The forest plots of
the aforementioned results are presented in Fig. S5, Fig.
S6, Fig. S7.
 | Table I.Summary of poorly differentiated
calculations. |
Table I.
Summary of poorly differentiated
calculations.
| Factor | Sensitivity | Specificity | DOR | Superiority | Relative
sensitivity | Relative
specificity |
|---|
| EpCAM | 0.2416
(0.1279–0.3857) | 0.7470
(0.5994–0.8637) | 1.0482
(0.3831–2.4093) | 1.1975
(0.1111–7.0000) | 1.0000
(1.0000–1.0000) | 1.0000
(1.0000–1.0000) |
| CD56 | 0.4184
(0.1401–0.7110) | 0.8061
(0.6057–0.9617) | 4.9086
(0.5793–21.2615) | 7.4498
(0.3333–13.0000) | 1.8182
(0.6226–3.4906) | 1.0834
(0.8490–1.3503) |
| CD13 | 0.3996
(0.1842–0.6656) | 0.6744
(0.3910–0.8729) | 1.9080
(0.3474–6.5866) | 3.2456
(0.1429–11.0000) | 1.7672
(0.7696–3.6111) | 0.9081
(0.5191–1.2265) |
| CD133 | 0.2710
(0.1613–0.4095) | 0.7602
(0.6164–0.8674) | 1.3134
(0.4898–2.8294) | 2.1189
(0.1429–9.0000) | 1.2062
(0.5679–2.3069) | 1.0242
(0.8099–1.2819) |
| CD44 | 0.2354
(0.0850–0.4720) | 0.6914
(0.4339–0.8955) | 0.9319
(0.1498–3.2421) | 1.0694
(0.0910–7.0000) | 1.0591
(0.3263–2.5968) | 0.9327
(0.5773–1.2952) |
| CK19 | 0.2533
(0.0478–0.5696) | 0.7691
(0.5282–0.9497) | 2.0494
(0.1496–8.8763) | 2.4342
(0.1111–11.0000) | 1.0911
(0.2137–2.6448) | 1.0334
(0.7408–1.3182) |
| CD90 | 0.3726
(0.0579–0.7633) | 0.2715
(0.0511–0.6145) | 0.3605
(0.0131–1.7778) | 0.4825
(0.0769–1.0000) | 1.6398
(0.2416–3.9625) | 0.3661
(0.0691–0.8366) |
 | Table II.Summary of calculation results of
microvascular invasion. |
Table II.
Summary of calculation results of
microvascular invasion.
| Factor | Sensitivity | Specificity | DOR | Superiority | Relative
sensitivity | Relative
specificity |
|---|
| EpCAM | 0.5597
(0.4677–0.6525) | 0.6086
(0.5248–0.6817) | 2.0197
(1.3994–2.8519) | 1.8716
(0.1429–9.0000) | 1.0000
(1.0000–1.0000) | 1.0000
(1.0000–1.0000) |
| CD133 | 0.5439
(0.3902–0.6864) | 0.5966
(0.4948–0.6903) | 1.8711
(0.9537–3.3679) | 1.3877
(0.0910–7.0000) | 0.9739
(0.7212–1.2079) | 0.9807
(0.8586–1.0935) |
| CD13 | 0.2416
(0.1086–0.4341) | 0.7941
(0.6543–0.8929) | 1.3831
(0.4679–3.4083) | 1.5087
(0.20000–5.0000) | 0.4325
(0.2006–0.7676) | 1.3086
(1.0730–1.5159) |
| K19 | 0.7960
(0.4480–0.9864) | 0.6119
(0.4211–0.7700) | 22.690
(1.1803–122.8203) | 8.4777
(0.2308–17.0000) | 1.4308
(0.7963–1.9356) | 1.0071
(0.7041–1.2810) |
| CD117 | 0.5954
(0.1309–0.9643) | 0.5729
(0.3774–0.7439) | 7.7117
(0.1944–34.0241) | 2.9064
(0.0588–15.0000) | 1.0691
(0.2287–1.7876) | 0.9422
(0.6302–1.2088) |
| CD155 | 0.4639
(0.1473–0.8173) | 0.5703
(0.2356–0.8565) | 1.5338
(0.2669–5.1778) | 1.4246
(0.0588–11.0000) | 0.8367
(0.2518–1.5430) | 0.9418
(0.3823–1.4403) |
| CD4 | 0.5766
(0.2351–0.8647) | 0.5575
(0.3436–0.7516) | 2.6428
(0.3581–10.3802) | 2.2501
(0.0667–13.0000) | 1.0343
(0.4459–1.6143) | 0.9170
(0.5851–1.2404) |
| CK19 | 0.6125
(0.4456–0.7741) | 0.6025
(0.4966–0.7009) | 2.6283
(1.1843–5.2354) | 2.9352
(0.1111–13.0000) | 1.0972
(0.7987–1.3774) | 0.9905
(0.8659–1.1148) |
| CD56 | 0.7344
(0.4580–0.9230) | 0.6070
(0.4770–0.7189) | 6.2781
(1.2031–20.2091) | 6.5486
(0.2–17.0000) | 1.3185
(0.8053–1.7407) | 0.9980
(0.8230–1.1559) |
| OV6 | 0.4301
(0.1966–0.6838) | 0.5517
(0.3934–0.6914) | 1.0879
(0.2755–2.8563) | 0.4685
(0.0588–3.0000) | 0.7686
(0.3629–1.2015) | 0.9065
(0.6771–1.1170) |
 | Table III.Summary of calculation results of
metastasis. |
Table III.
