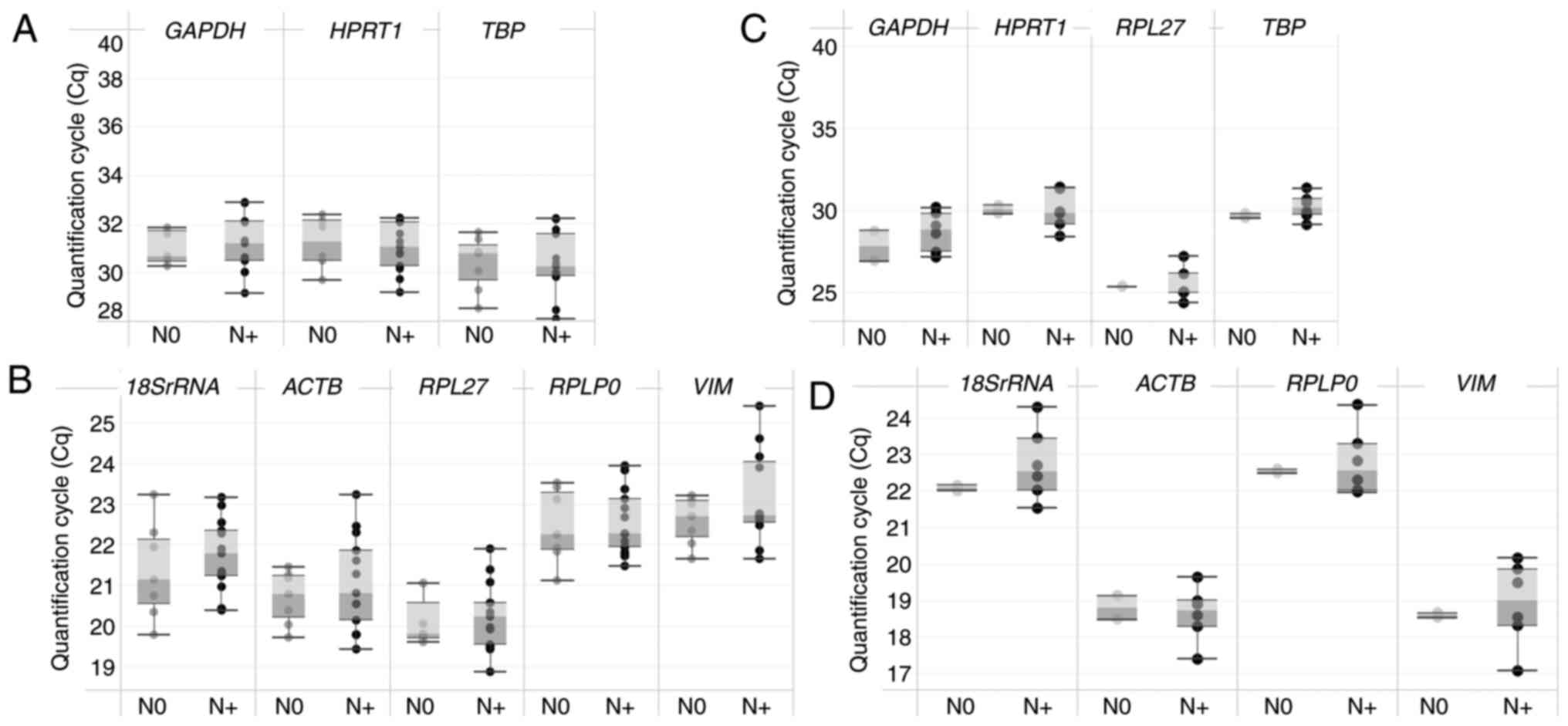
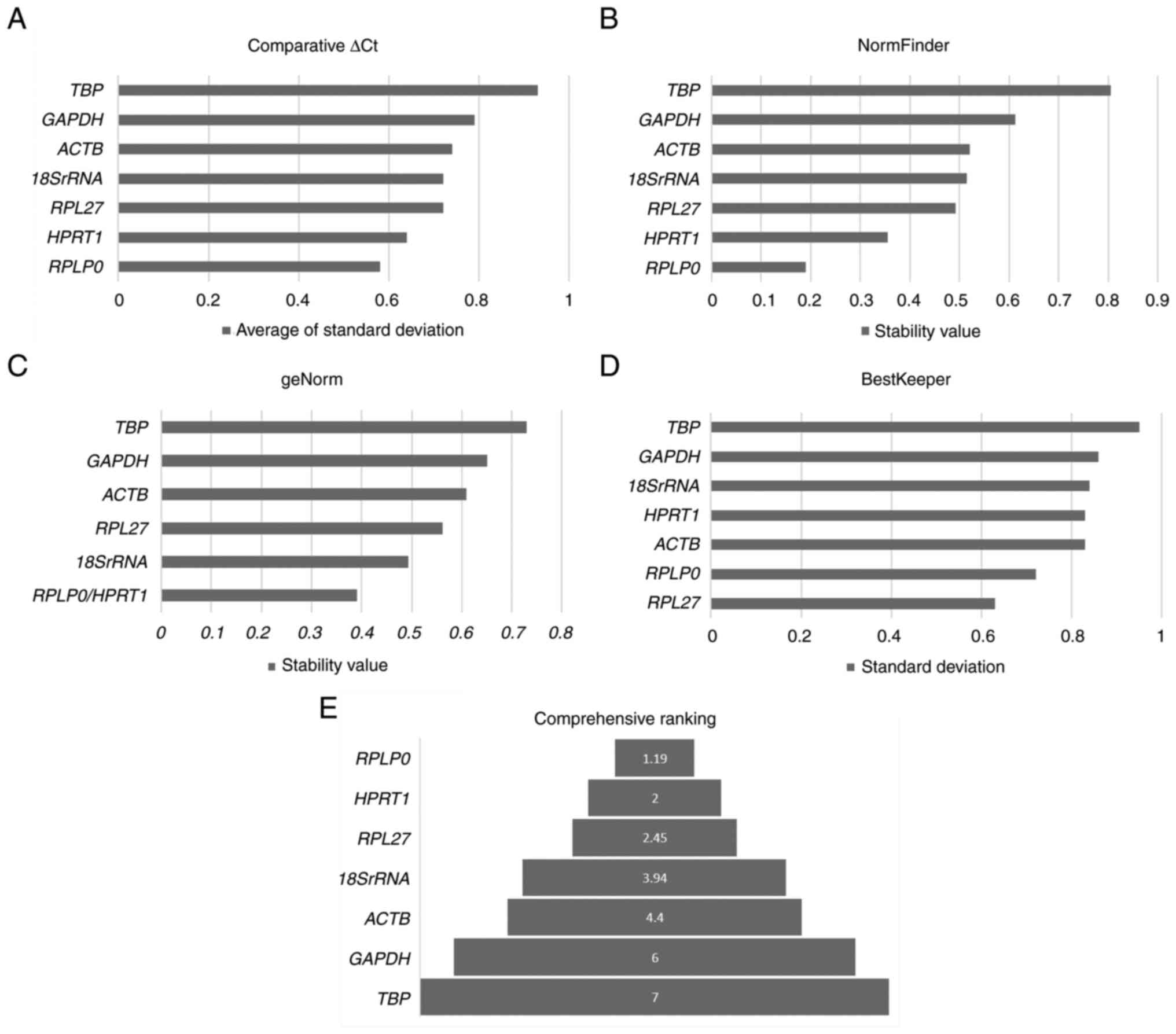
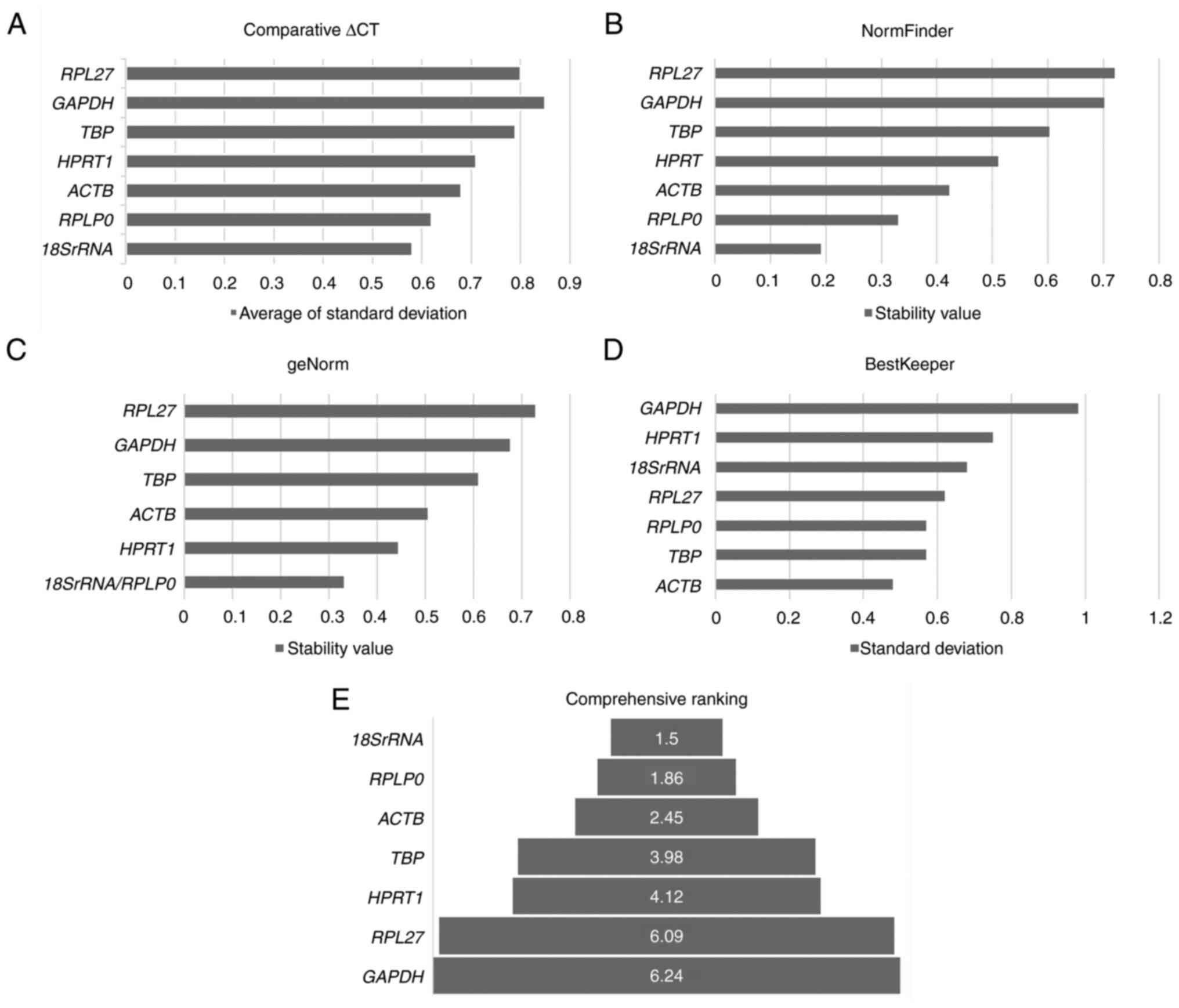
|
1
|
Huggett J, Dheda K, Bustin S and Zumla A:
Real-time RT-PCR normalisation; strategies and considerations.
Genes Immun. 6:279–284. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kozera B and Rapacz M: Reference genes in
real-time PCR. J Appl Genet. 54:391–406. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bustin SA, Benes V, Garson JA, Hellemans
J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL,
et al: The MIQE guidelines: Minimum information for publication of
quantitative real-time PCR experiments. Clin Chem. 55:611–622.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ferlay J, Ervik M, Lam F, Laversanne M,
Colombet M, Mery L, Piñeros M, Znaor A, Soerjomataram I and Bray F:
Global Cancer Observatory: Cancer Today. 2022 India fact sheet.
