Introduction
Biomarker profiling is a major component of
oncological research that enables the integration of molecular
patterns with the clinicopathological status of patients. Reverse
transcription-quantitative PCR (RT-qPCR), a valuable technique to
profile differential expression, helps understand the molecular
patterns in a disease condition, enabling biomarker development. An
accurate and economical technique, it is commonly used in the field
of molecular oncology. Based on the technology that provides
real-time quantifiable levels of biomarkers, it is extremely
important to use appropriate controls as the baseline for inferring
the results in RT-qPCR. Reference genes served as internal controls
to normalize the quantification cycle (Cq) in RT-qPCR, enabling the
accurate assessment of biomarker levels in the analyte. Hence, it
is extremely important to identify appropriate and reliable
reference genes (1,2). In addition to demonstrating minimum
variability under various physiological and disease conditions, a
reliable reference gene must be unaffected by experimental
conditions. The genes commonly used as reference genes are
housekeeping genes that are required for the normal functioning of
cells and are hence expected to be the least variable across
tissues/conditions. Given the possible heterogeneity in the
expression profiles of housekeeping genes, their utilization in a
particular cell/tissue type, disease condition and organism must be
evaluated. The Minimum Information for publication of Quantitative
Real-Time PCR Experiments guidelines, a set of guidelines necessary
for evaluating RT-qPCR experiments, mandates the evaluation of
reference genes suitable for a particular study type (3).
Oral squamous cell carcinoma (OSCC), with a
worldwide incidence of 389,846 individuals, is the second most
common cancer in the Indian sub-continent (4,5). Given
the challenges in staging the disease at diagnosis and improving
survival rates, biomarkers are a significant adjunct for early
diagnosis, cancer progression, relapse and prognosis, with RT-qPCR
being the most commonly used method for biomarker profiling
(6–9). Lymph node metastasis (LNM) is a
critical prognostic factor in OSCC and reduces survival by 50%
(10). Studies on the diagnosis,
intraoperative detection and inhibition of LNM routinely employ
RT-qPCR to document expression patterns, with housekeeping genes
serving as reference genes (11,12).
Previous studies on lymph nodes using RT-qPCR have mostly been
conducted in mouse models of non-cancerous conditions. In most
studies, normalization was performed using a single reference gene.
Although the utilization of multiple reference genes can improve
the resolution, interpretation and accuracy of the results, studies
investigating the comparative accuracy of these reference genes for
the accurate assessment of the expression profile in lymph nodes
and/or stromal cells derived from patients are lacking.
The present study aimed to address this gap and
identify appropriate reference genes that can be applied for
biomarker profiling in lymph node stromal cells and tissues derived
from patients with oral cancer. Based on literature review, the
known reference genes, 18S ribosomal RNA (18SrRNA),
ribosomal Protein Lateral Stalk Subunit P0 (RPLP0),
ribosomal Protein L27 (RPL27), TATA-box binding protein
(TBP), hypoxanthine phosphoribosyl-transferase 1
(HPRT1), beta-actin (ACTB),
glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH) and
vimentin (VIM) were evaluated for their variability and
applicability as reference genes in lymph node cells/tissues from
patients with OSCC.
Materials and methods
Patient selection and cell lines for
evaluation
The present study was approved [approval no.
NHH/MEC-CL-EL-6-2016-403(A-1)] by the Narayana Health Medical
Ethics Committee (Bangalore, India). A total of 14 patients with
treatment-naive OSCC who underwent neck dissection and gave written
consent to participate in the present study, were included (August.
2017-September, 2023). The mean age of the patients was 50 years
(SD, 14.75) and 35% (5/14) were females. Patients <18 years of
age and diagnosed with HIV/HBV/HCV were excluded from the present
study. Lymph node tissue samples were collected under the
supervision of surgeons and pathologists to obtain accurate
specimens without affecting the patient's diagnosis. The surgical
lymph node specimens identified by the surgeon were then evaluated
by a pathologist to ensure the metastatic status of the specimen
and then split into portions for histopathological evaluation.
Primary cultures of lymph node stromal cells (LNSCs) from lymph
node tissues previously established [approved by the Narayana
Health Medical Ethics Committee; approval no.
NHH/MEC-CL-EL-6-2016-403(A-1)] in the laboratory were also used in
the present study. LNSCs (passage <15) were cultured in
high-glucose Dulbecco's Minimum Essential Medium (DMEM-HG; cat. no.
AL007A; HiMedia) to 80–90% confluency for extraction, whereas lymph
node tissues were stored in RNAlater solution (cat. no. AM7021;
Ambion; Thermo Fisher Scientific, Inc.) at −80°C until
extraction.
RNA isolation, cDNA conversion and
RT-qPCR
The cells and tissues were lysed and RNA was eluted
from the column according to the manufacturer's instructions (cat.
no. 740933; Machery-Nagel GmbH). RNA was assessed using a Nanodrop
to measure yield and purity (A260/A280 ratio
>1.8; RNA integrity by electrophoresis). For cDNA conversion,
1,000 ng of total RNA was converted using high-capacity cDNA
reverse transcription Kit (Applied Biosystems™; cat. no.
4374966; Thermo Fisher Scientific, Inc.) in a 40 µl reaction
mixture. All reagents were thawed and the reaction mixture was
prepared on ice. The reaction was setup as per the manufacturer's
protocol (Step 1: 25°C for 10 min, Step 2: 37°C for 120 min, Step
3: 85°C for 5 min and Step 4: hold at 4°C). RT-qPCR was performed
using Kapa SYBR Fast (cat. no. KK4601; Kapa Biosystems; Roche
Diagnostics) on a Roche Light Cycler 480 II Real-Time PCR machine.
Reactions were performed in triplicate for 45 cycles using primers
specific to each gene (Table
I).
 | Table I.Primer sequence and efficiency. |
Table I.