Summary of calculation results of
metastasis.
| Factor | Sensitivity | Specificity | DOR | Superiority | Relative
sensitivity | Relative
specificity |
|---|
| EpCAM | 0.3342
(0.1960–0.5030) | 0.7584
(0.6061–0.8635) | 1.7251
(0.6697–3.3949) | 3.0422
(0.3333–9.0000) | 1.0000
(1.0000–1.0000) | 1.0000
(1.0000–1.0000) |
| CD133 | 0.3595
(0.1548–0.5944) | 0.8119
(0.6160–0.9376) | 3.2171
(0.7013–10.0291) | 5.6097
(0.3333–11.0000) | 1.1107
(0.4816–1.9226) | 1.0760
(0.8516–1.3590) |
| CD13 | 0.4204
(0.0760–0.8371) | 0.5746
(0.1655–0.9088) | 1.8698
(0.0967–9.3034) | 1.7618
(0.1111–9.0000) | 1.3327
(0.2223–3.0957) | 0.7645
(0.2204–1.2865) |
| CK19 | 0.3104
(0.1266–0.5686) | 0.7111
(0.4293–0.8661) | 1.4105
(0.2703–3.8446) | 1.8452
(0.1429–7.0000) | 0.9414
(0.4468–1.7433) | 0.9388
(0.5847–1.1276) |
| K19 | 0.1451
(0.0001–0.6825) | 0.7051
(0.3619–0.9237) | 1.0569
(0.0001–7.4208) | 1.1619
(0.0910–7.0000) | 0.4461
(0.0001–2.0763) | 0.9313
(0.4836–1.2655) |
| CD117 | 0.1434
(0.0001–0.6577) | 0.6964
(0.3587–0.9231) | 0.9043
(0.0001–6.1399) | 1.0187
(0.0910–7.0000) | 0.4417
(0.0001–1.9755) | 0.9199
(0.4848–1.2727) |
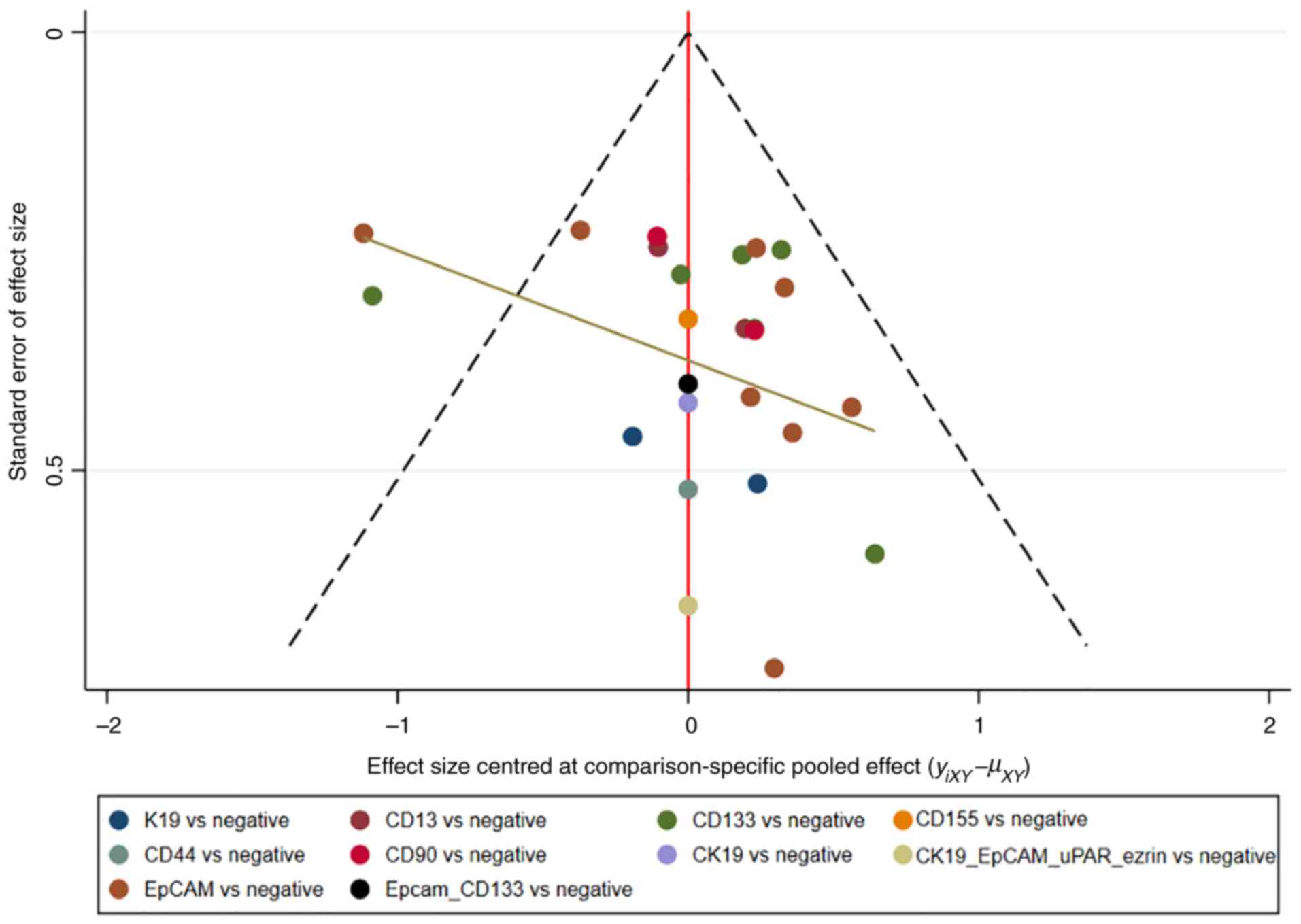
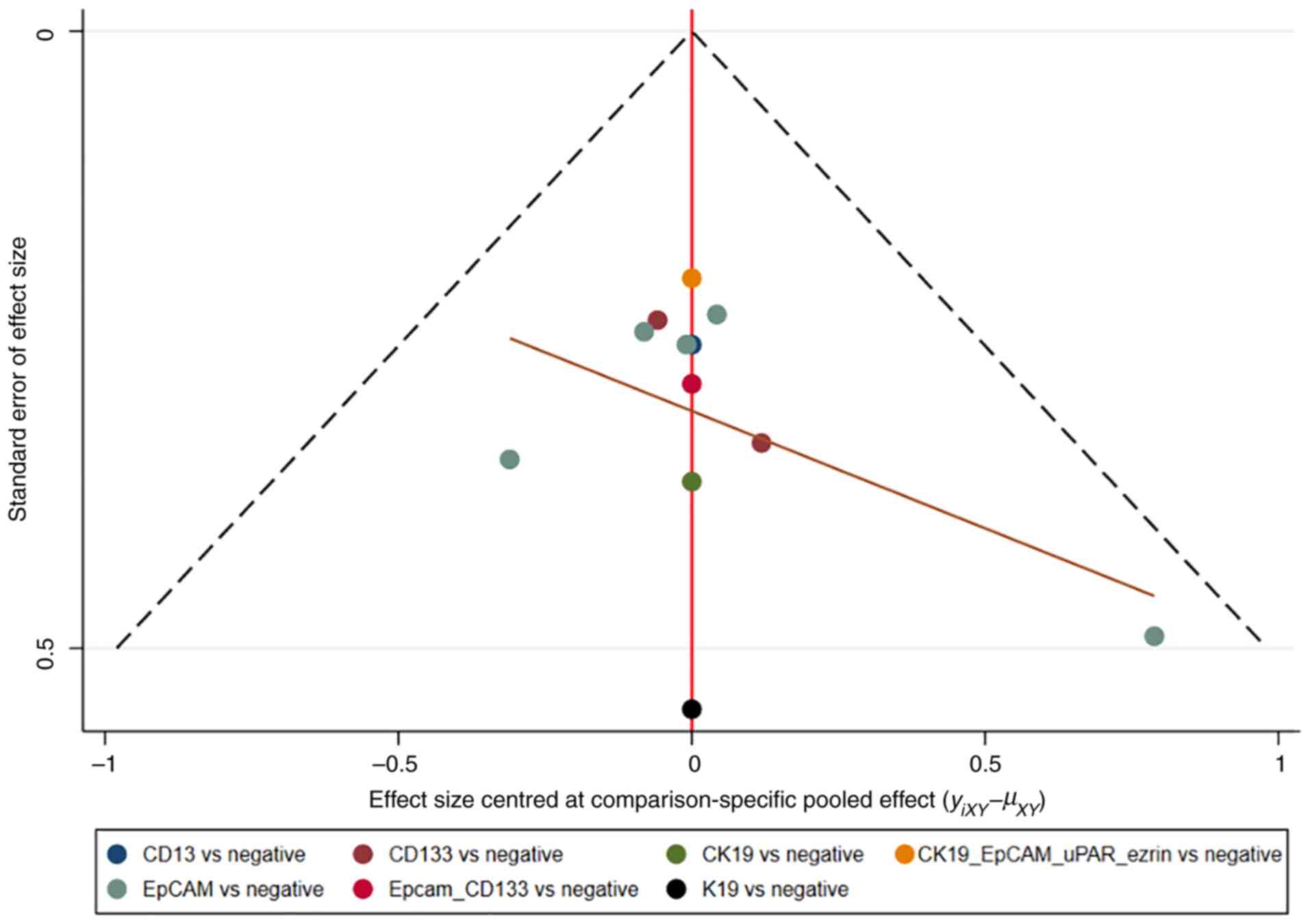
Assessment of consistency and
publication bias
The consistency and inconsistency models were
compared using DIC. Good consistency with DIC was indicated by
<5 points of the difference in all closed-loop models. In
addition, no evidence of publication bias was demonstrated in the
comparison-adjusted funnel plots (Fig.
11, Fig. 12, Fig. 13, Fig.
14).
Discussion
In the present study, literature involving both HCC
and CSC markers was retrieved, and 37 studies that met the
inclusion criteria were subjected to NMA. It was demonstrated that,
in terms of survival prognosis, simultaneous positive CK19 and
