Currently, pathological tissue biopsy is the gold
standard for diagnosing and monitoring a number of malignant
tumors, such as lung cancer, stomach cancer, colorectal cancer.
This method allows rapid assessment of the extent and nature of
lesions, provides relatively accurate pathological classification
and facilitates early detection and diagnosis. In addition, tissue
biopsies conducted during treatment can reflect disease progression
and treatment efficacy and provide clinicians with valuable
information to tailor treatment plans (1). However, use of insufficient or
inadequate tissue samples may lead to diagnostic bias, which may be
exacerbated by tumor heterogeneity (2). Furthermore, repeated invasive
procedures can cause discomfort for patients, particularly as the
disease advances.
The emergence of liquid biopsy has effectively
addressed several of these challenges. Liquid biopsy involves the
molecular analysis of liquid (non-tissue) samples to evaluate
physiological states (3). While
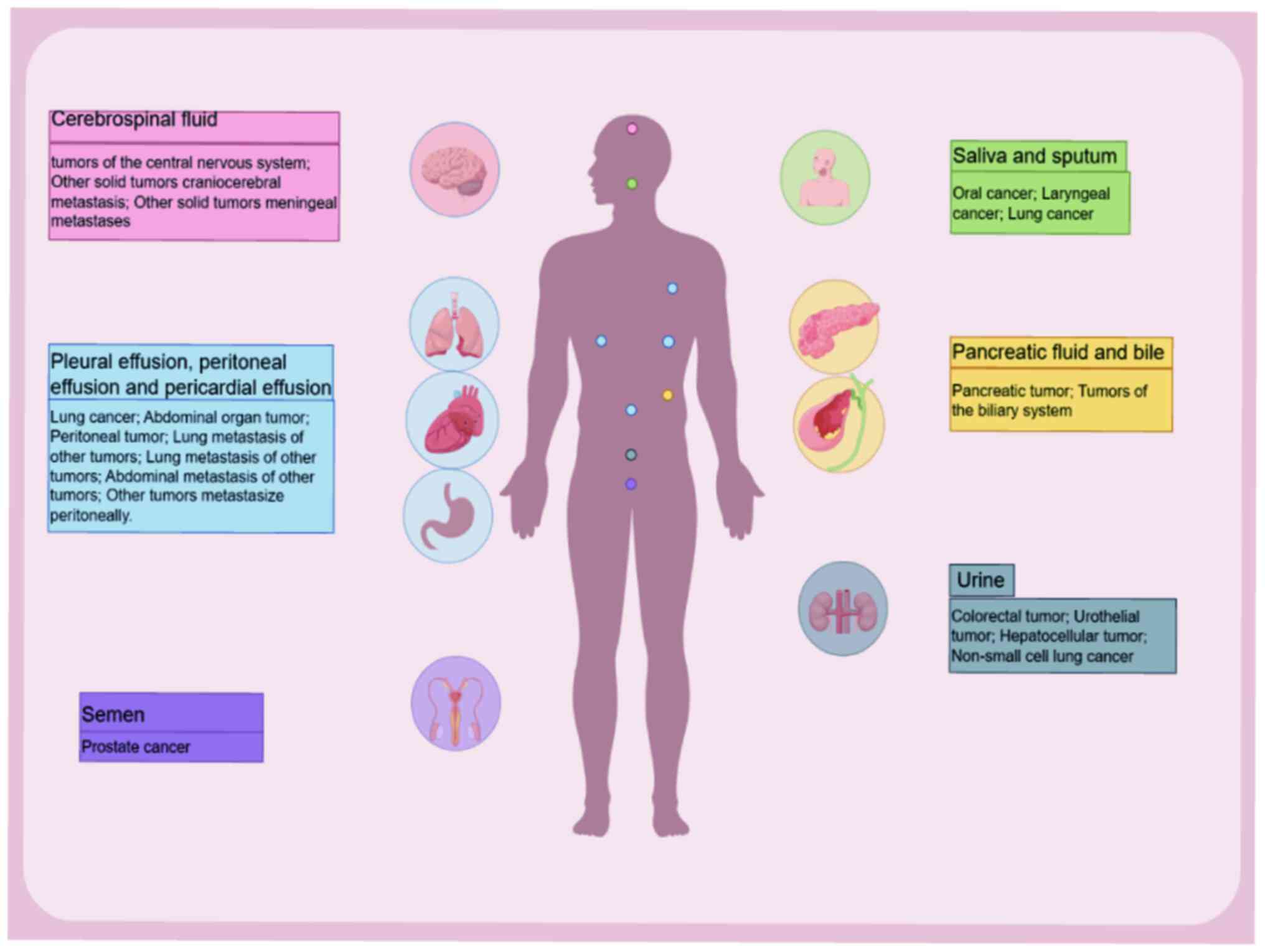
blood samples are the most commonly used, other bodily fluids such
as cerebrospinal fluid (CSF), saliva, pleural effusion, bile,
abdominal fluid and urine can also be utilized (4). Liquid biopsy primarily focuses on
analyzing circulating tumor cells (CTCs), circulating tumor DNA
(ctDNA), as well as circulating cell-free (cf)RNA, extracellular
vesicles and tumor-inducing platelets (5). ctDNA has emerged as a pivotal element
in clinical practice as it serves a key role in cancer diagnosis,
monitoring and treatment. In clinical treatment, physicians are
able manage treatment strategies by reference to ctDNA results. The
detection of ctDNA pre-treatment and pre- and post-operation guide
patient treatment plans (6).
However, there are limitations to ctDNA analysis in liquid biopsy,
including challenges in evaluating tumor pathology, detection
sensitivity and the absence of standardized protocols.
Circulating cfDNA refers to highly fragmented DNA
released from cells into the bloodstream that circulates freely in
human blood (7). First discovered
in 1948 (7), cfDNA has become a
major focus of medical research (8–10). It
is composed of both double- and single-stranded fragments (11–13),
typically 120–220 bp in length, with an average length of ~167 bp,
which is associated with the nucleosome (14).
In the bloodstream, cfDNA exists in three primary
forms: Free, bound to protein (such as nucleosomes and
lipoproteins) or associated with extracellular vesicles, such as
exosomes, apoptotic bodies and microvesicles (15,16).
The majority of plasma cfDNA is found in exosomes (17). The sources of cfDNA have been a
topic of debate and can be generally divided into two main
categories: Cellular destruction and active cellular release
(18,19). Potential sources of cfDNA include
cellular byproducts released during normal physiological processes,
exogenous DNA originating from dietary intake, blood transfusions
or infections, release from the nervous system, secretion into the
blood circulation due to factors such as stress, hereditary
conditions, degeneration or disease, fetal cellular material
transferred to the mother during pregnancy and systemic release due
to obesity and aging (19). Plasma
cfDNA from healthy individuals predominantly originates from white
blood cells (55%), red blood cell progenitors (30%) and vascular
endothelial (10%) and liver cells (1%) (19). In patients with cancer, however,
cfDNA is mainly derived from tumor tissue and surrounding cells
(18).
cfDNA is a key biomarker for various physiological
and pathological conditions and is associated with factors such as
aging (20) and physical or
psychological stress (21). In
addition, cfDNA serves as a key biomarker in several types of
cancer, including non-small cell lung cancer (NSCLC) (22), liver (23), breast (24,25),
pancreatic (26), oral (27) and colorectal cancer (28,29).
Previous studies suggest that cfDNA may also be associated with
xenotransplantation (30,31).
cfDNA encompasses short and long DNA fragments and
analysis of these fragments can provide valuable insight into a
health status. Increasing evidence shows that cfDNA is vital for
immune regulation, tumor-associated inflammation and the
maintenance of cell homeostasis (32,33).
cfDNA can impact cellular function and transformation, contribute
to tumor growth and metastasis and holds promise for early disease
detection (32,33). In healthy individuals, there is a
dynamic balance between production and clearance of cfDNA. cfDNA is
primarily cleared by the liver, spleen and kidney (34,35).
However, in patients with chronic inflammation or tumors, cfDNA
levels significantly increase due to impaired clearance and
subsequent accumulation (32).
ctDNA is a subset of cfDNA that consists of DNA
fragments that originate from tumor tissue and potentially other
sources, such as shedding cells from normal tissue, characterized
by genetic alterations that mirror those of the tumor (18,36).