International Agency for Research on Cancer; Lyon, France:
https://gco.iarc.who.int/media/globocan/factsheets/populations/356-india-fact-sheet.pdfMay
23–2024
|
|
6
|
Sajan T, Murthy S, Krishnankutty R and
Mitra J: A rapid, early detection of oral squamous cell carcinoma:
Real time PCR based detection of tetranectin. Mol Biol Res Commun.
8:33–40. 2019.PubMed/NCBI
|
|
7
|
Ueda S, Goto M, Hashimoto K, Hasegawa S,
Imazawa M, Takahashi M, Oh-Iwa I, Shimozato K, Nagao T and Nomoto
S: Salivary CCL20 level as a biomarker for oral squamous cell
carcinoma. Cancer Genomics Proteomics. 18:103–112. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lan T, Ge Q, Zheng K, Huang L, Yan Y,
Zheng L, Lu Y and Zheng D: FAT1 Upregulates in Oral squamous cell
carcinoma and promotes cell proliferation via cell cycle and DNA
repair. Front Oncol. 12:8700552022. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhou Q, Liu S, Kou Y, Yang P, Liu H,
Hasegawa T, Su R, Zhu G and Li M: ATP promotes oral squamous cell
carcinoma cell invasion and migration by activating the PI3K/AKT
pathway via the P2Y2-Src-EGFR axis. ACS Omega. 7:39760–39771. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bugshan A and Farooq I: Oral squamous cell
carcinoma: Metastasis, potentially associated malignant disorders,
etiology and recent advancements in diagnosis. F1000Res. 9:2292020.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yin P, Su Y, Chen S, Wen J, Gao F, Wu Y
and Zhang X: MMP-9 knockdown inhibits oral squamous cell carcinoma
lymph node metastasis in the nude mouse tongue-xenografted model
through the RhoC/Src pathway. Anal Cell Pathol (Amst).
2021:66833912021.PubMed/NCBI
|
|
12
|
Wei Y, Cheng X, Deng L, Dong H, Wei H, Xie
C, Tuo Y, Li G, Yu D and Cao Y: Expression signature and molecular
basis of CDH11 in OSCC detected by a combination of multiple
methods. BMC Med Genomics. 16:702023. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xie F, Wang J and Zhang B: RefFinder: A
web-based tool for comprehensively analyzing and identifying
reference genes. Funct Integr Genomics. 23:1252023. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xie F, Xiao P, Chen D, Xu L and Zhang B:
miRDeepFinder: A miRNA analysis tool for deep sequencing of plant
small RNAs. Plant Mol Biol. Jan 31–2012.(Epub ahead of print).
View Article : Google Scholar
|
|
15
|
Gil Z, Carlson DL, Boyle JO, Kraus DH,
Shah JP, Shaha AR, Singh B, Wong RJ and Patel SG: Lymph node
density is a significant predictor of outcome in patients with oral
cancer. Cancer. 115:5700–5710. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Thavarool SB, Muttath G, Nayanar S,
Duraisamy K, Bhat P, Shringarpure K, Nayak P, Tripathy JP, Thaddeus
A, Philip S and B S: Improved survival among oral cancer patients:
Findings from a retrospective study at a tertiary care cancer
centre in rural Kerala, India. World J Surg Oncol. 17:152019.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Geum DH, Roh YC, Yoon SY, Kim HG, Lee JH,
Song JM, Lee JY, Hwang DS, Kim YD, Shin SH, et al: The impact
factors on 5-year survival rate in patients operated with oral
cancer. J Korean Assoc Oral Maxillofac Surg. 39:207–216. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Acton SE, Farrugia AJ, Astarita JL,
Mourão-Sá D, Jenkins RP, Nye E, Hooper S, van Blijswijk J, Rogers
NC, Snelgrove KJ, et al: Dendritic cells control fibroblastic
reticular network tension and lymph node expansion. Nature.