Primer sequence and efficiency.
| S. No. | Primer (Accession
No.) |
Forward/Reverse | Sequence | Product length | Amplification
factor | Efficiency (%) |
|---|
| 1 | ACTB
(NM_001101.5) | Forward |
TCAAGATCATTGCTCCTCCTG | 101 | 2.01 | 101 |
|
|
| Reverse |
CTGCTTGCTGATCCACATCTG |
|
|
|
| 2 | HPRT1
(NM_000194.3) | Forward |
ATGAACCAGGTTATGACCTTGAT | 298 | 2.05 | 104.55 |
|
|
| Reverse |
CCTGTTGACTGGTCATTACAATA |
|
|
|
| 3 | RPLP0
(NM_001002.4) | Forward |
CCATTCTATCATCAACGGGTACAA | 75 | 2.01 | 100.53 |
|
|
| Reverse |
TCAGCAAGTGGGAAGGTGTAATC |
|
|
|
| 4 | 18SrRNA
(NM_022551.3) | Forward |
GAGGATGAGGTGGAACGTGT | 199 | 2.02 | 101.55 |
|
|
| Reverse |
AGAAGTGACGCAGCCCTCTA |
|
|
|
| 5 | GAPDH
(NM_002046.7) | Forward |
TCGACAGTCAGCCGCATCTTCTTT | 104 | 1.93 | 93.07 |
|
|
| Reverse |
GCCCAATACGACCAAATCCGTTGA |
|
|
|
| 6 | RPL27
(NM_000988.5) | Forward |
ACAATCACCTAATGCCCACA | 146 | 1.95 | 95.27 |
|
|
| Reverse |
GCCTGTCTTGTATCTCTCTTCAA |
|
|
|
| 7 | VIM
(NM_003380.5) | Forward |
AGGCAAAGCAGGAGTCCACTGA | 100 | 2.11 | 110.68 |
|
|
| Reverse |
ATCTGGCGTTCCAGGGACTCAT |
|
|
|
| 8 | TBP
(NM_003194.5) | Forward |
CCACTCACAGACTCTCACAAC | 127 | 1.96 | 96 |
|
|
| Reverse |
CTGCGGTACAATCCCAGAACT |
|
|
|
Primer efficiency
The efficiency of the primers specific for ACTB,
RPL27 and HPRT1 was evaluated. cDNA samples were
serially diluted (1:5 dilution) and RT-qPCR was performed using
seven dilutions. Average quantification cycle (Cq) values from
triplicate experiments were plotted against log10
(concentration). The slope of the regression line was used to
determine the amplification factor and efficiency using an online
tool from Thermo Fisher Scientific (qPCR Efficiency Calculator |
Thermo Fisher Scientific; https://www.thermofisher.com/uk/en/home/brands/thermo-scientific/molecular-biology/molecular-biology-learning-center/molecular-biology-resource-library/thermo-scientific-web-tools/qpcr-efficiency-calculator.html).
The primer efficiencies for VIM, TBP, GAPDH, RPLP0 and
18SrRNA were previously established in the laboratory.
Statistical analysis
Reference genes were assessed for amplicon nature,
melting temperature (Tm), expression range (Cq values) and
stability. The mean Cq was determined from triplicate measurements
for each sample. The mean Cq values across LNSCs/lymph node tissues
are presented as mean ± standard deviation (SD). The graphs were
plotted using Tableau Professional Edition (2022.2.0; Salesforce,
Inc.) and Microsoft Excel (Microsoft Corporation).
Stability analysis of the reference
genes
For stability analysis, the Cq values for these
reference genes were evaluated using the Reffinder tool (http://blooge.cn/RefFinder/), which analyzed the data
using multiple normalization methods including geNorm, NormFinder,
BestKeeper and the comparative ∆Ct methods. A comprehensive ranking
of different reference gene candidates was obtained based on these
four methods (13,14).
Results
Details of the patients and the
LNSCs
Lymph node tissues (N=20) were collected from 14
patients with treatment-naïve OSCC after obtaining written informed
consent. The mean age of the patients was 50 years (SD: 14.75) and
35% (5/14) were females (Table
II). Most patients (64.28%) chewed tobacco, smoked and consumed
alcohol. The patients were mostly diagnosed with tongue (57.14%)
and buccal mucosa (28.57%) tumors, with 35.71% having T1-T2 stage
tumors and 64.28% having T3-T4 stage tumors. The patients were
further distributed based on the status of nodal metastasis; 57.14%
of patients were diagnosed with nodal metastasis (N+ stage).
Primary LNSCs (n=8) from five patients with OSCC were assessed in
the present study. The average age of the patients was 57 years
(SD=7.92), with four out of five patients having smoking/tobacco
chewing risk habits.
 | Table II.Patient's demographics for lymph node
tissues (N=14 patients). |
Table II.
Patient's demographics for lymph node
tissues (N=14 patients).
| S. No. | Patient code | Sample code | Age | Sex | Tumor site | Pathologic T
stage | Risk habits |
|---|
| 1 | P1 | S1 | 67 | Male | Alveolus | T4bN2b | Tobacco chewing,
alcohol |
| 2 | P2 | S2 | 39 | Male | Tongue | T3N1 | Tobacco
chewing |
| 3 |
| S3 | 39 | Male | Tongue | T3N1 | Tobacco
chewing |
| 4 | P3 | S4 | 47 | Female | Tongue | T1N3b | No habits |
| 5 |
| S5 | 47 | Female | Tongue | T1N3b | No habits |
| 6 | P4 | S6 | 75 | Male | Tongue | T3N3b | Smoking, tobacco
chewing |
| 7 |
| S7 | 75 | Male | Tongue | T3N3b | Smoking, tobacco
chewing |
| 8 | P5 | S8 | 30 | Male | Retro molar
trigone | T4bN3b | Smoking |
| 9 |
| S9 | 30 | Male | Retro molar
trigone | T4bN3b | Smoking |
| 10 | P6 | S10 | 51 | Male | Tongue | T1N0 | Smoking, tobacco
chewing |
| 11 | P7 | S11 | 39 | Male | Tongue | T2N0 | Tobacco
chewing |
| 12 | P8 | S12 | 54 | Male | Tongue | T1N0 | No habits |
| 13 | P9 | S13 | 61 | Male | Buccal Mucosa | T3N0 | Smoking, tobacco
chewing |
| 14 | P10 | S14 | 48 | Female | Buccal Mucosa | T3N3b | Tobacco
chewing |
| 15 |
| S15 | 48 | Female | Buccal Mucosa | T3N3b | Tobacco
chewing |
| 16 | P11 | S16 | 50 | Female | Buccal Mucosa | T4aN3b | Tobacco
chewing |
| 17 | P12 | S17 | 21 | Female | Tongue | T3N0 | No habits |
| 18 | P13 | S18 | 72 | Female | Tongue | T3N3b | No habits |
| 19 | P14 | S19 | 46 | Male | Buccal Mucosa | T1N0 | No habits |
| 20 |
| S20 | 46 | Male | Buccal Mucosa | T1N0 | No habits |
Evaluation of expression levels
(quantification cycle) and specificity (melting curve)
Assessment of primer efficiency (Table I) indicated that the primers had
efficiencies that ranged from 93.07 to 110.68% with an
amplification factor of 1.93–2.11.
Reference genes were assessed based on their
expression levels (Cq values) and amplicon nature (melting curves)
across different cohorts. Comparison of the Cq values indicated
that lymph node tissues had lower expression of GAPDH, TBP
and HPRT1 (Cq range: ~28-33, Fig. 1A) and higher expression of
18SrRNA, RPLP0, ACTB, RPL27 and VIM (Cq range:
~18.5–26, Fig. 1B). Similarly,
LNSCs revealed lower expression of GAPDH, TBP, RPL27 and
HPRT1 (Cq range: ~24.5–32, Fig.