The increase in cfDNA levels in patients with cancer is primarily
attributable to the elevated levels of ctDNA (37). Typically, ctDNA fragments are ~140
bp in length and have a half-life >2 h, which makes their
dynamics variable (38,39). The detection rate of ctDNA in plasma
ranging from 0.01% to a majority in the cfDNA, which reflects
notable heterogeneity among different types of tumor (40,41).
Despite this variability, the quantity of ctDNA detected is usually
high compared with that of CTCs (42). The ability to detect ctDNA is
influenced by characteristics of the tumor. For example, smaller
solid tumors or those with low metabolic activity may be more
challenging to detect, which may lead to false-negative results
(6). Notably, ctDNA levels decrease
rapidly following radical tumor resection if there is no or minimal
residual tumor (43).
Acquiring brain tissue for diagnostic purposes is
both challenging and high-risk due to the unique structure and
function of the brain. Therefore, CSF serves an irreplaceable role.
While magnetic resonance imaging (MRI) is commonly used to monitor
CNS diseases (49), its predictive
capability is limited. Liquid biopsy that involves the detection of
ctDNA in CSF may serve as a novel detection method for CNS
diseases. The unique composition of CSF, along with the protective
blood-brain barrier, decreases interference from cfDNA, which
results in increased concentration of ctDNA and mutated allele
frequencies (MAF) in CSF. This improves the sensitivity and
accuracy of ctDNA mutation detection. Consequently, CSF biopsy may
be a promising diagnostic tool for the detection and monitoring of
brain tumors and CNS metastases (50–52).
A study has shown that carcinoembryonic antigen and
CSF ctDNA are effective biomarkers for distinguishing patients with
and without brain parenchymal tumor or CNS metastases (53). CSF ctDNA analysis has demonstrated
distinct mutational profiles in patients with bone marrow
metastasis (53). In a 2022 case
report (54), a patient with lung
adenocarcinoma experienced neurological symptoms, including
headache, nausea, aphasia, limb restlessness and sudden blindness
during treatment. While initial MRI and CSF cytology did not
indicate the presence of brain metastasis, CSF ctDNA analysis
identified the same EGFR mutation as that detected in the
lung tumor of the patient. Follow-up MRI scans 9 months later
confirmed brain metastasis, which suggested earlier MRI and CSF
cytology results were false negatives. This case underscores the
potential of CSF ctDNA as an early diagnostic biomarker as it may
detect brain metastasis before cytological or MRI evidence
emerges.
To the best of our knowledge, studies of ctDNA in
CSF have predominantly focused on adults (55–58),
with relatively few investigating its application in children
(59–61). Pages et al (62) assessed tumor reliability by
analyzing ctDNA in peripheral blood, CSF and urine samples from
children with brain tumors; ctDNA detection in this demographic was
limited by low tumor fraction (TF). In numerous cases, the actual
TF was >1%, with TF >0.1% being undetectable. Furthermore,
only a small percentage of high-grade tumors were available.
Therefore, uncertainty persists regarding the application of liquid
biopsy for pediatric tumors, indicating the need for further
research.
The clinical relevance of saliva as a diagnostic
tool is uncertain. A study from 2019 (63) suggested that saliva may not serve as
an adequate alternative sample for quantitative cfDNA testing due
to insufficient cfDNA concentration for diagnosis of NSCLC.
However, it was proposed that saliva may be beneficial as a
complementary method to cytology. Wang et al (64) made notable advancements by
optimizing the extraction technique for sputum samples to overcome
limitations posed by large amounts of mucus components and the low
yield of cancer cells. Super-amplification refractory mutation
system was used to analyze EGFR mutation status in cfDNA
derived from sputum samples; sputum could be a promising sample
type for detecting EGFR mutations, although its use for
diagnosing lung adenocarcinoma may be limited. Further research by
Wang et al (65) highlighted
saliva from patients with lung adenocarcinoma as a valuable
alternative source for detecting the EGFR exon 20 p.T790M
mutation, which is linked to resistance to EGFR targeted therapy
(65). In addition, a previous
study investigating head and neck cancer reported a high
concordance (93%) in ctDNA detection between saliva and blood
samples, as well as efficacy of ctDNA in saliva in predicting
patient outcomes (66). A
meta-analysis of 64 cases of malignant salivary gland carcinoma
found increased levels of ctDNA and CTCs in malignant cases
(67). According to the 2024 Expert
Consensus, ctDNA extracted from saliva, along with serum or plasma,
provides meaningful insight into tumor genetics and dynamics
(68).
Tumor supernatant, such as pleural, peritoneal and
pericardial effusion, are in proximity to tumors and may provide
distinct advantages over blood for ctDNA detection; for example, it
is easier to detect ctDNA of abdominal tumor with abdominal fluid A
previous study that compared mutant allele scores from 30
supernatant samples with those from paired formalin-fixed
paraffin-embedded cell blocks reported a variant concordance up to
90% (69), and similar results were
detected in both supernatant and FFPE samples in 74% of cases. This
suggests that supernatant may serve as a viable alternative to
traditional tissue biopsy (69).
In pancreaticobiliary tract tumors, obtaining tissue
biopsies can be challenging due to the occult nature of the
disease. Given their direct contact with tumor tissue, bile and
pancreatic fluid are ideal samples for liquid biopsies. Kinugasa
et al (72) compared levels
of ctDNA in tumor tissue with those in bile from 49 patients with
gallbladder cancer; ctDNA isolation from bile was a valuable
approach for diagnosing gallbladder cancer. The sensitivity of
ctDNA testing (58.3%) was higher compared with that of cytology
(45.8%), and there was a high mutation concordance between the two
methods. Further study has demonstrated consistent KRAS
mutations in ctDNA from bile, plasma and formalin-fixed
paraffin-embedded samples from patients with bile duct tumors
(39). Notably, only 18.8% of
plasma ctDNA samples test positive for KRAS mutations,
compared with a detection rate of 48.0% in bile ctDNA (39). Moreover, patients with KRAS
mutations detected in bile ctDNA exhibit significantly lower
survival rates compared with those with wild-type KRAS
(39).
In early-stage pancreatic ductal adenocarcinoma,
identification of mutations in plasma ctDNA is often challenging
(73). A study in 2023 compared the
detection rates of ctDNA sourced from pancreatic fluid with that in
plasma; DNA concentrations and the ratios of Alu247/Alu115 were
higher in pancreatic fluid compared with plasma, however, there was
no difference in the mutation detection rate between pancreas and
plasma (74). This limitation may
be attributable to the small sample size and influence of enzymes
present in pancreatic fluid, which underscores the need for further
investigation.
A recent prospective multi-center study reported
that measurement of DNA methylation in urine effectively
differentiates pathological types of bladder cancer and predicts
180-day recurrence-free survival with 100% accuracy (79). This represents a breakthrough in use
of urine DNA methylation for differentiation of pathological cancer
types. In addition, a study on ctDNA in urine during neoadjuvant
chemotherapy for bladder cancer demonstrated that monitoring tumor
DNA dynamics in urine, supernatant and plasma predicts treatment
response and outcome (80). Urinary
ctDNA has also been shown to detect the recurrence of upper urinary
tract urothelial carcinoma up to 60 days earlier than cystoscopy
(81). Kim et al (82) reported that binding urinary ctDNA
improves detection rate of hepatocellular carcinoma from 62 to 92%
(82). Increased DNA methylation
levels in urine have also been reported in patients with NSCLC.
Urine sampling is a non-invasive procedure that can
be performed by non-professionals. Patients can conveniently
collect samples at home, which can be sent to testing laboratories.
Unlike blood, which is subject to buffering and regulatory
mechanisms that may alter its properties (83), urine is excreted and may better
reflect bodily abnormality. In addition, urine has a lower presence
of contaminating proteins compared with blood, which simplifies DNA
extraction processes (76). The
ability to test large volumes of urine repeatedly enhances
sensitivity of ctDNA detection (84), which makes it a promising tool in
cancer diagnostics.