514:498–502. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li L, Wu J, Abdi R, Jewell CM and Bromberg
JS: Lymph node fibroblastic reticular cells steer immune responses.
Trends Immunol. 42:723–734. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Malhotra D, Fletcher AL, Astarita J,
Lukacs-Kornek V, Tayalia P, Gonzalez SF, Elpek KG, Chang SK,
Knoblich K, Hemler ME, et al: Transcriptional profiling of stroma
from inflamed and resting lymph nodes defines immunological
hallmarks. Nat Immunol. 13:499–510. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Martinez VG, Pankova V, Krasny L, Singh T,
Makris S, White IJ, Benjamin AC, Dertschnig S, Horsnell HL,
Kriston-Vizi J, et al: Fibroblastic reticular cells control conduit
matrix deposition during lymph node expansion. Cell Rep.
29:2810–2822.e5. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Riedel A, Helal M, Pedro L, Swietlik JJ,
Shorthouse D, Schmitz W, Haas L, Young T, da Costa ASH, Davidson S,
et al: Tumor-derived lactic acid modulates activation and metabolic
status of draining lymph node stroma. Cancer Immunol Res.
10:482–497. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Riedel A, Shorthouse D, Haas L, Hall BA
and Shields J: Tumor-induced stromal reprogramming drives lymph
node transformation. Nat Immun. 17:1118–1127. 2016. View Article : Google Scholar
|
|
24
|
Valencia J, Jiménez E, Martinez VG, Del
Amo BG, Hidalgo L, Entrena A, Fernández-Sevilla LM, Del Río F,
Varas A, Vicente Á and Sacedón R: Characterization of human
fibroblastic reticular cells as potential immunotherapeutic tools.
Cytotherapy. 19:640–653. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu F and Li S: Non-coding RNAs in skin
cancers:Biological roles and molecular mechanisms. Front Pharmacol.
13:9343962022. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu Q and Li S: Exosomal circRNAs: Novel
biomarkers and therapeutic targets for urinary tumors. Cancer Lett.
588:2167592024. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Guo X, Gao C, Yang DH and Li S: Exosomal
circular RNAs: A chief culprit in cancer chemotherapy resistance.
Drug Resist Updat. 67:1009372023. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li S, Kang Y and Zeng Y: Targeting tumor
and bone microenvironment: Novel therapeutic opportunities for
castration-resistant prostate cancer patients with bone metastasis.
Biochim Biophys Acta Rev Cancer. 1879:1890332024. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Dheda K, Huggett JF, Chang JS, Kim LU,
Bustin SA, Johnson MA, Rook GA and Zumla A: The implications of
using an inappropriate reference gene for real-time reverse
transcription PCR data normalization. Anal Biochem. 344:141–143.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Townsend MH, Felsted AM, Ence ZE, Piccolo
SR, Robison RA and O'Neill KL: Falling from grace: HPRT is not
suitable as an endogenous control for cancer-related studies. Mol
Cell Oncol. 6:15756912019.PubMed/NCBI
|
|
31
|
Wang J, Yu X, Cao X, Tan L, Jia B, Chen R
and Li J: GAPDH: A common housekeeping gene with an oncogenic role
in pan-cancer. Comput Struct Biotechnol J. 21:4056–4069. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Camara A, Cordeiro OG, Alloush F, Sponsel
J, Chypre M, Onder L, Asano K, Tanaka M, Yagita H, Ludewig B, et
al: Lymph node mesenchymal and endothelial stromal cells cooperate
via the RANK-RANKL cytokine axis to shape the sinusoidal macrophage
niche. Immunity. 50:1467–1481.e6. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Onder L, Mörbe U, Pikor N, Novkovic M,
Cheng HW, Hehlgans T, Pfeffer K, Becher B, Waisman A, Rülicke T, et
al: Lymphatic endothelial cells control initiation of lymph node
organogenesis. Immunity. 47:80–92.e4. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gu Y, Liu Y, Fu L, Zhai L, Zhu J, Han Y,
Jiang Y, Zhang Y, Zhang P, Jiang Z, et al: Tumor-educated B cells
selectively promote breast cancer lymph node metastasis by
HSPA4-targeting IgG. Nat Med. 25:312–322. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jiang L, Yilmaz M, Uehara M, Cavazzoni CB,
Kasinath V, Zhao J, Naini SM, Li X, Banouni N, Fiorina P, et al:
Characterization of leptin receptor+ stromal cells in
lymph node. Front Immunol. 12:7304382022. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kwok T, Medovich SC, Silva-Junior IA,
Brown EM, Haug JC, Barrios MR, Morris KA and Lancaster JN:
Age-associated changes to lymph node fibroblastic reticular cells.