1C) and higher expression of 18SrRNA, RPLP0, ACTB and
VIM (Cq range: ~17.4–24.5, Fig.
1D). The Cq values, when plotted between the metastatic (N+)
and non-metastatic (N0) patient groups for each primer,
demonstrated no significant differences between the cohorts
(Fig. 1) for lymph node tissues and
LNSCs. Furthermore, evaluation of the melting curves of the
amplicons across all the samples indicated that the amplified
products of VIM demonstrated multiple peaks, indicating more
than one product (Fig. S1).
VIM was excluded from further analysis.
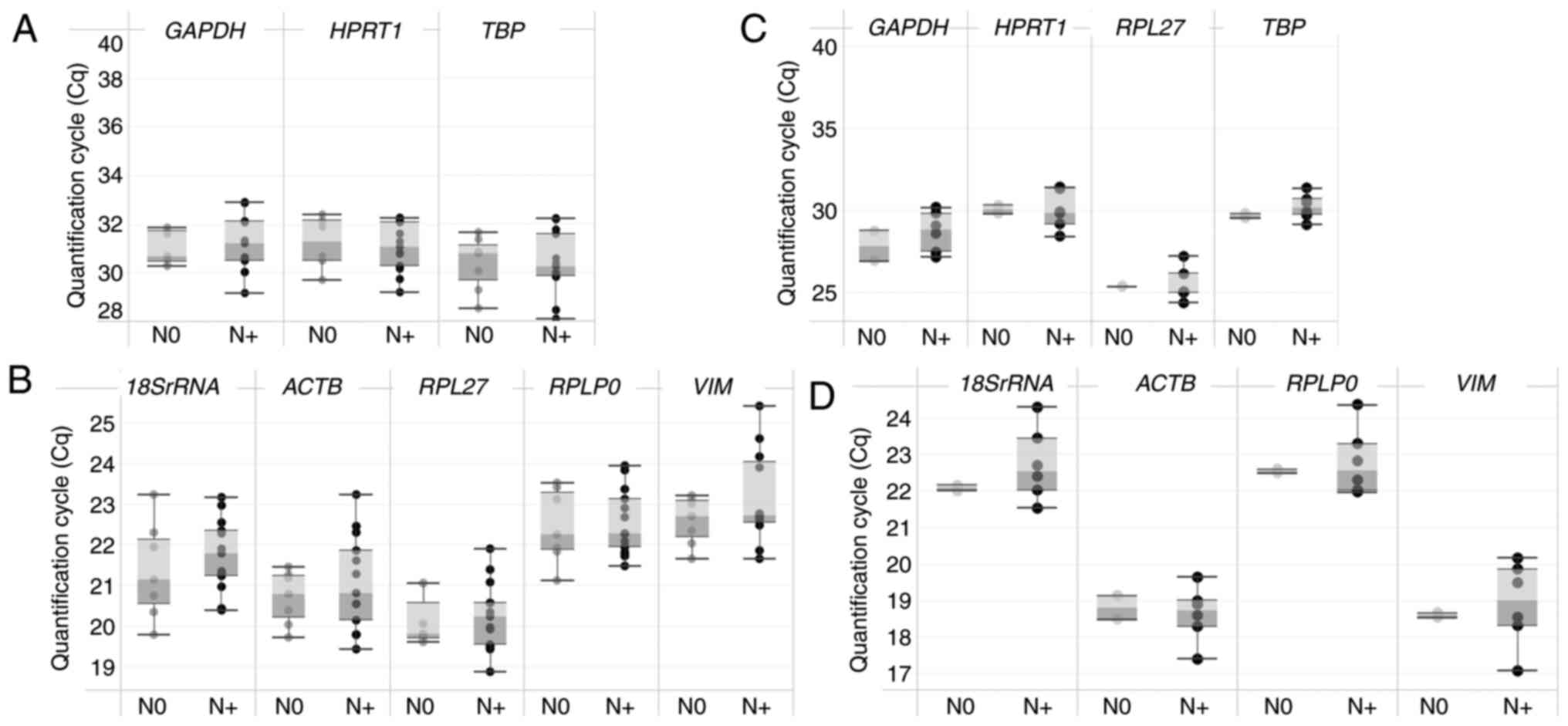 | Figure 1.Distribution of Cq values in lymph
node tissues and LNSCs. (A) A box and whisker plot depicting the Cq
distribution in lymph node tissues demonstrated comparatively lower
expression for GAPDH, TBP and HPRT1. (B) Higher
expression for RPL27, ACTB, 18SrRNA, RPLP0 and VIM.
(C) The scatter plot for Cq values for the reference gene with
LNSCs revealed varied distribution. (D) 18SrRNA, ACTB, RPLP0
& VIM demonstrated Cq ranging between 17–24. The box
represents the interquartile range and the whiskers represent the
minimum and maximum Cq values. LNSCs, lymph node stromal cells;
TBP, TATA-box binding protein; HPRT1, hypoxanthine
phosphoribosyl-transferase 1; RPL27, ribosomal protein L27; ACTB,
beta-actin; 18SrRNA, 18S ribosomal RNA; RPLP0, ribosomal protein
lateral stalk subunit P0; VIM, vimentin; N+, metastatic; N0,
non-metastatic. |
Stability analysis of reference genes
with lymph node tissues
The expression analysis of the genes with the lymph
node tissues revealed that the highest expression was observed in
18SrRNA, RPLP0, ACTB and RPL27 with Cq ranging
between 18.5 to 24 (Fig. 1A and B,
Table SI). The stability of these
genes across the 20 samples was further evaluated using Reffinder
(Fig. 2A-E, Table SII). The Comparative ∆Ct, geNorm
and NormFinder methods identified RPLP0, HPRT1, RPL27 and
18SrRNA as the most stable genes across the lymph nodes
irrespective of their metastatic status. All four methods
identified TBP and GAPDH as the least stable genes
(Table SII). A comprehensive
ranking combining all methods identified RPLP0, HPRT1, RPL27
and 18SrRNA as the most stable genes for RT-qPCR profiling
in lymph node tissues (Fig.
2E).