To the best of our knowledge, prostate-specific
antigen is the only well-established tumor marker for prostate
cancer. However, its specificity is limited, necessitating
exploration of additional diagnostic tools. ctDNA testing has
emerged as a valuable adjunct in diagnosis of prostate cancer
(85–87). While ctDNA is commonly detected in
advanced cancer such as pancreatic, ovarian, colorectal, bladder,
gastroesophageal, breast, hepatocellular and head and neck cancer
and melanoma, it is present in <50% of cases of primary brain,
kidney, prostate and thyroid cancer (40).
Semen may serve as a potential sample for prostate
cancer diagnostics. Significant differences in cfDNA levels have
been observed (83) in semen
samples from patients with prostate cancer, individuals with benign
prostatic hyperplasia and healthy individuals (88). Patients with prostate cancer exhibit
higher concentrations of cfDNA than healthy individuals in semen
alongside a distinctive size distribution of cfDNA fragments.
Notably, longer cfDNA fragments are significantly more prevalent in
semen from patients with prostate cancer compared with those with
benign hyperplasia or healthy individuals (69). Size and concentration of cfDNA
fragments in semen is associated with tumor burden and treatment
response (88). As a direct source
of prostate disease-specific molecules, semen represents a
promising body fluid for identification of prostate cancer
biomarkers and may provide important insight for early diagnosis
(Fig. 1) (89).
ctDNA provides insight for cancer diagnosis and
treatment. To harness the potential of ctDNA, accurate detection
methods are key. The main techniques for ctDNA detection include
DNA sequencing, PCR-based methods and DNA-based hybridization
strategies (90–92). At present there is no universally
accepted standard of detection (93).
TAm-Seq is a labeled amplicon deep sequencing method
that integrates efficient library preparation with advanced
statistical analysis. This technique enables the sequencing of
~6,000 nucleotides and in-depth analysis (103). A notable implementation of TAm-Seq
is InVisionFirst® (Neogenomics Laboratories).
Liquid biopsy platform, which is designed to detect
both hotspot mutations and entire coding regions across 35
cancer-associated genes. Leveraging enhanced TAm-Seq techniques, it
identifies low-frequency mutations in ctDNA by amplifying highly
fragmented DNA (104). Further
improvements of Tm-Seq involves optimization of the amplification
process by splitting it into two steps. The initial step involves
limited cycle pre-amplification using all primer sets to capture
the starting molecules present in the template. This is followed by
a single amplification step to purify and isolate the target
sequence. This refined approach enables detection of cancer
mutations in ctDNA at allelic frequencies as low as 2% with
sensitivity and specificity of >97% (105). TAm-Seq method demonstrates its
utility in clinical settings by routinely detecting ctDNA not only
at the time of diagnosis, but also post-treatment (106).
Microdroplet ddPCR, also known as third-generation
PCR, uses sample allocation, restricted dilution and statistical
data processing based on Poisson distributions to accurately and
reliably quantify nucleic acids. This technique divides mixed
nucleic acid molecules and PCR solution into small droplets. By
using microfluidic loops and surfactant chemistry, sample DNA is
randomly assigned to isolated droplets, generating 20,000 droplets.
The template is amplified and product is detected based on specific
fluorescent labeling (97,118,119). ddPCR is well-suited for studying
specific single-gene hotspot mutations that may be found in CSF
samples, achieving a limit of detection as low as 0.01%/reaction
(119,120). It can measure mutations that
constitute 0.01% of a sample (39),
offering greater sensitivity compared with qPCR (97). However, compared with NGS, ddPCR has
narrower reference range (90.0% of operable mutations) and does not
cover certain variants, such as estrogen receptor 1 mutation
(121). In addition, ddPCR
requires specialized personnel to operate, which adds to overall
complexity and operational costs (39,118,122).
BEAMing is a digital PCR method that enhances the
capability of ddPCR by incorporating pre-amplifications of DNA
using conventional PCR and target-specific primers (97). PCR products amplified by BEAMing
molecules are linked to single magnetic beads and the mutation
sites extended via fluorescent probes or primers. By counting
fluorescently labelled beads, BEAMing allows the quantitative
detection of mutant alleles (119,123). Taniguchi et al (123) used BEAMing to monitor disease
progression in patients with lung cancer undergoing EGFR targeted
therapy, which effectively determined the proportion of
T790M-positive alleles in cancer cells, regardless of potential
contamination from normal cell DNA.
An ultra-fast monitoring method known as thermal
coupling exponential amplification test has been recently reported
(124). This technique combines
exponential amplification reaction (EXPAR) with Thermus
thermophilus argonaute-coupling), from the thermophilic
bacterium Thermus thermophilus, to quickly and accurately
detect ctDNA in ~16 min (124). A
previous study on tumor threshold changes in mouse models (seven
Kirsten rat sarcoma-2 virus (KRAS) point mutations) indicated that
this method holds significant potential for monitoring tumor load
and evaluating chemotherapy response (121). TtAgo-CEAR assay leverages rapid,
specific cleavage function of TtAgo and the high amplification
efficiency of EXPAR to identify common hotspot mutations in
KRAS (124).
Traditional methods for detecting and quantifying
ctDNA, such as PCR and NGS, are well-established but have
limitations (125–127). These methods are not suitable for
detecting short ctDNA fragments (<100 bp). Moreover, they can be
costly, require complex instrumentation, time-consuming due to
multiple reaction steps and prone to false positives (128). By contrast, hybrid chain reaction
is an isothermal, enzyme-free amplification technique that allows
indefinite amplification of signals. This method provides
advantages for the detection of small molecules and shows potential
for ctDNA detection (129). In
2021, researchers successfully employed a hydrogel-based hybrid
chain reaction to amplify small amounts of exosomal microRNA from
urine samples, achieving a 35-fold increase in detection
sensitivity and effectively distinguishing patients with prostate
cancer from normal controls (130). A novel device known as the hybrid
chain reactor-driven laboratory fiber optic device has been
introduced for ultra-fast and sensitive detection of ctDNA in whole
blood. This method is time-efficient, straightforward and
cost-effective, as it enables real-time monitoring of ctDNA changes
(128) and represents a promising
direction for advancing detection capability.