Front Aging. 3:8389432022. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tsaur I, Renninger M, Hennenlotter J,
Oppermann E, Munz M, Kuehs U, Stenzl A and Schilling D: Reliable
housekeeping gene combination for quantitative PCR of lymph nodes
in patients with prostate cancer. Anticancer Res. 33:5243–5248.
2013.PubMed/NCBI
|
|
38
|
Alvarenga HG and Marti L: Multifunctional
roles of reticular fibroblastic cells: More than meets the eye? J
Immunol Res. 2014:4020382014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
de Jong TA, Semmelink JF, Denis SW, Bolt
JW, Maas M, van de Sande MGH, Houtkooper RHL and van Baarsen LGM:
Lower metabolic potential and impaired metabolic flexibility in
human lymph node stromal cells from patients with rheumatoid
arthritis. Cells. 12:12022. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liang L, Zhang X, Su X, Zeng T, Suo D, Yun
J, Wang X, Guan XY and Li Y: Fibroblasts in metastatic lymph nodes
confer cisplatin resistance to ESCC tumor cells via PI16.
Oncogenesis. 12:502023. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Pérez-Gómez JM, Porcel-Pastrana F, De La
Luz-Borrero M, Montero-Hidalgo AJ, Gómez-Gómez E, Herrera-Martínez
AD, Guzmán-Ruiz R, Malagón MM, Gahete MD and Luque RM: LRP10, PGK1
and RPLP0: Best reference genes in periprostatic adipose tissue
under obesity and prostate cancer conditions. Int J Mol Sci.
24:151402023. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Asiabi P, Ambroise J, Giachini C, Coccia
ME, Bearzatto B, Chiti MC, Dolmans MM and Amorim CA: Assessing and
validating housekeeping genes in normal, cancerous, and polycystic
human ovaries. J Assist Reprod Genet. 37:2545–2553. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Liu LL, Zhao H, Ma TF, Ge F, Chen CS and
Zhang YP: Identification of valid reference genes for the
normalization of RT-qPCR expression studies in human breast cancer
cell lines treated with and without transient transfection. PLoS
One. 10:e01170582015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Geigges M, Gubser PM, Unterstab G,
Lecoultre Y, Paro R and Hess C: Reference genes for expression
studies in human CD8+ naïve and effector memory t cells
under resting and activating conditions. Sci Rep. 10:94112020.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Panagodimou E, Koika V, Markatos F,
Kaponis A, Adonakis G, Georgopoulos NA and Markantes GK: Expression
stability of ACTB, 18S, and GAPDH in human placental tissues from
subjects with PCOS and controls: GAPDH expression is increased in
PCOS. Hormones (Athens). 21:329–333. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Freitag D, Koch A, Lawson McLean A, Kalff
R and Walter J: Validation of reference genes for expression
studies in human meningiomas under different experimental settings.
Mol Neurobiol. 55:5787–5797. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Janik ME, Szwed S, Grzmil P, Kaczmarek R,
Czerwiński M and Hoja-Łukowicz D: RT-qPCR analysis of human
melanoma progression-related genes-A novel workflow for selection
and validation of candidate reference genes. Int J Biochem Cell
Biol. 101:12–28. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Townsend MH, Robison RA and O'Neill KL: A
review of HPRT and its emerging role in cancer. Med Oncol.