EpCAM had the strongest predictive effect on OS, indicating that
patients with CK19 and EpCAM positive HCC had a lower OS rate. K19
positivity also had the strongest predictor of RFS and DFS,
suggesting that low rates of RFS and DFS in patients with HCC were
associated with K19 positivity. Moreover, simultaneous positivity
of EpCAM and CD90 had the strongest predictive effect on the
recurrence rate, indicating that patients with CD90 positive HCC
had a higher recurrence rate. In terms of diagnostic value, CD56,
K19 and CD133 were the strongest predictors of poor
differentiation, microvascular invasion and metastasis,
respectively. In summary, CSC markers may have a certain diagnostic
or predictive value in patients with HCC.
The association between the CSC markers CK19, CD133
and EpCAM with the prognosis of patients with HCC has been assessed
in previous studies. For example, the association between CK19
overexpression with decreased OS was reported in a meta-analysis of
2,943 patients with HCC (58).
Nuclear transcription factors (spalt-like transcription factor 4,
activator protein 1 and activator protein 1) activate the CK19
promoter in response to extracellular stimuli that are transmitted
to the cytoplasm via the TGF-β, MAKP/JNK and MEK-ERK1/2 signaling
pathways, further inducing the CK19 expression-associated
regulatory network (59). The
Wnt/β-catenin pathway has been reported to be one of the most
important pathways associated with CSC markers, including CD133 and
EpCAM, which triggers angiogenesis, tumor invasion and metastasis
by enhancing the secretion of angiogenic factors (10). EpCAM is involved in several
biological processes of tumors, such as proliferation, invasion,
tumorigenesis and metastasis, primarily through the Wnt/β-catenin
pathway and other pathways (60).