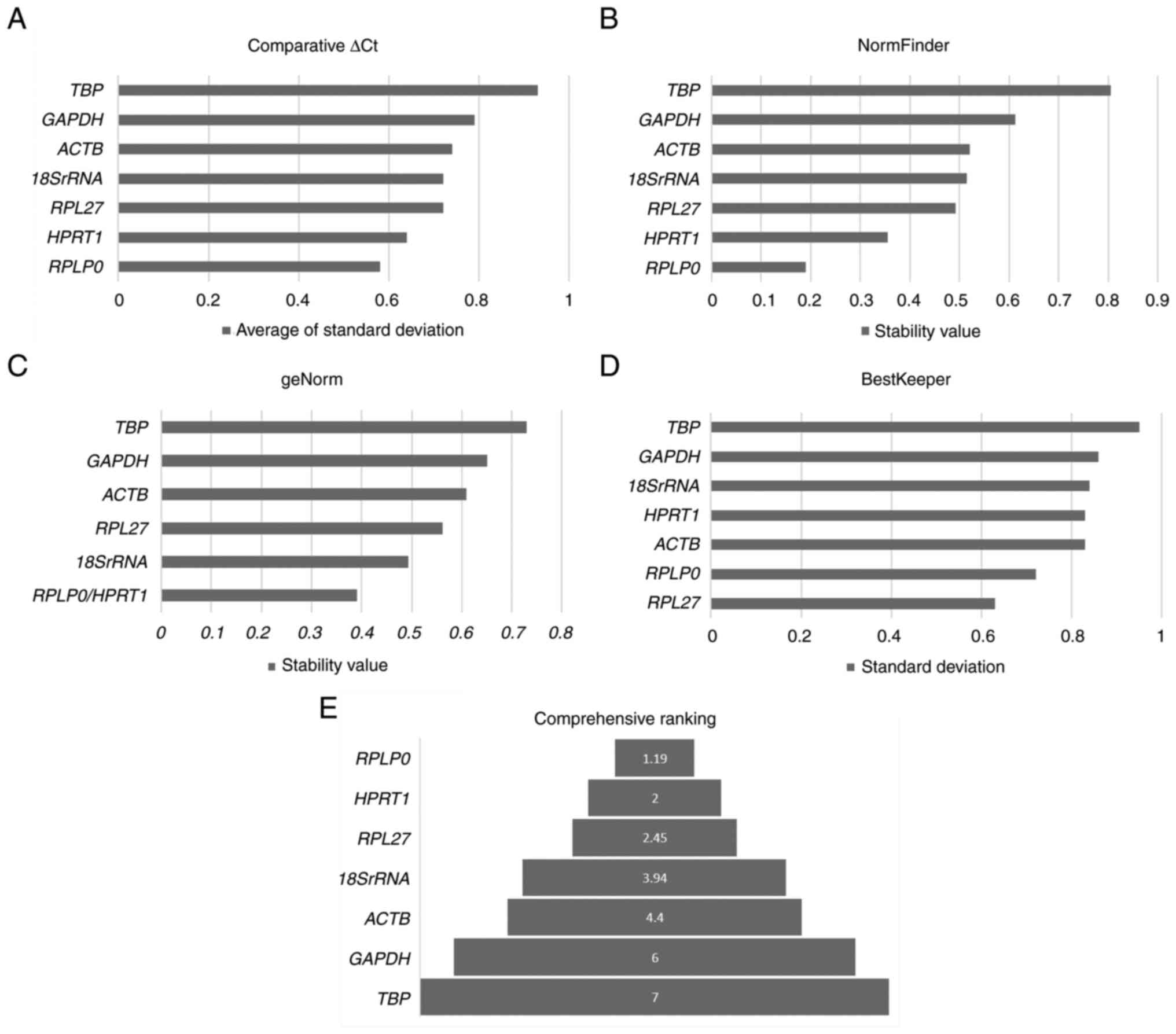 | Figure 2.Ranking for reference genes for lymph
node tissues. (A-D) The analysis of reference gene stability with
four different methods, namely (A) comparative ∆Ct, (B) NormFinder,
(C) geNorm and (D) BestKeeper are demonstrated. The stability
analysis with comparative ∆Ct, geNorm and NormFinder identified
RPLP0 and HPRT1 as the most stable genes except
BestKeeper which identified RPL27 and RPLP0 as the
most stable. (E) The comprehensive ranking analysis identifies
RPLP0, HPRT1, RPL27 and 18SrRNA as the most stable
genes, appropriate to be used as reference genes. RPLP0, ribosomal
protein lateral stalk subunit P0; HPRT1, hypoxanthine
phosphoribosyl-transferase 1; RPL27, ribosomal protein L27;
18SrRNA, 18S ribosomal RNA; ACTB, beta-actin. |
Evaluation of reference genes
stability with LNSCs
The RT-qPCR analysis with the lymph node cells
revealed that ACTB had the highest expression with Cq
ranging between 17 to 20 followed by RPLP0 and
18SrRNA (Cq ranging between ~21.5 to 24), whereas Cq values
of GAPDH, TBP, RPL27 and HPRT1 ranged between 24 to
30 (Fig. 1C and D, Table SIII).
The Cq values were further analyzed for expression
stability using Reffinder (Fig.
3A-E, Table SIV). The
comparative ∆Ct, NormFinder and geNorm methods identified ACTB,
18SrRNA and RPLP0 as the most stable genes and
GAPDH and RPL27 as the least stable genes. BestKeeper
identified ACTB, TBP and RPLP0 as the most stable
genes (Table SIV). Comprehensive
ranking using Reffinder identified 18SrRNA, RPLP0, ACTB and
TBP as the four most stable genes for LNSC expression
(Fig. 3E).