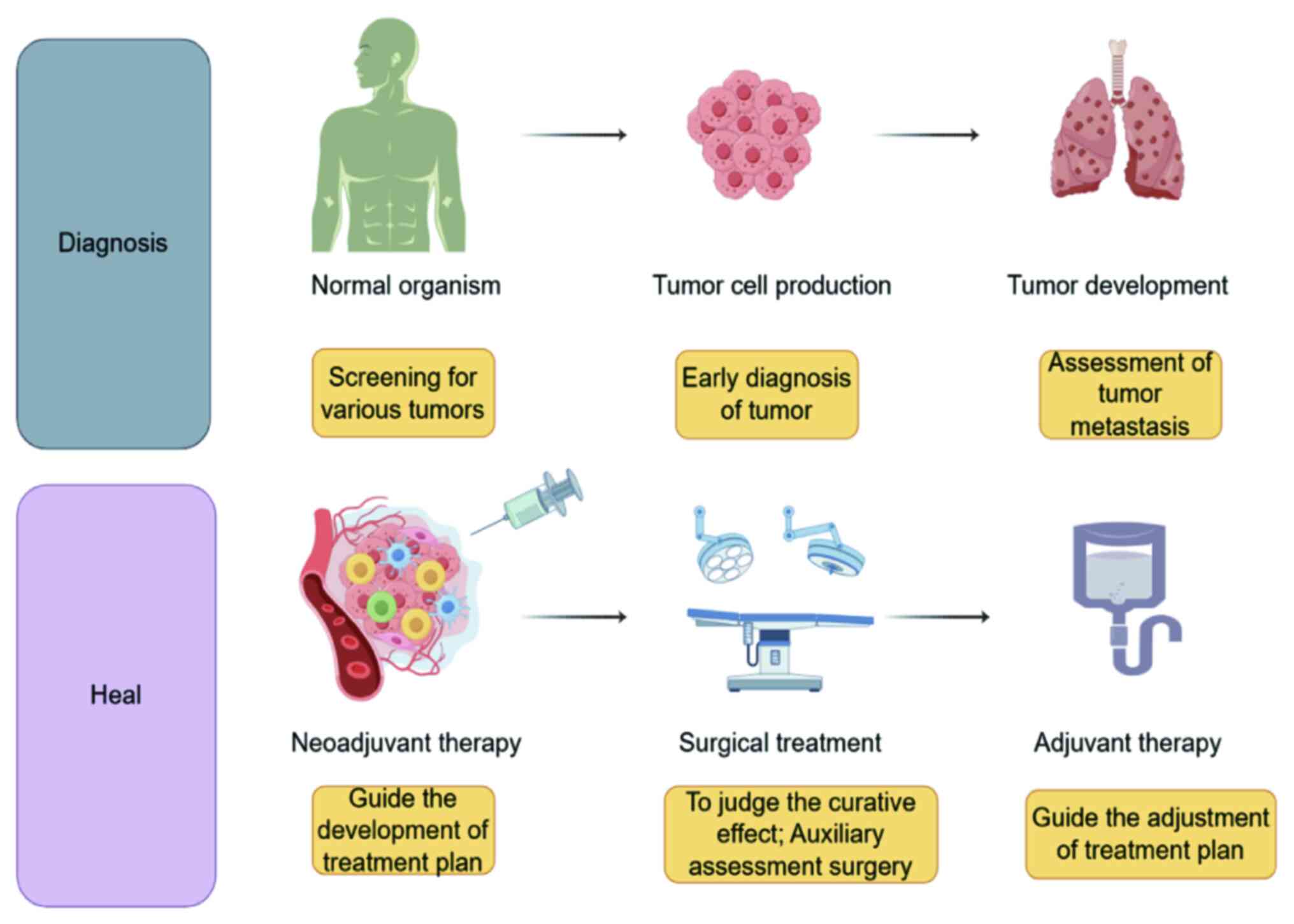
ctDNA serves a key role in the screening and early
diagnosis of various solid types of tumors, particularly among
asymptomatic individuals. Phallen et al (94) demonstrated a strong correlation
between plasma somatic mutations and tumor changes in patients with
stage I or II colorectal, ovarian and breast cancer. This suggested
that ctDNA analysis may be instrumental in both early detection and
ongoing disease management. In a study of esophageal
adenocarcinoma, baseline ctDNA levels were used to identify
patients with locally advanced disease at higher risk of relapse
(106). This highlights potential
of ctDNA not only as a biomarker for early diagnosis but also as a
prognostic tool for tailoring treatment and monitoring disease
progression.
The role of ctDNA as a prognostic marker has gained
recognition in recent years (131–133). ctDNA is detected in various solid
tumors, with its concentration associated with the stage of the
disease (134). In a prospective
phase II clinical trial, ctDNA was dynamically monitored every
three treatment cycles in five patients with solid tumors
undergoing immune checkpoint inhibitor (ICI) treatment. The ctDNA
levels were associated with tumor status, demonstrating predictive
value both at baseline and following treatment (135). The presence of ctDNA following
surgery is strongly indicative of tumor recurrence (6). Gale et al (136) demonstrated that ctDNA could
identify residual lesions and predict recurrence in patients with
NSCLC. Pre- and post-treatment ctDNA testing is shown to identify
patients with NSCLC at high risk for recurrence (136). Monitoring ctDNA levels at
baseline, during neoadjuvant and adjuvant therapy and after radical
therapy allows clinicians to assess drug response and refine
treatment regimens. This dynamic longitudinal monitoring ultimately
improves prognostic evaluation and informs clinical
decision-making, contributing to improved patient management and
outcome.
Response Evaluation Criteria in Solid Tumors
(RECIST1.1) guidelines are key for assessing tumor progression
(137). However, they also have
limitations, particularly concerning pseudo-progression, which
refers to the transient appearance of space-occupying lesions and
edema following treatment. This can often mimic disease progression
on radiographical imaging but typically resolves or changes after
4–8 weeks of follow-up (138). In
clinical practice, clinicians rely on RECIST guidelines to evaluate
disease progression and make treatment decisions. However, ctDNA
detection can offer earlier and more accurate indication of disease
status compared with imaging, potentially reducing follow-up time
and offering better guidance for clinical treatment. Similarly, the
emerging concept of ‘hyperprogression’ describes a rapid
acceleration in tumor growth that can be induced by ICIs (139). To the best of our knowledge, no
studies have reported the association between ctDNA and
hyperprogressive disease. Future research should explore this
association and its implications.
Detection of ctDNA provides extensive information
and enables analysis of minimal residual lesions. Early detection
of residual ctDNA following local radical treatment can indicate
MRD and identify patients at higher risk of recurrence or
metastasis (140). Due to limited
sensitivity of CTC detection, it is rarely used for MRD evaluation
(141). Over the past two decades,
early detection of MRD in children with acute lymphoblastic
leukemia has significantly improved risk stratification, enhanced
treatment for high-risk patients and decreased treatment intensity
for those at low risk (142). The
potential for MRD detection is established in other malignancies,
including acute myeloid (143,144) and chronic lymphocytic leukemia
(145), NSCLC (146,147), multiple myeloma (148–151), breast cancer (152), melanoma (153), head and neck squamous cell
carcinoma (154), follicular
lymphoma (155), urothelial
carcinoma (156) and colorectal
cancer (157).
A promising application of ctDNA is its ability to
inform decisions regarding treatment escalation and de-escalation
(158). Patients with a positive
MRD result may be candidates for intensified adjuvant therapy,
while those with a negative MRD result may potentially benefit from
a reduction in treatment intensity (43). It has been suggested that a key
treatment endpoint for colorectal cancer should be complete
clearance of ctDNA (43).
Currently, a multicenter, prospective, randomized clinical trial is
underway to evaluate efficacy of ctDNA-guided adjuvant chemotherapy
strategies compared to standard care (158). This aims to assess whether
ctDNA-guided treatment adjustments yield superior outcomes in terms
of three-year disease-free survival for patients with high-risk
stage II and III colorectal cancer (Fig. 2) (158).
A notable challenge associated with ctDNA-based
liquid biopsy is limited detection capability. The mutation
abundance of ctDNA is often lower compared with that in localized
tumor tissues, and its detectability is influenced by factors
including tumor type and load, anatomical location, cellular
turnover and disease stage (141).
In the context of early cancer detection, ctDNA levels are
particularly low, often causing MAF to fall below detection limits
of current methods (159). Thus,
improving sensitivity of existing detection methods is key.
Employing a combination of DNA analysis from liquid biopsy and
tissue samples may improve the overall sensitivity and diagnostic
accuracy (160).
Discrepancies in results can arise from the diverse
standards and interpretations employed by different ctDNA testing
methods and laboratories. There is need for the establishment of
standardized testing protocols and interpretative guidelines. It is
also important to select the appropriate sampling methods, as
improper collection of cfDNA from body fluids can lead to missed
detection of ctDNA even with appropriate testing methods (100).
Another barrier is the high cost associated with
ctDNA detection technologies and equipment, which hampers
comprehensive clinical monitoring and may affect treatment
decisions. Furthermore, there is currently no evidence to suggest
that ctDNA can fully replace traditional pathological testing.
Addressing these challenges is key for refining the role of ctDNA
in clinical practice.
ctDNA may serve a key role tumor treatment. Despite
existing challenges, ongoing research may advance the utility of
ctDNA in clinical practice. As improvements in detection
sensitivity, standardization of testing protocols and cost
reduction are realized, ctDNA may enhance patient care by guiding
treatment decisions, monitoring therapeutic response and improving
outcomes.
Not applicable.
The present study was supported by Shaanxi Provincial Cancer
Hospital, grant no. 2022SF-282, cphcf-2022-218); and
24YXYJ0179).
Not applicable.
QG and ZYZ conceived the study. QG, ZYZ, SL, JM and
ZZ wrote and reviewed the manuscript. Data authentication is not
applicable. All authors have read and approved the final
manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Schoop R, Roode LM, de Boer LL and
Dashtbozorg B: Framework for deep learning based Multi-Modality
image registration of snapshot and pathology images. IEEE J Biomed
Health Inform. Aug 16–2024.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pritzker K and Nieminen HJ: Needle biopsy
adequacy in the era of precision medicine and value-based health
care. Arch Pathol Lab Med. 143:1399–1415. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nikanjam M, Kato S and Kurzrock R: Liquid
biopsy: Current technology and clinical applications. J Hematol
Oncol. 15:1312022. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tivey A, Church M, Rothwell D, Dive C and
Cook N: Circulating tumour DNA-looking beyond the blood. Nat Rev
Clin Oncol. 19:600–612. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Alix-Panabieres C and Pantel K: Liquid
biopsy: From discovery to clinical application. Cancer Discov.