Ma et al (61) performed a
meta-analysis that reported that CD133 and EpCAM were notably
associated with a low survival rate, including OS and DFS rates,
which is consistent with the findings in the present study.
Furthermore, the present study revealed that positivity of CD133
and EpCAM could predict the survival rate of patients with HCC.
Kawai et al (62) reported that K19-positive cells are
implicated in epithelial-mesenchymal transition (EMT) and
TGF-β/Smad signaling, and that TGF-β/Smad signaling confers a high
proliferation capacity in K19-positive cells. K19-positive patients
with HCC were also reported to have a higher TGFβR1 expression and
shorter RFS than K19-negative patients. Yokomichi et al
(63) also reported that the RFS
rate was lower in K19-positive patients with HCC than in
K19-negative patients with HCC, suggesting a poor prognosis
resulting from extrahepatic recurrence. Bae et al (64) reported that K19 is a prognostic
factor for HCC as its expression is related to a high tumor grade
and high AFP levels. They concluded that K19 positivity was notably
associated with a worse prognosis in HCC. Moreover, Kim et
al (65) reported that
K19-positive patients with HCC had markedly increased expressions
of EMT-related proteins and mRNAs, with stronger tumor
invasiveness, in comparison with K19-negative patients with HCC,
and K19 positivity was an important independent predictor of a low
DFS rate in patients with HCC. These results are consistent with
the findings in the present study, which indicated that K19
positivity could predict the RFS and DFS rates of patients with
HCC.
EpCAM is considered to be the most representative
marker of LCSC, and EpCAM-positive HCC cells exhibit stem cell-like
characteristics, including self-renewal, differentiation, strong
invasiveness and tumorigenicity (66). Shousha et al (67) reported that the baseline and
post-treatment levels of serum EpCAM were higher in patients with
recurrence, suggesting that EpCAM positivity was associated with
recurrence in patients with liver cancer. Luo et al
(68) reported that the Notch
pathway may enhance the CSC characteristics of CD90-positive CSCs,
increasing the recurrence rate of CD90-positive patients with HCC,
which is associated with a poor prognosis of HCC. Moreover, the
Notch signaling pathway is an evolutionarily conserved pathway that
can facilitate self-renewal, differentiation, proliferation,
survival, angiogenesis and migration of CSCs (68). In the present study, NMA on the
association between the expression of CSC markers and HCC
recurrence was performed, and it was demonstrated that EpCAM
positivity and simultaneous positivity of EpCAM and CD90 could
predict the recurrence rate in patients with HCC, similar to the
findings of Shousha et al (67) and Luo et al (68). The findings further provide a
reliable basis for the prediction of HCC recurrence by EpCAM
positivity and simultaneous positivity of EpCAM and CD90.
The present study revealed that CD56 had the highest
superiority index for predicting poor differentiation, with higher
sensitivity and specificity. Liu et al (69) reported that CD90 had higher tissue
specificity in HCC and higher sensitivity in predicting poorly
differentiated HCC than CD133, EpCAM and CD44. This is a
potentially promising target for patient classification and
differentiation therapy, and the findings of Liu et al are
inconsistent with the results of the present paper. Moreover, Liu
et al analyzed the effect of four CSC markers, CD90, CD133,
EpCAM and CD44, on poor differentiation in HCC, and the present
study analyzed the effect of seven CSS markers, CD56, CK19, CD90,
CD13, CD133, EpCAM and CD44, on poor differentiation in HCC.
Subsequently, the superiority indexes of the four CSC markers
included in the study of Liu et al were further compared.
CD133 had the highest superiority index in the present study. Such
a difference may be attributed to the incomplete conformity in the
types of CSC markers: Namely, the present study assessed more types
of CSC markers, whereas CD56 was not studied by Liu et al.
Moreover, the analytical methods were different: A direct
comparison was made by Liu et al whereas a pooled analysis
was performed by both direct and indirect comparisons in the
present study, so that the CSC markers that were the most effective
in predicting poor differentiation could be identified. In
addition, the differences in the findings may be due to the
heterogeneity of the study populations, which were not the same in
the two studies, with differences between individuals within the
populations in terms of sex, age, disease duration and tumor
stage.
Kim et al (65) reported that microvascular invasion
occurred more frequently in K19-positive patients than in
K19-negative patients with HCC. In the present study, K19 was
demonstrated to be the strongest predictor of microvascular
invasion, consistent with the view of Takano et al (70), which reported that K19 was
associated with HCC progression and adverse clinical outcomes by
directly enhancing cancer cell survival, invasion and angiogenesis.
In the present study, K19 positivity was associated with
microvascular invasion in patients with HCC, consistent with the
results by Takano et al and Kim et al which reported
that K19 expression was positively associated with tumor
angiogenesis and invasion. Therefore, K19 is a possible new target
for the treatment of K19-positive HCC.
TGF-β can activate the expression of CD133, and
inhibit the expressions of DNA methyltransferase (DNMT)1 and
DNMT3β, thereby maintaining DNA methylation. TGF-β-induced
CD133-positive cells may initiate tumor development in vivo
(71). Yan et al (72) reported that CD133-positive HCC cells
possessed stronger migration and invasion capacity in vitro,
and enhanced metastasis capacity in vivo and in human HCC
specimens. In the present study, the results revealed that CD133
was the strongest predictor of metastasis; however, Zhong et
al (73) reported that the
expression of CD133 was not associated with metastasis. The reason
for such a difference may be attributed to different study
populations, regions, time and analytical methods. Therefore, it is
suggested that further studies are performed to confirm the
clinical significance of these CSC markers in the diagnosis and
prognostic prediction of patients with HCC.