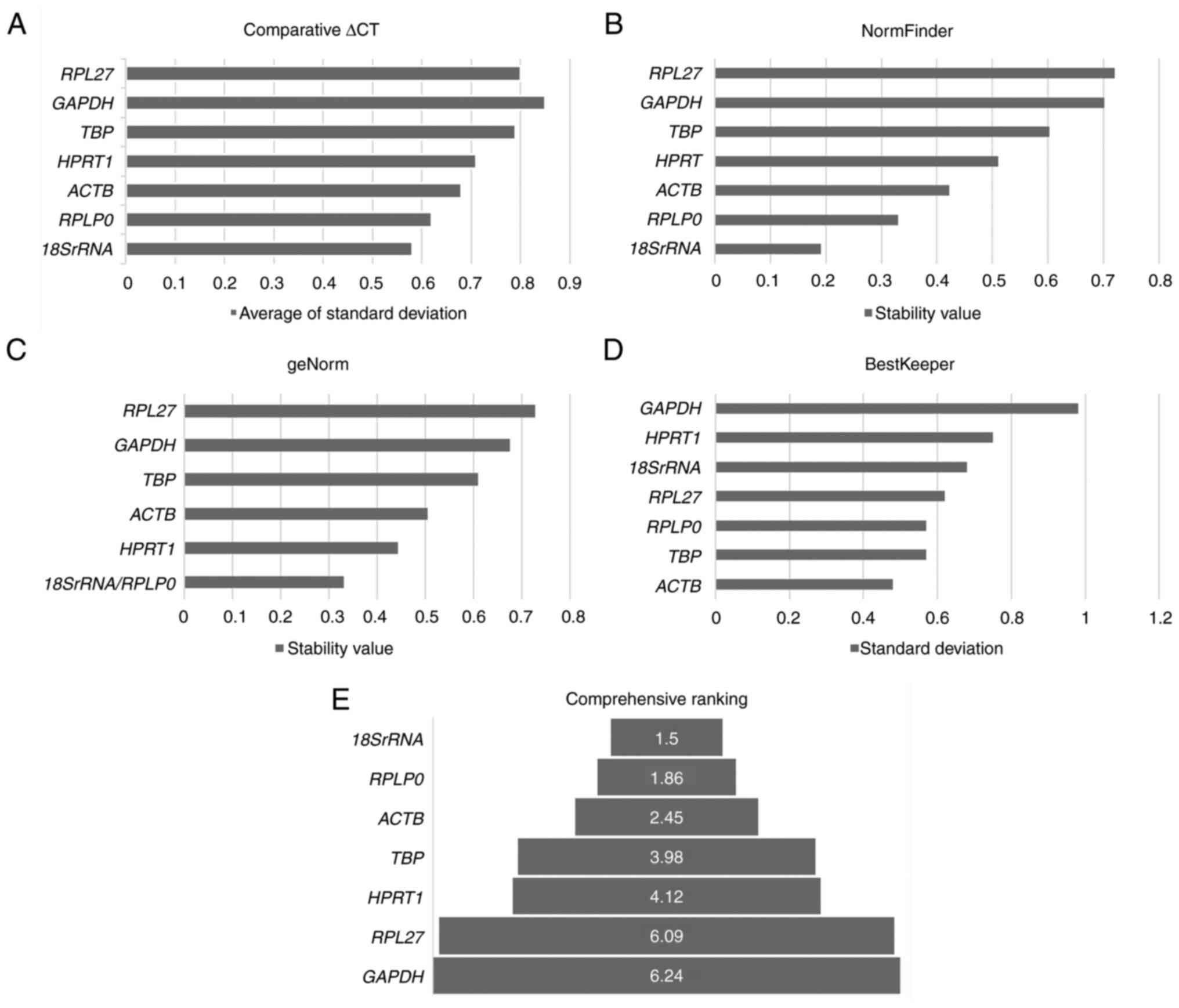 | Figure 3.Ranking for reference genes for
LNSCs. The analysis was carried out with eight LNSCs developed
in-house. (A-E) The comprehensive analysis with (A) comparative
∆Ct, (B) NormFinder, (C) geNorm, and (D) BestKeeper methods ranked
(E) 18SrRNA, RPLP0, ACTB and TBP as the most stable
genes that can serve as appropriate reference genes for LNSCs.
LNSCs, lymph node stromal cells; 18SrRNA, 18S ribosomal RNA; RPLP0,
ribosomal protein lateral stalk subunit P0; ACTB, beta-actin; TBP,
TATA-box binding protein; HPRT1, hypoxanthine
phosphoribosyl-transferase 1; RPL27, ribosomal protein L27; TBP,
TATA-box binding protein. |
Discussion
Carcinogenesis is a complex process involving
various pathways composed of distinct molecular markers.
Delineating these complex pathways, identifying markers that
specify these processes and are clinically relevant is challenging.
Oral cancer, with high incidence (3,89,846) and mortality (1,88,438) worldwide (4), has added heterogeneity owing to the
site, mode of metastasis and differential response to treatment.
The incidence and mortality of cancers of lip and oral cavity rank
16 and 15th respectively among the top 32 cancers worldwide. LNM,
the most common pattern of metastasis, notably affects the
prognosis of oral OSCC, reducing five-year survival rates to 30–59%
(15–17). LNSCs, which form the major component
of lymph nodes, play a crucial role in tumor-stromal interactions
(18–24). Furthermore, metastasis, a process
central to the prognostic outcomes in patients, is extremely
complex with multiple cellular (tumor cells, stromal cells,
extra-cellular vesicles) and molecular players including
ncRNAs/miRNAs (25–28). A comprehensive mechanistic and an
accurate understanding of the underlying molecular patterns and
representative biomarkers is crucial. Molecular profiling employing
lymph node tissues and cells has been the focus of numerous
studies. Expression profiling at the transcript level is a major
strategy, and RT-qPCR is an easy, accurate and quantitative tool
for targeted expression profiling. Adequate normalization using
reference genes is the key to obtaining reliable RT-qPCR results.
In the present study, eight commonly used reference genes were
evaluated and selected from the literature for their relevance as
reference genes in the RT-qPCR-based profiling of lymph node
tissues and cultured LNSCs.
Housekeeping genes are usually the chosen subset of
reference genes; however, multiple studies have identified
inaccurate usage of some of these reference genes, further
emphasizing the need for the validation of reference genes in
specific tissues, cells, diseases and experimental conditions
(2,29–31).
Given the cellular and tissue-level heterogeneity in samples,
established guidelines recommend the validation of genes within the
context of the tissue type and experimental design. In the present
study, it was revealed that in lymph node tissues RPLP0, HPRT1,
RPL27 and 18SrRNA were the stable genes. These genes
remained stable in lymph node tissues, regardless of metastatic or
non-metastatic nodes, and their expression was not affected by
presence or absence of tumor cells. Multiple studies have utilized
RT-qPCR profiling to study various aspects of lymph node
organogenesis in mouse tissues; however, normalization was
performed using one housekeeping gene (GAPDH, ACTB, TBP)
(32–36). Studies on human lymph node tissues
have been comparatively fewer. HPRT1 and TBP have
been identified as reliable reference genes for molecular profiling
in metastatic and non-metastatic pelvic lymph node tissues from
patients with prostate cancer (37).
In the present study, reference genes were assessed
in two cohorts of patient-derived LNSCs and lymph node tissues.
LNSCs comprised multiple cell types, including fibroblastic
reticular cells and double-negative cells (20,38).
Cells derived from both metastatic/non-metastatic nodes were
evaluated; 18SrRNA, RPLP0, ACTB and TBP were the most
stable genes for expression profiling in nodal stromal cells. A
study on LNSCs of patients with rheumatoid arthritis revealed that
the metabolic landscape, including genes involved in glucose, fatty
acid and glutamine metabolism, was altered in patients with
established disease, as well as in high-risk individuals.
Normalization of the target genes was performed using the geometric
mean of RPLP0 and POLR2G as reference genes (39). There are few RT-qPCR studies of
LNSCs from patients with cancer (40); however, an evaluation of the best
combination of reference genes has not been performed. Given the
cellular heterogeneity and the effect of tumor cell cues,
identification of appropriate reference genes is crucial, and to
the best of the authors' knowledge, the present study is the first
study reporting the same in patient-derived LNSCs.
In the present study, gene stability revealed a
differential pattern in the two cohorts, re-emphasizing the
inherent heterogeneity and need for context-dependent reference
gene evaluation. The two common genes that were stable in cultured
LNSCs as well as in lymph node tissues were 18SrRNA and the
ribosomal protein RPLP0, which corroborates with other
studies wherein the ribosomal family of proteins has been efficient
as reference genes (41–45). In tissues, RPLP0, HPRT1,
RPL27 and 18SrRNA were the most stable genes; however,
the results further indicated differences in expression levels.