11:858–873. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cohen SA, Liu MC and Aleshin A: Practical
recommendations for using ctDNA in clinical decision making.
Nature. 619:259–268. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mandel P and Metais P: Nuclear acids in
human blood plasma. C R Seances Soc Biol Fil. 142:241–243. 1948.(In
French). PubMed/NCBI
|
|
8
|
Hobbs KJ, Cooper BL, Dembek K and Sheats
MK: Investigation of extracted plasma cell-free DNA as a biomarker
in foals with sepsis. Vet Sci. 11:3462024.PubMed/NCBI
|
|
9
|
Zhang LX, Jiang YZ, Qiu LJ and Huang DP:
Quantitative detection and integrality analysis of plasma
circulating Cell-free DNA in multiple myeloma. Zhongguo Shi Yan Xue
Ye Xue Za Zhi. 32:1106–1111. 2024.(In Chinese). PubMed/NCBI
|
|
10
|
Joshi J, Raval A, Desai U, Upadhyay V,
Bhavsar M, Shah K, Rawal R, Panchal H and Shah F: EGFR mutation
analysis in Non-small cell lung carcinoma patients: A liquid biopsy
approach. Indian J Clin Biochem. 36:51–58. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Volik S, Alcaide M, Morin RD and Collins
C: Cell-free DNA (cfDNA): Clinical significance and utility in
cancer shaped by emerging technologies. Mol Cancer Res. 14:898–908.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tan EM, Schur PH, Carr RI and Kunkel HG:
Deoxybonucleic acid (DNA) and antibodies to DNA in the serum of
patients with systemic lupus erythematosus. J Clin Invest.
45:1732–1740. 1966. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Koffler D, Agnello V, Winchester R and
Kunkel HG: The occurrence of single-stranded DNA in the serum of
patients with systemic lupus erythematosus and other diseases. J
Clin Invest. 52:198–204. 1973. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Giacona MB, Ruben GC, Iczkowski KA, Roos
TB, Porter DM and Sorenson GD: Cell-free DNA in human blood plasma:
Length measurements in patients with pancreatic cancer and healthy
controls. Pancreas. 17:89–97. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Szilagyi M, Pos O, Marton E, Buglyo G,
Soltesz B, Keseru J, Penyige A, Szemes T and Nagy B: Circulating
cell-free nucleic acids: Main characteristics and clinical
application. Int J Mol Sci. 21:68272020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Biro O, Fothi A, Alasztics B, Nagy B,
Orban TI and Rigo JJ: Circulating exosomal and Argonaute-bound
microRNAs in preeclampsia. Gene. 692:138–144. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fernando MR, Jiang C, Krzyzanowski GD and
Ryan WL: New evidence that a large proportion of human blood plasma
cell-free DNA is localized in exosomes. PLoS One. 12:e01839152017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Aucamp J, Bronkhorst AJ, Badenhorst C and
Pretorius PJ: The diverse origins of circulating cell-free DNA in
the human body: A critical re-evaluation of the literature. Biol
Rev Camb Philos Soc. 93:1649–1683. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Moss J, Magenheim J, Neiman D, Zemmour H,
Loyfer N, Korach A, Samet Y, Maoz M, Druid H, Arner P, et al:
Comprehensive human cell-type methylation atlas reveals origins of
circulating cell-free DNA in health and disease. Nat Commun.
9:50682018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Teo YV, Capri M, Morsiani C, Pizza G,
Faria A, Franceschi C and Neretti N: Cell-free DNA as a biomarker
of aging. Aging Cell. 18:e128902019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hummel EM, Hessas E, Muller S, Beiter T,
Fisch M, Eibl A, Wolf OT, Giebel B, Platen P, Kumsta R and Moser
DA: Cell-free DNA release under psychosocial and physical stress
conditions. Transl Psychiatry. 8:2362018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ai B, Liu H, Huang Y and Peng P:
Circulating cell-free DNA as a prognostic and predictive biomarker
in non-small cell lung cancer. Oncotarget. 7:44583–44595. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kim DY, Cho EH, Kim JS, Chie EK and Kang
HC: Plasma Circulating Cell-free DNa in advanced hepatocellular
carcinoma patients treated with radiation therapy. In Vivo.
37:2306–2313. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gianni C, Palleschi M, Merloni F, Di Menna
G, Sirico M, Sarti S, Virga A, Ulivi P, Cecconetto L, Mariotti M
and De Giorgi U: Cell-Free DNA Fragmentomics: A promising biomarker
for diagnosis, prognosis and prediction of response in breast
cancer. Int J Mol Sci. 23:141972022. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Al SN, Messaoudi SA, Babu SR, Chaudhary
AB, Alsharm AA, Alrefaei AF, Kadasah S, Abu-Elmagd M, Assidi M,
Buhmeida A, et al: Utility of circulating Cell-free DNA in
assessing microsatellite instability and loss of Heterozygosity in
breast cancer using human identification approach. Genes (Basel).
13:5902022. View Article : Google Scholar
|
|
26
|
Bahado-Singh RO, Turkoglu O, Aydas B and
Vishweswaraiah S: Precision oncology: Artificial intelligence,
circulating cell-free DNA, and the minimally invasive detection of
pancreatic cancer-A pilot study. Cancer Med. 12:19644–19655. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lin LH, Chang KW, Kao SY, Cheng HW and Liu
CJ: Increased plasma circulating Cell-Free DNA could be a potential
marker for oral cancer. Int J Mol Sci. 19:33032018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fang Q, Yuan Z, Hu H, Zhang W, Wang G and
Wang X: Genome-wide discovery of circulating cell-free DNA
methylation biomarkers for colorectal cancer detection. Clin
Epigenetics. 15:1192023. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Eskander NS, Mansour L, Abdelaal A, Saad E
and Mohamed D: Circulating cell free DNA integrity index as a
biomarker for response to chemotherapy in patients with metastatic
colorectal carcinoma. Asian Pac J Cancer Prev. 23:339–348. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Heidrich I and Pantel K: Liquid biopsy:
Blood-based analyses of circulating cell-free DNA in xenografts.
EMBO Mol Med. 14:e163262022. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kroeze A, Cornelissen AS, Pascutti MF,
Verheij M, Bulder I, Klarenbeek S, Ait SA, Hazenberg MD, Nur E, van
der Schoot CE, et al: Cell-free DNA levels are increased in acute
graft-versus-host disease. Eur J Haematol. 109:271–281. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kustanovich A, Schwartz R, Peretz T and
Grinshpun A: Life and death of circulating cell-free DNA. Cancer
Biol Ther. 20:1057–1067. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Stewart CM and Tsui D: Circulating
cell-free DNA for non-invasive cancer management. Cancer Genet.
228–229. 169–179. 2018.
|
|
34
|
Yu SC, Lee SW, Jiang P, Leung TY, Chan KC,
Chiu RW and Lo YM: High-resolution profiling of fetal DNA clearance
from maternal plasma by massively parallel sequencing. Clin Chem.
59:1228–1237. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Butler TM, Spellman PT and Gray J:
Circulating-tumor DNA as an early detection and diagnostic tool.
Curr Opin Genet Dev. 42:14–21. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jahr S, Hentze H, Englisch S, Hardt D,
Fackelmayer FO, Hesch RD and Knippers R: DNA fragments in the blood
plasma of cancer patients: Quantitations and evidence for their
origin from apoptotic and necrotic cells. Cancer Res. 61:1659–1665.
2001.PubMed/NCBI
|
|
37
|
Miller AM and Karajannis MA: Current role
and future potential of CSF ctDNA for the diagnosis and clinical
management of pediatric central nervous system tumors. J Natl Compr
Canc Netw. 20:1363–1369. 2022.PubMed/NCBI
|
|
38
|
Diehl F, Schmidt K, Choti MA, Romans K,
Goodman S, Li M, Thornton K, Agrawal N, Sokoll L, Szabo SA, et al:
Circulating mutant DNA to assess tumor dynamics. Nat Med.