To the best of our knowledge, the NMA in the present
study is the first to compare the diagnostic value of CSC markers
for HCC and their associations with survival prognosis, and
identify the CSC marker with the best predictive effect by indirect
comparisons. The findings provide valuable information for the
diagnostic value of CSC markers for HCC and their associations with
survival prognosis. However, the NMA has certain limitations: i)
The accuracy and applicability of the findings may be affected due
to the limited number of studies on certain outcome measures and
CSC markers; ii) the follow-up time varied greatly across studies,
and no subgroup or regression analysis could be performed due to
the limited number of studies; and iii) the present study was
restricted only to studies published in English, thus introducing
selection bias. Therefore, more high-quality randomized controlled
trials are required to confirm the findings of the present
study.
In conclusion, no single CSC marker possesses the
best predictive effect on all indexes of survival prognosis and
diagnostic value of patients with HCC. In terms of survival
prognosis, simultaneous positivity of CK19 and EpCAM displayed the
strongest predictive effect on the OS rate, indicating that it was
associated with low OS rate in patients with HCC; K19 positivity
displayed the strongest predictive effect on the RFS and DFS rates,
indicating that it was associated with low RFS and DFS rates in
patients with HCC; simultaneous positivity of EpCAM and CD90 had
the strongest predictive effect on the recurrence rate, indicating
that it was associated with high recurrence rate in patients with
HCC. In terms of diagnostic value, CD56, K19 and CD133 were the
strongest predictors of poor differentiation, microvascular
invasion and metastasis, respectively. However, due to certain
limitations of existing clinical studies and evidence, more
rigorous study designs with a larger sample sizes and longer
follow-up times are needed to confirm the aforementioned findings
in the future.
Furthermore, CSC markers possess great potential for
predicting the prognosis of patients with HCC, identifying the
patients at high risk of recurrence and metastasis, providing
targeted therapies, and monitoring the therapeutic response in
patients with HCC. Finally, CSC markers can open up new roads for
targeted therapies in HCC, and the following directions for future
research are suggested: i) The biological properties of CSC
markers; and ii) how to enhance the specificity of targeted
therapies to eliminate CSCs in HCC without adverse effects on
normal cells as CSCs and normal stem cells in HCC share many
similarities.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present work was supported by the Science and Health Joint
Project of the Hunan Provincial Natural Science Foundation of China
(grant no. 2021JJ70051).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
ZO, SF, JY, JH and WZ contributed to the study
conception and design. ZO, SF and JY performed data acquisition,
data analysis and manuscript preparation. JH and WZ assisted with
data acquisition, data analysis and statistical analysis. The
initial draft and revisions of the manuscript were written by ZO
and WZ. ZO and WZ confirm the authenticity of all the raw data. All
authors commented on previous versions of the manuscript. All
authors read and approved the final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD, Wagle NS and Jemal
A: Cancer statistics, 2023. CA Cancer J Clin. 73:17–48. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yoh T, Seo S, Taura K, Iguchi K, Ogiso S,
Fukumitsu K, Ishii T, Kaido T and Uemoto S: Surgery for recurrent
hepatocellular carcinoma: Achieving Long-term survival. Ann Surg.
273:792–799. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lou J, Zhang L, Lv S, Zhang C and Jiang S:
Biomarkers for hepatocellular carcinoma. Biomark Cancer. 9:1–9.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Debnath P, Dalal K, Dalal B, Athalye S,
Chandnani S, Jain S, Shukla A, Rathi P and Shankarkumar A:
Characterization of circulating tumor cells using imaging flow
cytometry in liver disease patients. J Clin Exp Hepatol.
13:608–617. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fang X, Yan Q, Liu S and Guan XY: Cancer
stem cells in hepatocellular carcinoma: Intrinsic and extrinsic
molecular mechanisms in stemness regulation. Int J Mol Sci.
23:123272022. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
He G, Dhar D, Nakagawa H, Font-Burgada J,
Ogata H, Jiang Y, Shalapour S, Seki E, Yost SE, Jepsen K, et al:
Identification of liver cancer progenitors whose malignant
progression depends on autocrine IL-6 signaling. Cell. 155:384–396.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nio K, Yamashita T and Kaneko S: The
evolving concept of liver cancer stem cells. Mol Cancer. 16:42017.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wu K, Ding J, Chen C, Sun W, Ning BF, Wen
W, Huang L, Han T, Yang W, Wang C, et al: Hepatic transforming
growth factor beta gives rise to tumor-initiating cells and
promotes liver cancer development. Hepatology. 56:2255–2267. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jeng KS, Chang CF, Sheen IS, Jeng CJ and
Wang CH: Cellular and molecular biology of cancer stem cells of
hepatocellular carcinoma. Int J Mol Sci. 24:14172023. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tseeleesuren D, Hsiao HH, Kant R, Huang
YC, Tu HP, Lai CC, Huang SF and Yen CH: The Expression and
prognostic value of cancer stem cell markers, NRF2, and its target
genes in TAE/TACE-Treated hepatocellular carcinoma. Medicina
(Kaunas). 58:2122022. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sekar V, Veerabathiran R, Pandian A and
Sivamani G: Targeting liver cancer stem cell through EpCAM therapy
targeted with chemotherapy endorse enhanced progression in
hepatocellular carcinoma. Egyptian Liver J. 13:292023. View Article : Google Scholar
|
|
13
|
Hutton B, Salanti G, Caldwell DM, Chaimani
A, Schmid CH, Cameron C, Ioannidis JP, Straus S, Thorlund K, Jansen
JP, et al: The PRISMA extension statement for reporting of
systematic reviews incorporating network meta-analyses of health
care interventions: Checklist and explanations. Ann Intern Med.