While HPRT1 demonstrated a low expression (31±0.95),
RPLP0 (22.5±0.8), 18SrRNA (21.6±0.97) and
RPL27 (20.2±0.75), had a higher expression level indicating
that they are more suitable as reference genes. The cultured LNSCs
further revealed that in addition to 18SrRNA (22.5±0.83) and
RPLP0 (22.7±0.73), ACTB (18.6±0.62) and TBP
(30.0±0.67) were the stable genes; TBP being unsuitable
considering the low level of expression. Thus, the present study
indicated that, in addition to stability, the expression levels of
the chosen reference should also be considered during
selection.
Among these genes, HPRT1 and TBP
demonstrated an inverse pattern of stability between stromal cells
and nodal tissue. This may be due to inherent differences in cell
types, considering that lymph node tissues are composed of multiple
types of LNSCs, lymphocytes, macrophages, dendritic cells and
endothelial cells. The use of HPRT1 as a candidate reference
gene has provided contradictory evidence. Studies on meningioma and
melanoma have determined HPRT1 as the most stable
housekeeping gene under experimental conditions (46,47).
However, another study identified HPRT1 unsuitable as a
reference gene for cancer-related studies and proposed its
discontinuation, especially in the context of tumor-normal tissue
comparisons (30). A review of
HPRT1 further described its changing role from being a
regulator of nucleotide synthesis in normal cells to becoming an
accessory in cancer cells by helping them bypass nucleotide
synthesis (48). In the present
study, although HPRT1 was stably expressed in the lymph node
tissues of patients with OSCC, its stability was poor in LNSCs.
Furthermore, its low expression in tissues precludes its
candidature as a reference gene. Similar to HPRT1, the
evidence for TBP, another commonly used reference gene,
contradicts studies on its stability and utility as a reference
gene in various cancer cell lines (49–51).
In the present study, although TBP expression was stable in LNSCs,
it revealed lower expression range that differed from that of other
stable genes. Hence, the decision to use HPRT1 or TBP
as reference genes should be carefully evaluated, depending on the
experimental design and specific cancer type under study.
Furthermore, the present study provides strong evidence towards the
need to evaluate the reference gene while comparing parent tissue
and derived primary cells.
GAPDH was the least stable gene in both the
lymph node tissues and cultured LNSCs. GAPDH is the most
popular and accepted reference standard for RT-qPCR assays
(52–55). However, multiple studies have
questioned the use of GAPDH as a reference gene because of
its involvement in cell proliferation, migration and glycolysis,
and studies have also reported that the gene is oncogenic (31,56–59).
The present study corroborated the finding that GAPDH is
unsuitable for the normalization of LNSC and lymph node tissues
from patients with OSCC.
Biomarkers for the detection and prediction of nodal
metastasis as well as toward delineating the underlying mechanisms
of metastatic progression continue to be challenging (25,28,60–62).
The present study identified and validated a panel of reference
genes that could be used in lymph node stromal cells and tissues.
The need for tissue/cell/context-dependent selection of reference
genes for RT-qPCR-based expression profiling was further emphasized
by the identification of a different panel of genes for the lymph
node tissue and node tissue-derived cells. The panel of genes
recommended in the present study will be invaluable towards
acquiring accurate expression data from lymph nodes and LNSCs of
patients with oral cancer.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Mazumdar Shaw Medical Foundation (Bangalore, India)
provided the laboratory facilities for the study.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study are included
in the figures and tables of this article.
Authors' contributions
BLJ, AS and MAK conceptualized the present study.
BLJ, SNZ, NBS, VBR, YD, VS, VP, AS and MAK developed methodology.
BLJ performed formal analysis. BLJ and AS interpreted the data. BLJ
and AS wrote the original draft. BLJ, AS and MAK wrote, reviewed
and edited the manuscript. BLJ and AS confirm the authenticity of
all the raw data in the study. All the authors read and approved
the final version of the manuscript.
Ethics approval and consent to
participate
The present study was approved [approval no. NHH/
MEC-CL-EL-6-2016-403(A-1)] by the Narayana Health Medical Ethics
Committee (Bangalore, India). Samples were collected after
obtaining written informed consent from the patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Huggett J, Dheda K, Bustin S and Zumla A:
Real-time RT-PCR normalisation; strategies and considerations.
Genes Immun. 6:279–284. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kozera B and Rapacz M: Reference genes in
real-time PCR. J Appl Genet. 54:391–406. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bustin SA, Benes V, Garson JA, Hellemans
J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL,
et al: The MIQE guidelines: Minimum information for publication of
quantitative real-time PCR experiments. Clin Chem. 55:611–622.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ferlay J, Ervik M, Lam F, Laversanne M,
Colombet M, Mery L, Piñeros M, Znaor A, Soerjomataram I and Bray F:
Global Cancer Observatory: Cancer Today. 2022 India fact sheet.
International Agency for Research on Cancer; Lyon, France:
https://gco.iarc.who.int/media/globocan/factsheets/populations/356-india-fact-sheet.pdfMay
23–2024
|
|
6
|
Sajan T, Murthy S, Krishnankutty R and
Mitra J: A rapid, early detection of oral squamous cell carcinoma:
Real time PCR based detection of tetranectin. Mol Biol Res Commun.
8:33–40. 2019.PubMed/NCBI
|
|
7
|
Ueda S, Goto M, Hashimoto K, Hasegawa S,
Imazawa M, Takahashi M, Oh-Iwa I, Shimozato K, Nagao T and Nomoto
S: Salivary CCL20 level as a biomarker for oral squamous cell
carcinoma. Cancer Genomics Proteomics. 18:103–112. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lan T, Ge Q, Zheng K, Huang L, Yan Y,
Zheng L, Lu Y and Zheng D: FAT1 Upregulates in Oral squamous cell
carcinoma and promotes cell proliferation via cell cycle and DNA
repair. Front Oncol. 12:8700552022. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhou Q, Liu S, Kou Y, Yang P, Liu H,
Hasegawa T, Su R, Zhu G and Li M: ATP promotes oral squamous cell
carcinoma cell invasion and migration by activating the PI3K/AKT
pathway via the P2Y2-Src-EGFR axis. ACS Omega. 7:39760–39771. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bugshan A and Farooq I: Oral squamous cell
carcinoma: Metastasis, potentially associated malignant disorders,
etiology and recent advancements in diagnosis. F1000Res. 9:2292020.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yin P, Su Y, Chen S, Wen J, Gao F, Wu Y
and Zhang X: MMP-9 knockdown inhibits oral squamous cell carcinoma
lymph node metastasis in the nude mouse tongue-xenografted model
through the RhoC/Src pathway. Anal Cell Pathol (Amst).