14:985–990. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Han JY, Ahn KS, Kim TS, Kim YH, Cho KB,
Shin DW, Baek WK, Suh SI, Jang BC and Kang KJ: Liquid biopsy from
Bile-Circulating tumor DNA in patients with biliary tract cancer.
Cancers (Basel). 13:45812021. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Bettegowda C, Sausen M, Leary RJ, Kinde I,
Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM, et al:
Detection of circulating tumor DNA in early- and late-stage human
malignancies. Sci Transl Med. 6:224ra242014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Diehl F, Li M, Dressman D, He Y, Shen D,
Szabo S, Diaz LJ, Goodman SN, David KA, Juhl H, et al: Detection
and quantification of mutations in the plasma of patients with
colorectal tumors. Proc Natl Acad Sci USA. 102:16368–16373. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Dawson SJ, Tsui DW, Murtaza M, Biggs H,
Rueda OM, Chin SF, Dunning MJ, Gale D, Forshew T, Mahler-Araujo B,
et al: Analysis of circulating tumor DNA to monitor metastatic
breast cancer. N Engl J Med. 368:1199–1209. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Sato S, Nakamura Y, Oki E and Yoshino T:
Molecular residual Disease-guided adjuvant treatment in resected
colorectal cancer: Focus on CIRCULATE-Japan. Clin Colorectal
Cancer. 22:53–58. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yi K, Wang X, Filippov SK and Zhang H:
Emerging ctDNA detection strategies in clinical cancer
theranostics. Smart Med. 2:e202300312023. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Cheng F, Su L and Qian C: Circulating
tumor DNA: A promising biomarker in the liquid biopsy of cancer.
Oncotarget. 7:48832–48841. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Lee TH, Montalvo L, Chrebtow V and Busch
MP: Quantitation of genomic DNA in plasma and serum samples: Higher
concentrations of genomic DNA found in serum than in plasma.
Transfusion. 41:276–282. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Heger JM, Mattlener J, Schneider J, Godel
P, Sieg N, Ullrich F, Lewis RI, Bucaciuc-Mracica T, Schwarz RF,
Ruess D, et al: Entirely noninvasive outcome prediction in central
nervous system lymphomas using circulating tumor DNA. Blood.
143:522–534. 2023. View Article : Google Scholar
|
|
48
|
Werner B, Warton K and Ford CE:
Transcending Blood-Opportunities for alternate liquid biopsies in
oncology. Cancers (Basel). 14:13092022. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Seyhan AA: Circulating liquid biopsy
biomarkers in glioblastoma: Advances and challenges. Int J Mol Sci.
25:79742024. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zheng MM, Li YS, Jiang BY, Tu HY, Tang WF,
Yang JJ, Zhang XC, Ye JY, Yan HH, Su J, et al: Clinical utility of
cerebrospinal fluid Cell-Free DNA as liquid biopsy for
leptomeningeal metastases in ALK-Rearranged NSCLC. J Thorac Oncol.
14:924–932. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wu J, Liu Z, Huang T, Wang Y, Song MM,
Song T, Long G, Zhang X, Li X and Zhang L: Cerebrospinal fluid
circulating tumor DNA depicts profiling of brain metastasis in
NSCLC. Mol Oncol. 17:810–824. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
De Mattos-Arruda L, Mayor R, Ng C, Weigelt
B, Martinez-Ricarte F, Torrejon D, Oliveira M, Arias A, Raventos C,
Tang J, et al: Cerebrospinal fluid-derived circulating tumour DNA
better represents the genomic alterations of brain tumours than
plasma. Nat Commun. 6:88392015. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Wang Y, Luo N, Gao Y, Wu Y, Qin X, Qi Y,
Sun T, Tao R, Qi C, Liu B and Yuan S: The joint detection of CEA
and ctDNA in cerebrospinal fluid: An auxiliary tool for the
diagnosis of leptomeningeal metastases in cancer. J Cancer Res Clin
Oncol. 149:1679–1690. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Bai Y, Yu Q, Liu N, Liu J, Wang D, Liu X
and Yuan S: Case report: Cerebrospinal fluid-derived circulating
tumor DNA diagnoses and guides the treatment of a lung
adenocarcinoma case with leptomeningeal metastasis. Front Oncol.
12:9449632022. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
van der Wel J, Boelens MC, Jebbink M,
Smulders SA, Maas KW, Luitse M, Compter A, Boltjes R, Sol N,
Monkhorst K, et al: Osimertinib-induced DNA resistance mutations in
cerebrospinal fluid of EGFR mutated NSCLC patients developing
leptomeningeal metastases: ORA-LM study. Neuro Oncol. Aug
7–2024.doi: 10.1093/neuonc/noae138 (Epub ahead of print).
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Azad TD, Nanjo S, Jin MC, Chabon JJ, Kurtz
DM, Chaudhuri AA, Connolly ID, Hui AB, Liu CL, Merriott D, et al:
Quantification of cerebrospinal fluid tumor DNA in lung cancer
patients with suspected leptomeningeal carcinomatosis. NPJ Precis
Oncol. 8:1212024. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Valerius AR, Webb MJ, Hammad N, Sener U
and Malani R: Cerebrospinal fluid liquid biopsies in the evaluation
of adult gliomas. Curr Oncol Rep. 26:377–390. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Dai L, Liu Z, Zhu Y and Ma L: Genome-wide
methylation analysis of circulating tumor DNA: A new biomarker for
recurrent glioblastom. Heliyon. 9:e143392023. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Kojic M, Maybury MK, Waddell N,
Koufariotis LT, Addala V, Millar A, Wood S, Pearson JV, Hansford
JR, Hassall T, et al: Efficient detection and monitoring of
pediatric brain malignancies with liquid biopsy based on
patient-specific somatic mutation screening. Neuro Oncol.
25:1507–1517. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Izquierdo E, Proszek P, Pericoli G,
Temelso S, Clarke M, Carvalho DM, Mackay A, Marshall LV, Carceller
F, Hargrave D, et al: Droplet digital PCR-based detection of
circulating tumor DNA from pediatric high grade and diffuse midline
glioma patients. Neurooncol Adv. 3:vdab0132021.PubMed/NCBI
|
|
61
|
Li J, Zhao S, Lee M, Yin Y, Li J, Zhou Y,
Ballester LY, Esquenazi Y, Dashwood RH, Davies P, et al: Reliable
tumor detection by whole-genome methylation sequencing of cell-free
DNA in cerebrospinal fluid of pediatric medulloblastoma. Sci Adv.
6:eabb54272020. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Pages M, Rotem D, Gydush G, Reed S,
Rhoades J, Ha G, Lo C, Fleharty M, Duran M, Jones R, et al: Liquid
biopsy detection of genomic alterations in pediatric brain tumors
from cell-free DNA in peripheral blood, CSF, and urine. Neuro
Oncol. 24:1352–1363. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Ding S and Song X, Geng X, Liu L, Ma H,
Wang X, Wei L, Xie L and Song X: Saliva-derived cfDNA is applicable
for EGFR mutation detection but not for quantitation analysis in
non-small cell lung cancer. Thorac Cancer. 10:1973–1983. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Wang Z, Zhang L, Li L, Li X, Xu Y, Wang M,
Liang L, Jiao P, Li Y, He S, et al: Sputum Cell-Free DNA: Valued
surrogate sample for detection of EGFR mutation in patients with
advanced lung adenocarcinoma. J Mol Diagn. 22:934–942. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Wang Z, Li X, Zhang L, Xu Y, Wang M, Liang
L, Jiao P, Li Y, He S, Du J, et al: Sputum cell-free DNA: Valued
surrogate sample for the detection of EGFR exon 20 p.T790M mutation
in patients with advanced lung adenocarcinoma and acquired
resistance to EGFR-TKIs. Cancer Med. 10:3323–3331. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Ferrier ST, Tsering T, Sadeghi N, Zeitouni
A and Burnier JV: Blood and saliva-derived ctDNA is a marker of
residual disease after treatment and correlates with recurrence in
human papillomavirus-associated head and neck cancer. Cancer Med.