2:162:777–784. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Eriksen MB and Frandsen TF: The impact of
patient and interventioncomparison and outcome (PICO) as a search
strategy tool on literature search quality: A systematic review. J
Med Libr Assoc. 106:420–431. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Whiting PF, Rutjes AW, Westwood ME,
Mallett S, Deeks JJ, Reitsma JB, Leeflang MM, Sterne JA and Bossuyt
PM: QUADAS-2: QUADAS-2: A revised tool for the quality assessment
of diagnostic accuracy studies. Ann Intern Med. 155:529–536. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Brennan B, Kirton L, Marec-Bérard P,
Gaspar N, Laurence V, Martín-Broto J, Sastre A, Gelderblom H, Owens
C, Fenwick N, et al: Comparison of two chemotherapy regimens in
patients with newly diagnosed Ewing sarcoma (EE2012): An
open-label, randomised, phase 3 trial. Lancet. 400:1513–1521. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Veroniki AA, Straus SE, Fyraridis A and
Tricco AC: The rank-heat plot is a novel way to present the results
from a network meta-analysis including multiple outcomes. J Clin
Epidemiol. 76:193–199. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ye J, Hu Y, Chen X, Chang C and Li K:
Comparative effects of different nutritional supplements on
inflammation and nutritional status, and clinical outcomes in
colorectal cancer patients: A systematic review and network
meta-analysis. Nutrients. 15:27722023. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Nyaga VN, Aerts M and Arbyn M: ANOVA model
for network meta-analysis of diagnostic test accuracy data. Stat
Methods Med Res. 27:1766–1784. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang S, Chen Y, Zhou F, Wang L and Luo Q:
Effect of care bundles for acute kidney injury: A systematic review
and meta-analysis. PLoS One. 19:e03021792024. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bally S, Cottin J, Gagnieu MC, Lega JC,
Verstuyft C, Rheims S, Lesca G, Cucherat M and Grenet G:
Publication bias in pharmacogenetics of adverse reaction to
antiseizure drugs: An umbrella review and a meta-epidemiological
study. PLoS One. 17:e02788392022. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kumagai A, Kondo F, Sano K, Inoue M, Fujii
T, Hashimoto M, Watanabe M, Soejima Y, Ishida T, Tokairin T, et al:
Immunohistochemical study of hepatocyte and cholangiocyte stem cell
markers of hepatocellular carcinoma: The second report:
Relationship with tumor size and cell differentiation. J
Hepatobiliary Pancreat Sci. 23:414–421. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Seino S, Tsuchiya A, Watanabe Y, Kawata Y,
Kojima Y, Ikarashi S, Yanai H, Nakamura K, Kumaki D, Hirano M, et
al: Clinical outcome of hepatocellular carcinoma can be predicted
by the expression of hepatic progenitor cell markers and serum
tumour markers. Oncotarget. 9:21844–21860. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
von Felden J, Schulze K, Krech T, Ewald F,
Nashan B, Pantel K, Lohse AW, Riethdorf S and Wege H: Circulating
tumor cells as liquid biomarker for high HCC recurrence risk after
curative liver resection. Oncotarget. 8:89978–89987. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ziol M, Sutton A, Calderaro J, Calderaro
J, Barget N, Aout M, Leroy V, Blanc JF, Sturm N, Bioulac-Sage P, et
al: ESM-1 expression in stromal cells is predictive of recurrence
after radiofrequency ablation in early hepatocellular carcinoma. J
Hepatol. 59:1264–1270. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Schulze K, Gasch C, Staufer K, Nashan B,
Lohse AW, Pantel K, Riethdorf S and Wege H: Presence of
EpCAM-positive circulating tumor cells as biomarker for systemic
disease strongly correlates to survival in patients with
hepatocellular carcinoma. Int J Cancer. 133:2165–2171. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Matthai SM and Ramakrishna B: Cancer stem
cells in hepatocellular carcinoma-An immuno-histochemical study
with histopathological association. Indian J Med Res. 142:391–398.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xu M, Qian G, Xie F, Shi C, Yan L, Yu L,
Zheng T, Wei L and Yang J: Expression of epithelial cell adhesion
molecule associated with elevated ductular reactions in
hepatocellar carcinoma. Clin Res Hepatol Gastroenterol. 38:699–705.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hwang HS, Yoo JE, Han DH, Choi JS, Lee JG,
Joo DJ, Kim MS, Kim SI, Choi GH and Park YN: Circulating cancer
stem cells expressing EpCAM/CD90 in hepatocellular carcinoma: A
pilot study for predicting tumor recurrence after living donor
liver transplantation. Gut Liver. 16:443–455. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang J, Qi YP, Ma N, Lu F, Gong WF, Chen
B, Ma L, Zhong JH, Xiang BD and Li LQ: Overexpression of epcam and
CD133 correlates with poor prognosis in Dual-phenotype
hepatocellular carcinoma. J Cancer. 11:3400–3406. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Krause J, von Felden J, Casar C, Fründt
TW, Galaski J, Schmidt C, Jung C, Ittrich H, Weidemann SA, Krech T,
et al: Hepatocellular carcinoma: Intratumoral EpCAM-positive cancer
stem cell heterogeneity identifies high-risk tumor subtype. BMC
Cancer. 20:11302020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Abdelgawad IA: Epithelial cell adhesion
molecule mRNA can be a potential marker to predict metastasis in
hepatocellular carcinoma patients. Asian Pac J Cancer Prev.
21:861–866. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yamanaka C, Wada H, Eguchi H, Hatano H,
Gotoh K, Noda T, Yamada D, Asaoka T, Kawamoto K, Nagano H, et al:
Clinical significance of CD13 and epithelial mesenchymal transition
(EMT) markers in hepatocellular carcinoma. Jpn J Clin Oncol.