35:892018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Gorji-Bahri G, Moradtabrizi N and Hashemi
A: Uncovering the stability status of the reputed reference genes
in breast and hepatic cancer cell lines. PLoS One. 16:e02596692021.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Irie N, Warita K, Tashiro J, Zhou Y,
Ishikawa T, Oltvai ZN and Warita T: Expression of housekeeping
genes varies depending on mevalonate pathway inhibition in cancer
cells. Heliyon. 9:e180172023. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Ge WL, Shi GD, Huang XM, Zong QQ, Chen Q,
Meng LD, Miao Y, Zhang JJ and Jiang KR: Optimization of internal
reference genes for qPCR in human pancreatic cancer research.
Transl Cancer Res. 9:2962–2971. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zainuddin A, Chua KH, Abdul Rahim N and
Makpol S: Effect of experimental treatment on GAPDH mRNA expression
as a housekeeping gene in human diploid fibroblasts. BMC Mol Biol.
11:592010. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Thiel CS, Huge A, Hauschild S, Tauber S,
Lauber BA, Polzer J, Paulsen K, Lier H, Engelmann F, Schmitz B, et
al: Stability of gene expression in human T cells in different
gravity environments is clustered in chromosomal region 11p15.4.
NPJ Microgravity. 3:222017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Panina Y, Germond A, Masui S and Watanabe
TM: Validation of common housekeeping genes as reference for qPCR
gene expression analysis during iPS reprogramming process. Sci Rep.
8:87162018. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Molina CE, Jacquet E, Ponien P,
Muñoz-Guijosa C, Baczkó I, Maier LS, Donzeau-Gouge P, Dobrev D,
Fischmeister R and Garnier A: Identification of optimal reference
genes for transcriptomic analyses in normal and diseased human
heart. Cardiovasc Res. 114:247–258. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Liu K, Tang Z, Huang A, Chen P, Liu P,
Yang J, Lu W, Liao J, Sun Y, Wen S, et al:
Glyceraldehyde-3-phosphate dehydrogenase promotes cancer growth and
metastasis through upregulation of SNAIL expression. Int J Oncol.
50:252–262. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Ramos D, Pellín-Carcelén A, Agustí J,
Murgui A, Jordá E, Pellín A and Monteagudo C: Deregulation of
glyceraldehyde-3-phosphate dehydrogenase expression during tumor
progression of human cutaneous melanoma. Anticancer Res.
35:439–444. 2015.PubMed/NCBI
|
|
58
|
Shen C, Li W and Wang Y: Research on the
oncogenic role of the house-keeping gene GAPDH in human tumors.
Transl Cancer Res. 12:525–535. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Ouyang X, Zhu R, Lin L, Wang X, Zhuang Q
and Hu D: GAPDH is a novel ferroptosis-related marker and
correlates with immune microenvironment in lung adenocarcinoma.
Metabolites. 13:1422023. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Cao LM, Zhong NN, Li ZZ, Huo FY, Xiao Y,
Liu B and Bu LL: Lymph node metastasis in oral squamous cell
carcinoma: Where we are and where we are going. Clin Transl Disc.
3:e2272023. View Article : Google Scholar
|
|
61
|
Tsai TY, Iandelli A, Marchi F, Huang Y,
Tai SF, Hung SY, Kao HK and Chang KP: The prognostic value of lymph
node burden in oral cavity cancer: Systematic review and
meta-analysis. Laryngoscope. 132:88–95. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Ji H, Hu C, Yang X, Liu Y, Ji G, Ge S,
Wang X and Wang M: Lymph node metastasis in cancer progression:
Molecular mechanisms, clinical significance and therapeutic
interventions. Signal Transduct Target Ther. 8:3672023. View Article : Google Scholar : PubMed/NCBI
|