12:15777–15787. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Britze TE, Jakobsen KK, Gronhoj C and von
Buchwald C: A systematic review on the role of biomarkers in liquid
biopsies and saliva samples in the monitoring of salivary gland
cancer. Acta Otolaryngol. 143:709–713. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Gupta S, Singh B, Abhishek R, Gupta S and
Sachan M: The emerging role of liquid biopsy in oral squamous cell
carcinoma detection: Advantages and challenges. Expert Rev Mol
Diagn. 24:311–331. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Perrone ME, Alvarez R, Vo TT, Chung MW,
Chhieng DC, Paulson VA, Colbert BG, Q Konnick E and Huang EC:
Validating cell-free DNA from supernatant for molecular diagnostics
on cytology specimens. Cancer Cytopathol. 129:956–965. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Yang SR, Mooney KL, Libiran P, Jones CD,
Joshi R, Lau HD, Stehr H, Berry GJ, Zehnder JL, Long SR, et al:
Targeted deep sequencing of cell-free DNA in serous body cavity
fluids with malignant, suspicious, and benign cytology. Cancer
Cytopathol. 128:43–56. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Leick KM, Kazarian AG, Rajput M,
Tomanek-Chalkley A, Miller A, Shrader HR, Mccarthy A, Coleman KL,
Kasi PM and Chan C: Peritoneal Cell-free tumor DNA as biomarker for
peritoneal surface malignancies. Ann Surg Oncol. 27:5065–5071.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Kinugasa H, Nouso K, Ako S, Dohi C,
Matsushita H, Matsumoto K, Kato H and Okada H: Liquid biopsy of
bile for the molecular diagnosis of gallbladder cancer. Cancer Biol
Ther. 19:934–938. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Takai E, Totoki Y, Nakamura H, Morizane C,
Nara S, Hama N, Suzuki M, Furukawa E, Kato M, Hayashi H, et al:
Clinical utility of circulating tumor DNA for molecular assessment
in pancreatic cancer. Sci Rep. 5:184252015. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Levink I, Jansen M, Azmani Z, van Ijcken
W, van Marion R, Peppelenbosch MP, Cahen DL, Fuhler GM and Bruno
MJ: Mutation analysis of pancreatic juice and plasma for the
detection of pancreatic cancer. Int J Mol Sci. 24:131162023.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Fitzgerald JM, Ramchurren N, Rieger K,
Levesque P, Silverman M, Libertino JA and Summerhayes IC:
Identification of H-ras mutations in urine sediments complements
cytology in the detection of bladder tumors. J Natl Cancer Inst.
87:129–133. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Jain S, Lin SY, Song W and Su YH:
Urine-based liquid biopsy for nonurological cancers. Genet Test Mol
Biomarkers. 23:277–283. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Su YH, Wang M, Block TM, Landt O, Botezatu
I, Serdyuk O, Lichtenstein A, Melkonyan H, Tomei LD and Umansky S:
Transrenal DNA as a diagnostic tool: Important technical notes. Ann
N Y Acad Sci. 1022:81–89. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Su YH, Wang M, Brenner DE, Norton PA and
Block TM: Detection of mutated K-ras DNA in urine, plasma, and
serum of patients with colorectal carcinoma or adenomatous polyps.
Ann N Y Acad Sci. 1137:197–206. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Xiao Y, Ju L, Qian K, Jin W, Wang G, Zhao
Y, Jiang W, Liu N, Wu K, Peng M, et al: Non-invasive diagnosis and
surveillance of bladder cancer with driver and passenger DNA
methylation in a prospective cohort study. Clin Transl Med.
12:e10082022. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Christensen E, Nordentoft I,
Birkenkamp-Demtroder K, Elbaek SK, Lindskrog SV, Taber A, Andreasen
TG, Strandgaard T, Knudsen M, Lamy P, et al: Cell-Free urine and
plasma DNA mutational analysis predicts neoadjuvant chemotherapy
response and outcome in patients with muscle-invasive bladder
cancer. Clin Cancer Res. 29:1582–1591. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Tamura D, Abe M, Hiraki H, Sasaki N,
Yashima-Abo A, Ikarashi D, Kato R, Kato Y, Maekawa S, Kanehira M,
et al: Postoperative recurrence detection using individualized
circulating tumor DNA in upper tract urothelial carcinoma. Cancer
Sci. 115:529–539. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Kim AK, Hamilton JP, Lin SY, Chang TT,
Hann HW, Hu CT, Lou Y, Lin YJ, Gade TP, Park G, et al: Urine DNA
biomarkers for hepatocellular carcinoma screening. Br J Cancer.
126:1432–1438. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Adrogue HJ and Madias NE: Assessing
Acid-base status: Physiologic versus physicochemical approach. Am J
Kidney Dis. 68:793–802. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Dermody SM, Bhambhani C, Swiecicki PL,
Brenner JC and Tewari M: Trans-renal cell-free tumor DNA for
Urine-based liquid biopsy of cancer. Front Genet. 13:8791082022.
View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Alahdal M, Perera RA, Moschovas MC, Patel
V and Perera RJ: Current advances of liquid biopsies in prostate
cancer: Molecular biomarkers. Mol Ther Oncolytics. 30:27–38. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Fonseca NM, Maurice-Dror C, Herberts C, Tu
W, Fan W, Murtha AJ, Kollmannsberger C, Kwan EM, Parekh K, Schonlau
E, et al: Prediction of plasma ctDNA fraction and prognostic
implications of liquid biopsy in advanced prostate cancer. Nat
Commun. 15:18282024. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Tolmeijer SH, Boerrigter E, Van Erp NP and
Mehra N: Using early on-treatment circulating tumor DNA
measurements as response assessment in metastatic castration
resistant prostate cancer. Oncotarget. 15:421–423. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Ponti G, Maccaferri M, Manfredini M,
Micali S, Torricelli F, Milandri R, Del PC, Ciarrocchi A, Ruini C,
Benassi L, et al: Quick assessment of cell-free DNA in seminal
fluid and fragment size for early non-invasive prostate cancer
diagnosis. Clin Chim Acta. 497:76–80. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Ponti G, Maccaferri M, Percesepe A, Tomasi
A and Ozben T: Liquid biopsy with cell free DNA: New horizons for
prostate cancer. Crit Rev Clin Lab Sci. 58:60–76. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Yu B and Ma W: Biomarker discovery in
hepatocellular carcinoma (HCC) for personalized treatment and
enhanced prognosis. Cytokine Growth Factor Rev. Aug 24–2024.doi:
10.1016/j.cytogfr.2024.08.006 (Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Zhu L, Xu R, Yang L, Shi W, Zhang Y, Liu
J, Li X, Zhou J and Bing P: Minimal residual disease (MRD)
detection in solid tumors using circulating tumor DNA: A systematic
review. Front Genet. 14:11721082023. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Li S, Li H, Li X, Zhu M, Li H and Xia F:
Hybridization Chain Reaction-amplified electrochemical DNA-based
sensors enable calibration-free measurements of nucleic acids
directly in whole blood. Anal Chem. 93:8354–8361. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Ho HY, Chung KK, Kan CM and Wong SC:
Liquid biopsy in the clinical management of cancers. Int J Mol Sci.
25:85942024. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Phallen J, Sausen M, Adleff V, Leal A,
Hruban C, White J, Anagnostou V, Fiksel J, Cristiano S, Papp E, et
al: Direct detection of early-stage cancers using circulating tumor
DNA. Sci Transl Med. 9:eaan24152017. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Bittla P, Kaur S, Sojitra V, Zahra A,
Hutchinson J, Folawemi O and Khan S: Exploring Circulating tumor
DNA (CtDNA) and its role in early detection of cancer: A systematic
review. Cureus. 15:e457842023.PubMed/NCBI
|
|
96
|
Yu W, Hurley J, Roberts D, Chakrabortty
SK, Enderle D, Noerholm M, Breakefield XO and Skog JK:
Exosome-based liquid biopsies in cancer: Opportunities and
challenges. Ann Oncol. 32:466–477. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Kemper M, Krekeler C, Menck K, Lenz G,
Evers G, Schulze AB and Bleckmann A: Liquid Biopsies in Lung
Cancer. Cancers (Basel). 15:14302023. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Lin C, Liu X, Zheng B, Ke R and Tzeng CM:
Liquid biopsy, ctDNA diagnosis through NGS. Life (Basel).