48:52–60. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Shen J, Wang WS, Zhu XL and Ni CF: High
epithelial cell adhesion molecule-positive circulating tumor cell
count predicts poor survival of patients with unresectable
hepatocellular carcinoma treated with transcatheter arterial
chemoembolization. J Vasc Interv Radiol. 29:1678–1684. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Noh CK, Wang HJ, Kim CM, Kim J, Yoon SY,
Lee GH, Cho HJ, Yang MJ, Kim SS, Hwang JC, et al: EpCAM as a
predictive marker of tumor recurrence and survival in patients who
underwent surgical resection for hepatocellular Carcinoma.
Anticancer Res. 38:4101–4109. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lima LDP, Machado CJ, Rodrigues JBSR,
Vasconcellos LS, Junior EP, Vidigal PVT and Resende V:
Immunohistochemical coexpression of epithelial cell adhesion
molecule and alpha-fetoprotein in hepatocellular carcinoma. Can J
Gastroenterol Hepatol. 2018:59708522018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ko CJ, Li CJ, Wu MY and Chu PY:
Overexpression of epithelial cell adhesion molecule as a predictor
of poor outcome in patients with hepatocellular carcinoma. Exp Ther
Med. 16:4810–4816. 2018.PubMed/NCBI
|
|
38
|
Dai XM, Yang SL, Zheng XM, Chen GG, Chen J
and Zhang T: CD133 expression and α-fetoprotein levels define novel
prognostic subtypes of HBV-associated hepatocellular carcinoma: A
long-term follow-up analysis. Oncol Lett. 15:2985–2991.
2018.PubMed/NCBI
|
|
39
|
Dai XM, Huang T, Yang SL, Zheng XM, Chen
GG and Zhang T: Peritumoral EpCAM is an independent prognostic
marker after curative resection of HBV-Related hepatocellular
carcinoma. Dis Markers. 2017:84953262017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chen YL, Lin PY, Ming YZ, Huang WC, Chen
RF, Chen PM and Chu PY: The effects of the location of cancer stem
cell marker CD133 on the prognosis of hepatocellular carcinoma
patients. BMC Cancer. 17:4742017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhao Q, Zhou H, Liu Q, Cao Y, Wang G, Hu
A, Ruan L, Wang S, Bo Q, Chen W, et al: Prognostic value of the
expression of cancer stem cell-related markers CD133 and CD44 in
hepatocellular carcinoma: From patients to patient-derived tumor
xenograft models. Oncotarget. 7:47431–47443. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Nam SJ, Yeo HY, Chang HJ, Kim BH, Hong EK
and Park JW: A new cell block method for multiple
immunohistochemical analysis of circulating tumor cells in patients
with liver cancer. Cancer Res Treat. 48:1229–1242. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ye F, Jing YY, Guo SW, Yu GF, Fan QM, Qu
FF, Gao L, Yang Y, Wu D, Meng Y, et al: Proliferative ductular
reactions correlate with hepatic progenitor cell and predict
recurrence in HCC patients after curative resection. Cell Biosci.
4:502014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Chan AWH, Tong JHM, Chan SL, Lai PBS and
To KF: Expression of stemness markers (CD133 and EpCAM) in
prognostication of hepatocellular carcinoma. Histopathology.
64:935–950. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wong E, Srivastava S, Yeoh KG, Teh M and
Salto-Tellez M: Clinical and biological relevance of thy-1/CD90
protein overexpression in human hepatocellular carcinoma. J Onco
Pathol. 1:1–9. 2013.
|
|
46
|
Sun YF, Xu Y, Yang XR, Guo W, Zhang X, Qiu
SJ, Shi RY, Hu B, Zhou J and Fan J: Circulating stem cell-like
epithelial cell adhesion molecule-positive tumor cells indicate
poor prognosis of hepatocellular carcinoma after curative
resection. Hepatology. 57:1458–1468. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhao RC, Zhou J, Chen KF, Gong J, Liu J,
He JY, Guan P, Li B and Qin Y: The prognostic value of combination
of CD90 and OCT4 for hepatocellular carcinoma after curative
resection. Neoplasma. 63:288–298. 2016.PubMed/NCBI
|
|
48
|
Sung JJ, Noh SJ, Bae JS, Park HS, Jang KY,
Chung MJ and Moon WS: Immunohistochemical expression and clinical
significance of suggested stem cell markers in hepatocellular
carcinoma. J Pathol Transl Med. 50:52–57. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Rhee H, Nahm JH, Kim H, Choi GH, Yoo JE,
Lee HS, Koh MJ and Park YN: Poor outcome of hepatocellular
carcinoma with stemness marker under hypoxia: Resistance to
transarterial chemoembolization. Mod Pathol. 29:1038–1049. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Fatourou E, Koskinas J, Karandrea D,
Palaiologou M, Syminelaki T, Karanikolas M, Felekouras E, Antoniou
E, Manesis EK, Delladetsima J and Tiniakos D: Keratin 19 protein
expression is an independent predictor of survival in human
hepatocellular carcinoma. Eur J Gastroenterol Hepatol.
27:1094–1102. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yilmaz G, Akyol G, Cakir A and Ilhan M:
Investigation of diagnostic utility and expression profiles of stem
cell markers (CD133 and CD90) in hepatocellular carcinoma, small
cell dysplasia, and cirrhosis. Pathol Res Pract. 210:419–425. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Tovuu LO, Imura S, Utsunomiya T, Morine Y,
Ikemoto T, Arakawa Y, Mori H, Hanaoka J, Kanamoto M, Sugimoto K, et
al: Role of CD44 expression in non-tumor tissue on intrahepatic
recurrence of hepatocellular carcinoma. Int J Clin Oncol.
18:651–656. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Lee Y, Park H, Lee H, Cho JY, Yoon YS,
Choi YR, Han HS, Jang ES, Kim JW, Jeong SH, et al: The
clinicopathological and prognostic significance of the gross
classification of hepatocellular carcinoma. J Pathol Transl Med.