2021:66833912021.PubMed/NCBI
|
|
12
|
Wei Y, Cheng X, Deng L, Dong H, Wei H, Xie
C, Tuo Y, Li G, Yu D and Cao Y: Expression signature and molecular
basis of CDH11 in OSCC detected by a combination of multiple
methods. BMC Med Genomics. 16:702023. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xie F, Wang J and Zhang B: RefFinder: A
web-based tool for comprehensively analyzing and identifying
reference genes. Funct Integr Genomics. 23:1252023. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xie F, Xiao P, Chen D, Xu L and Zhang B:
miRDeepFinder: A miRNA analysis tool for deep sequencing of plant
small RNAs. Plant Mol Biol. Jan 31–2012.(Epub ahead of print).
View Article : Google Scholar
|
|
15
|
Gil Z, Carlson DL, Boyle JO, Kraus DH,
Shah JP, Shaha AR, Singh B, Wong RJ and Patel SG: Lymph node
density is a significant predictor of outcome in patients with oral
cancer. Cancer. 115:5700–5710. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Thavarool SB, Muttath G, Nayanar S,
Duraisamy K, Bhat P, Shringarpure K, Nayak P, Tripathy JP, Thaddeus
A, Philip S and B S: Improved survival among oral cancer patients:
Findings from a retrospective study at a tertiary care cancer
centre in rural Kerala, India. World J Surg Oncol. 17:152019.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Geum DH, Roh YC, Yoon SY, Kim HG, Lee JH,
Song JM, Lee JY, Hwang DS, Kim YD, Shin SH, et al: The impact
factors on 5-year survival rate in patients operated with oral
cancer. J Korean Assoc Oral Maxillofac Surg. 39:207–216. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Acton SE, Farrugia AJ, Astarita JL,
Mourão-Sá D, Jenkins RP, Nye E, Hooper S, van Blijswijk J, Rogers
NC, Snelgrove KJ, et al: Dendritic cells control fibroblastic
reticular network tension and lymph node expansion. Nature.
514:498–502. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li L, Wu J, Abdi R, Jewell CM and Bromberg
JS: Lymph node fibroblastic reticular cells steer immune responses.
Trends Immunol. 42:723–734. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Malhotra D, Fletcher AL, Astarita J,
Lukacs-Kornek V, Tayalia P, Gonzalez SF, Elpek KG, Chang SK,
Knoblich K, Hemler ME, et al: Transcriptional profiling of stroma
from inflamed and resting lymph nodes defines immunological
hallmarks. Nat Immunol. 13:499–510. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Martinez VG, Pankova V, Krasny L, Singh T,
Makris S, White IJ, Benjamin AC, Dertschnig S, Horsnell HL,
Kriston-Vizi J, et al: Fibroblastic reticular cells control conduit
matrix deposition during lymph node expansion. Cell Rep.
29:2810–2822.e5. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Riedel A, Helal M, Pedro L, Swietlik JJ,
Shorthouse D, Schmitz W, Haas L, Young T, da Costa ASH, Davidson S,
et al: Tumor-derived lactic acid modulates activation and metabolic
status of draining lymph node stroma. Cancer Immunol Res.
10:482–497. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Riedel A, Shorthouse D, Haas L, Hall BA
and Shields J: Tumor-induced stromal reprogramming drives lymph
node transformation. Nat Immun. 17:1118–1127. 2016. View Article : Google Scholar
|
|
24
|
Valencia J, Jiménez E, Martinez VG, Del
Amo BG, Hidalgo L, Entrena A, Fernández-Sevilla LM, Del Río F,
Varas A, Vicente Á and Sacedón R: Characterization of human
fibroblastic reticular cells as potential immunotherapeutic tools.
Cytotherapy. 19:640–653. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu F and Li S: Non-coding RNAs in skin
cancers:Biological roles and molecular mechanisms. Front Pharmacol.
13:9343962022. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu Q and Li S: Exosomal circRNAs: Novel
biomarkers and therapeutic targets for urinary tumors. Cancer Lett.
588:2167592024. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Guo X, Gao C, Yang DH and Li S: Exosomal
circular RNAs: A chief culprit in cancer chemotherapy resistance.
Drug Resist Updat. 67:1009372023. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li S, Kang Y and Zeng Y: Targeting tumor
and bone microenvironment: Novel therapeutic opportunities for
castration-resistant prostate cancer patients with bone metastasis.
Biochim Biophys Acta Rev Cancer. 1879:1890332024. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Dheda K, Huggett JF, Chang JS, Kim LU,
Bustin SA, Johnson MA, Rook GA and Zumla A: The implications of
using an inappropriate reference gene for real-time reverse
transcription PCR data normalization. Anal Biochem. 344:141–143.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Townsend MH, Felsted AM, Ence ZE, Piccolo
SR, Robison RA and O'Neill KL: Falling from grace: HPRT is not
suitable as an endogenous control for cancer-related studies. Mol
Cell Oncol. 6:15756912019.PubMed/NCBI
|
|
31
|
Wang J, Yu X, Cao X, Tan L, Jia B, Chen R
and Li J: GAPDH: A common housekeeping gene with an oncogenic role
in pan-cancer. Comput Struct Biotechnol J. 21:4056–4069. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Camara A, Cordeiro OG, Alloush F, Sponsel
J, Chypre M, Onder L, Asano K, Tanaka M, Yagita H, Ludewig B, et
al: Lymph node mesenchymal and endothelial stromal cells cooperate
via the RANK-RANKL cytokine axis to shape the sinusoidal macrophage
niche. Immunity. 50:1467–1481.e6. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Onder L, Mörbe U, Pikor N, Novkovic M,
Cheng HW, Hehlgans T, Pfeffer K, Becher B, Waisman A, Rülicke T, et
al: Lymphatic endothelial cells control initiation of lymph node
organogenesis. Immunity. 47:80–92.e4. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gu Y, Liu Y, Fu L, Zhai L, Zhu J, Han Y,
Jiang Y, Zhang Y, Zhang P, Jiang Z, et al: Tumor-educated B cells
selectively promote breast cancer lymph node metastasis by
HSPA4-targeting IgG. Nat Med. 25:312–322. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jiang L, Yilmaz M, Uehara M, Cavazzoni CB,
Kasinath V, Zhao J, Naini SM, Li X, Banouni N, Fiorina P, et al:
Characterization of leptin receptor+ stromal cells in
lymph node. Front Immunol. 12:7304382022. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kwok T, Medovich SC, Silva-Junior IA,
Brown EM, Haug JC, Barrios MR, Morris KA and Lancaster JN:
Age-associated changes to lymph node fibroblastic reticular cells.
Front Aging. 3:8389432022. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tsaur I, Renninger M, Hennenlotter J,
Oppermann E, Munz M, Kuehs U, Stenzl A and Schilling D: Reliable
housekeeping gene combination for quantitative PCR of lymph nodes
in patients with prostate cancer. Anticancer Res. 33:5243–5248.
2013.PubMed/NCBI
|
|
38
|
Alvarenga HG and Marti L: Multifunctional
roles of reticular fibroblastic cells: More than meets the eye? J
Immunol Res. 2014:4020382014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
de Jong TA, Semmelink JF, Denis SW, Bolt
JW, Maas M, van de Sande MGH, Houtkooper RHL and van Baarsen LGM:
Lower metabolic potential and impaired metabolic flexibility in
human lymph node stromal cells from patients with rheumatoid
arthritis. Cells. 12:12022. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liang L, Zhang X, Su X, Zeng T, Suo D, Yun
J, Wang X, Guan XY and Li Y: Fibroblasts in metastatic lymph nodes
confer cisplatin resistance to ESCC tumor cells via PI16.
Oncogenesis. 12:502023. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Pérez-Gómez JM, Porcel-Pastrana F, De La
Luz-Borrero M, Montero-Hidalgo AJ, Gómez-Gómez E, Herrera-Martínez
AD, Guzmán-Ruiz R, Malagón MM, Gahete MD and Luque RM: LRP10, PGK1
and RPLP0: Best reference genes in periprostatic adipose tissue
under obesity and prostate cancer conditions. Int J Mol Sci.
24:151402023. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Asiabi P, Ambroise J, Giachini C, Coccia
ME, Bearzatto B, Chiti MC, Dolmans MM and Amorim CA: Assessing and
validating housekeeping genes in normal, cancerous, and polycystic
human ovaries. J Assist Reprod Genet. 37:2545–2553. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Liu LL, Zhao H, Ma TF, Ge F, Chen CS and
Zhang YP: Identification of valid reference genes for the
normalization of RT-qPCR expression studies in human breast cancer
cell lines treated with and without transient transfection. PLoS
One. 10:e01170582015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Geigges M, Gubser PM, Unterstab G,
Lecoultre Y, Paro R and Hess C: Reference genes for expression
studies in human CD8+ naïve and effector memory t cells
under resting and activating conditions. Sci Rep. 10:94112020.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Panagodimou E, Koika V, Markatos F,
Kaponis A, Adonakis G, Georgopoulos NA and Markantes GK: Expression
stability of ACTB, 18S, and GAPDH in human placental tissues from
subjects with PCOS and controls: GAPDH expression is increased in
PCOS. Hormones (Athens). 21:329–333. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Freitag D, Koch A, Lawson McLean A, Kalff
R and Walter J: Validation of reference genes for expression
studies in human meningiomas under different experimental settings.
Mol Neurobiol. 55:5787–5797. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Janik ME, Szwed S, Grzmil P, Kaczmarek R,
Czerwiński M and Hoja-Łukowicz D: RT-qPCR analysis of human
melanoma progression-related genes-A novel workflow for selection
and validation of candidate reference genes. Int J Biochem Cell
Biol. 101:12–28. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Townsend MH, Robison RA and O'Neill KL: A
review of HPRT and its emerging role in cancer. Med Oncol.
35:892018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Gorji-Bahri G, Moradtabrizi N and Hashemi
A: Uncovering the stability status of the reputed reference genes
in breast and hepatic cancer cell lines. PLoS One. 16:e02596692021.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Irie N, Warita K, Tashiro J, Zhou Y,
Ishikawa T, Oltvai ZN and Warita T: Expression of housekeeping
genes varies depending on mevalonate pathway inhibition in cancer
cells. Heliyon. 9:e180172023. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Ge WL, Shi GD, Huang XM, Zong QQ, Chen Q,
Meng LD, Miao Y, Zhang JJ and Jiang KR: Optimization of internal
reference genes for qPCR in human pancreatic cancer research.
Transl Cancer Res. 9:2962–2971. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zainuddin A, Chua KH, Abdul Rahim N and
Makpol S: Effect of experimental treatment on GAPDH mRNA expression
as a housekeeping gene in human diploid fibroblasts. BMC Mol Biol.
11:592010. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Thiel CS, Huge A, Hauschild S, Tauber S,
Lauber BA, Polzer J, Paulsen K, Lier H, Engelmann F, Schmitz B, et
al: Stability of gene expression in human T cells in different
gravity environments is clustered in chromosomal region 11p15.4.
NPJ Microgravity. 3:222017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Panina Y, Germond A, Masui S and Watanabe
TM: Validation of common housekeeping genes as reference for qPCR
gene expression analysis during iPS reprogramming process. Sci Rep.
8:87162018. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Molina CE, Jacquet E, Ponien P,
Muñoz-Guijosa C, Baczkó I, Maier LS, Donzeau-Gouge P, Dobrev D,
Fischmeister R and Garnier A: Identification of optimal reference
genes for transcriptomic analyses in normal and diseased human
heart. Cardiovasc Res. 114:247–258. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Liu K, Tang Z, Huang A, Chen P, Liu P,
Yang J, Lu W, Liao J, Sun Y, Wen S, et al:
Glyceraldehyde-3-phosphate dehydrogenase promotes cancer growth and
metastasis through upregulation of SNAIL expression. Int J Oncol.
50:252–262. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Ramos D, Pellín-Carcelén A, Agustí J,
Murgui A, Jordá E, Pellín A and Monteagudo C: Deregulation of
glyceraldehyde-3-phosphate dehydrogenase expression during tumor
progression of human cutaneous melanoma. Anticancer Res.
35:439–444. 2015.PubMed/NCBI
|
|
58
|
Shen C, Li W and Wang Y: Research on the
oncogenic role of the house-keeping gene GAPDH in human tumors.
Transl Cancer Res. 12:525–535. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Ouyang X, Zhu R, Lin L, Wang X, Zhuang Q
and Hu D: GAPDH is a novel ferroptosis-related marker and
correlates with immune microenvironment in lung adenocarcinoma.
Metabolites. 13:1422023. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Cao LM, Zhong NN, Li ZZ, Huo FY, Xiao Y,
Liu B and Bu LL: Lymph node metastasis in oral squamous cell
carcinoma: Where we are and where we are going. Clin Transl Disc.
3:e2272023. View Article : Google Scholar
|
|
61
|
Tsai TY, Iandelli A, Marchi F, Huang Y,
Tai SF, Hung SY, Kao HK and Chang KP: The prognostic value of lymph
node burden in oral cavity cancer: Systematic review and
meta-analysis. Laryngoscope. 132:88–95. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Ji H, Hu C, Yang X, Liu Y, Ji G, Ge S,
Wang X and Wang M: Lymph node metastasis in cancer progression:
Molecular mechanisms, clinical significance and therapeutic
interventions. Signal Transduct Target Ther. 8:3672023. View Article : Google Scholar : PubMed/NCBI
|

