52:85–92. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Xu J, Zhang Y, Wang Y, Tao X, Cheng L, Wu
S and Tao Y: Correlation of KAI1, CD133 and vasculogenic mimicry
with the prediction of metastasis and prognosis in hepatocellular
carcinoma. Int J Clin Exp Pathol. 11:3638–3646. 2018.PubMed/NCBI
|
|
55
|
Jin AL, Zhang CY, Zheng WJ, Xian JR, Yang
WJ, Liu T, Chen W, Li T, Wang BL, Pan BS, et al: CD155/SRC complex
promotes hepatocellular carcinoma progression via inhibiting the
p38 MAPK signalling pathway and correlates with poor prognosis.
Clin Transl Med. 12:e7942022. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Yu GF, Lin X, Luo RC and Fang WY: Nuclear
CD133 expression predicts poor prognosis for hepatocellular
carcinoma. Int J Clin Exp Pathol. 11:2092–2099. 2018.PubMed/NCBI
|
|
57
|
Zhou Y, Wang B, Wu J, Zhang C, Zhou Y,
Yang X, Zhou J, Guo W and Fan J: Association of preoperative EpCAM
circulating tumor cells and peripheral treg cell levels with early
recurrence of hepatocellular carcinoma following radical hepatic
resection. BMC Cancer. 16:5062016. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Sun DW, Zhang YY, Sun XD, Chen YG, Qiu W,
Ji M and Lv GY: Prognostic value of cytokeratin 19 in
hepatocellular carcinoma: A meta-analysis. Clin Chim Acta.
448:161–169. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Zhuo JY, Lu D, Tan WY, Zheng SS, Shen YQ
and Xu X: CK19-positive hepatocellular carcinoma is a
characteristic subtype. J Cancer. 11:5069–5077. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Zhan T, Rindtorff N and Boutros M: Wnt
signaling in cancer. Oncogene. 36:1461–1473. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Ma YC, Yang JY and Yan LN: Relevant
markers of cancer stem cells indicate a poor prognosis in
hepatocellular carcinoma patients: A meta-analysis. Eur J
Gastroenterol Hepatol. 25:1007–1016. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Kawai T, Yasuchika K, Ishii T, Katayama H,
Yoshitoshi EY, Ogiso S, Kita S, Yasuda K, Fukumitsu K, Mizumoto M,
et al: Keratin 19, a cancer stem cell marker in human
hepatocellular carcinoma. Clin Cancer Res. 21:3081–3091. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Yokomichi N, Nishida N, Umeda Y, Taniguchi
F, Yasui K, Toshima T, Mori Y, Nyuya A, Tanaka T, Yamada T, et al:
Heterogeneity of epigenetic and epithelial mesenchymal transition
marks in hepatocellular carcinoma with keratin 19 proficiency.
Liver Cancer. 8:239–254. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Bae JS, Choi HN, Noh SJ, Park BH, Jang KY,
Park CK and Moon WS: Expression of K19 and K7 in dysplastic nodules
and hepatocellular carcinoma. Oncol Lett. 4:213–220. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Kim H, Choi GH, Na DC, Ahn EY, Kim GI, Lee
JE, Cho JY, Yoo JE, Choi JS and Park YN: Human hepatocellular
carcinomas with ‘Stemness’-Related marker expression: Keratin 19
expression and a poor prognosis. Hepatology. 54:1707–1717. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Park DJ, Sung PS, Kim JH, Lee GW, Jang JW,
Jung ES, Bae SH, Choi JY and Yoon SK: EpCAM-high liver cancer stem
cells resist natural killer cell-mediated cytotoxicity by
upregulating CEACAM1. J Immunother Cancer. 8:e0003012020.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Shousha HI, Fouad R, Elbaz TM, Sabry D,
Mahmoud Nabeel M, Hosni Abdelmaksoud A, Mahmoud Elsharkawy A,
Soliman ZA, Habib G, Abdelaziz AO, et al: Predictors of recurrence
and survival of hepatocellular carcinoma: A prospective study
including transient elastography and cancer stem cell markers. Arab
J Gastroenterol. 21:95–101. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Luo J, Wang P, Wang R, Wang J, Liu M,
Xiong S, Li Y and Cheng B: The Notch pathway promotes the cancer
stem cell characteristics of CD90+ cells in hepatocellular
carcinoma. Oncotarget. 7:9526–9538. 2016.
|
|
69
|
Liu R, Shen Y, Nan K, Mi B, Wu T, Guo J,
Li M, Lv Y and Guo H: Association between expression of cancer stem
cell markers and poor differentiation of hepatocellular carcinoma:
A Meta-Analysis (PRISMA). Medicine (Baltimore). 94:e13062015.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Takano M, Shimada K, Fujii T, Morita K,
Takeda M, Nakajima Y, Nonomura A, Konishi N and Obayashi C: Keratin
19 as a key molecule in progression of human hepatocellular
carcinomas through invasion and angiogenesis. BMC Cancer.
16:9032016. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
You H, Ding W and Rountree CB: Epigenetic
regulation of cancer stem cell marker CD133 by transforming growth
factor-beta. Hepatology. 51:1635–1644. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Yan M, Li H, Zhu M, Zhao F, Zhang L, Chen
T, Jiang G, Xie H, Cui Y, Yao M, et al: G protein-coupled receptor
87 (GPR87) promotes the growth and metastasis of CD133+ cancer
stem-like cells in hepatocellular carcinoma. PLoS One.
8:e610562013. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Zhong C, Wu JD, Fang MM and Pu LY:
Clinicopathological significance and prognostic value of the
expression of the cancer stem cell marker CD133 in hepatocellular
carcinoma: A meta-analysis. Tumour Biol. 36:7623–7630. 2015.
View Article : Google Scholar : PubMed/NCBI
|