11:8902021.PubMed/NCBI
|
|
99
|
Fernandes M, Cruz-Martins N, Souto MC,
Guimaraes S, Pereira RJ, Justino A, Pina MJ, Magalhaes A, Queiroga
H, Machado JC, et al: Clinical application of Next-generation
sequencing of plasma Cell-free DNA for genotyping untreated
advanced Non-small cell lung cancer. Cancers (Basel). 13:27072021.
View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Roberto TM, Jorge MA, Francisco GV, Noelia
T, Pilar RG and Andres C: Strategies for improving detection of
circulating tumor DNA using next generation sequencing. Cancer
Treat Rev. 119:1025952023. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Grada A and Weinbrecht K: Next-generation
sequencing: Methodology and application. J Invest Dermatol.
133:e112013. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Cheng ML, Pectasides E, Hanna GJ, Parsons
HA, Choudhury AD and Oxnard GR: Circulating tumor DNA in advanced
solid tumors: Clinical relevance and future directions. CA Cancer J
Clin. 71:176–190. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Ma M, Zhu H, Zhang C, Sun X, Gao X and
Chen G: ‘Liquid biopsy’-ctDNA detection with great potential and
challenges. Ann Transl Med. 3:2352015.PubMed/NCBI
|
|
104
|
Gale D, Lawson A, Howarth K, Madi M,
Durham B, Smalley S, Calaway J, Blais S, Jones G, Clark J, et al:
Development of a highly sensitive liquid biopsy platform to detect
clinically-relevant cancer mutations at low allele fractions in
cell-free DNA. PLoS One. 13:e01946302018. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Forshew T, Murtaza M, Parkinson C, Gale D,
Tsui DW, Kaper F, Dawson SJ, Piskorz AM, Jimenez-Linan M, Bentley
D, et al: Noninvasive identification and monitoring of cancer
mutations by targeted deep sequencing of plasma DNA. Sci Transl
Med. 4:136ra682012. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Cabalag CS, Yates M, Corrales MB, Yeh P,
Wong SQ, Zhang BZ, Fujihara KM, Chong L, Hii MW, Dawson SJ, et al:
Potential clinical utility of a targeted circulating tumor DNA
Assay in esophageal adenocarcinoma. Ann Surg. 276:e120–e126. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Newman AM, Bratman SV, To J, Wynne JF,
Eclov NC, Modlin LA, Liu CL, Neal JW, Wakelee HA, Merritt RE, et
al: An ultrasensitive method for quantitating circulating tumor DNA
with broad patient coverage. Nat Med. 20:548–554. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Azad TD, Chaudhuri AA, Fang P, Qiao Y,
Esfahani MS, Chabon JJ, Hamilton EG, Yang YD, Lovejoy A, Newman AM,
et al: Circulating tumor DNA analysis for detection of minimal
residual disease after chemoradiotherapy for localized esophageal
cancer. Gastroenterology. 158:494–505. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Noguchi T, Sakai K, Iwahashi N, Matsuda K,
Matsukawa H, Yahata T, Toujima S, Nishio K and Ino K: Changes in
the gene mutation profiles of circulating tumor DNA detected using
CAPP-Seq in neoadjuvant chemotherapy-treated advanced ovarian
cancer. Oncol Lett. 19:2713–2720. 2020.PubMed/NCBI
|
|
110
|
Jung D, Jain P, Yao Y and Wang M: Advances
in the assessment of minimal residual disease in mantle cell
lymphoma. J Hematol Oncol. 13:1272020. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Satyal U, Srivastava A and Abbosh PH:
Urine biopsy-liquid gold for molecular detection and surveillance
of bladder cancer. Front Oncol. 9:12662019. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Taylor K, Zou J, Magalhaes M, Oliva M,
Spreafico A, Hansen AR, Mcdade SS, Coyle VM, Lawler M, Elimova E,
et al: Circulating tumour DNA kinetics in recurrent/metastatic head
and neck squamous cell cancer patients. Eur J Cancer. 188:29–38.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Aoude LG, Brosda S, Ng J, Lonie JM, Belle
CJ, Patel K, Koufariotis LT, Wood S, Atkinson V, Smithers BM, et
al: Circulating tumor DNA: A promising biomarker for predicting
recurrence in patients with BRAF-Negative melanoma. J Mol Diagn.
25:771–781. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Grassi T, Harris FR, Smadbeck JB, Murphy
SJ, Block MS, Multinu F, Schaefer KJ, Zhang P, Karagouga G, Liu MC,
et al: Personalized tumor-specific DNA junctions to detect
circulating tumor in patients with endometrial cancer. PLoS One.
16:e02523902021. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Mansson CT, Vad-Nielsen J, Meldgaard P,
Nielsen AL and Sorensen BS: EGFR transcription in non-small-cell
lung cancer tumours can be revealed in ctDNA by cell-free chromatin
immunoprecipitation (cfChIP). Mol Oncol. 15:2868–2876. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Jafri H, Mushtaq S, Baig S, Bhatty A and
Siraj S: Comparison of KRAS gene in circulating tumor DNA levels vs
histological grading of colorectal cancer patients through liquid
biopsy. Saudi J Gastroenterol. 29:371–375. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
FDA, . Summary of Safety and Effectiveness
Data (SSED) P150047. Cobas EGFR Mutation Test v2®.
2016.
|
|
118
|
Biglari N, Soltani-Zangbar MS, Mohammadian
J, Mehdizadeh A and Abbasi K: ctDNA as a novel and promising
approach for cancer diagnosis: A focus on hepatocellular carcinoma.
EXCLI J. 22:752–780. 2023.PubMed/NCBI
|
|
119
|
Hickman RA, Miller AM and Arcila ME:
Cerebrospinal fluid: A unique source of circulating tumor DNA with
broad clinical applications. Transl Oncol. 33:1016882023.
View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Rimelen V, Ahle G, Pencreach E, Zinniger
N, Debliquis A, Zalmai L, Harzallah I, Hurstel R, Alamome I, Lamy
F, et al: Tumor cell-free DNA detection in CSF for primary CNS
lymphoma diagnosis. Acta Neuropathol Commun. 7:432019. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Venetis K, Pepe F, Pescia C, Cursano G,
Criscitiello C, Frascarelli C, Mane E, Russo G, Taurelli SB,
Troncone G, et al: ESR1 mutations in HR+/HER2-metastatic breast
cancer: Enhancing the accuracy of ctDNA testing. Cancer Treat Rev.
121:1026422023. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Teh SY, Lin R, Hung LH and Lee AP: Droplet
microfluidics. Lab Chip. 8:198–220. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Taniguchi K, Uchida J, Nishino K, Kumagai
T, Okuyama T, Okami J, Higashiyama M, Kodama K, Imamura F and Kato
K: Quantitative detection of EGFR mutations in circulating tumor
DNA derived from lung adenocarcinomas. Clin Cancer Res.
17:7808–7815. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Fang J, Yuan C, Luo X, He Z and Fu W: A
Thermus thermophilus argonaute-coupling exponential
amplification assay for ultrarapid analysis of circulating tumor
DNA. Talanta. 266:1250342024. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Cappello F, Angerilli V, Munari G, Ceccon
C, Sabbadin M, Pagni F, Fusco N, Malapelle U and Fassan M:
FFPE-Based NGS approaches into clinical practice: The limits of
glory from a pathologist viewpoint. J Pers Med. 12:1026422022.
View Article : Google Scholar
|
|
126
|
Mantilla WA, Sanabria-Salas MC, Baldion
AM, Sua LF, Gonzalez DM and Lema M: NGS in lung, breast, and
unknown primary cancer in colombia: A multidisciplinary consensus
on challenges and opportunities. JCO Glob Oncol. 7:1012–1023. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Lin YH, Liao XJ, Chang W and Chiou CC:
Ultrafast DNA amplification using microchannel Flow-through PCR
device. Biosensors (Basel). 12:3032022. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Xu J, Han X, Xu W, Liu J, Zhu A, Song D
and Long F: Development of a hybridization chain reaction-powered
lab-on-fiber device for ultrafast point-of-care testing of
circulating tuor DNA in whole blood. Talanta. 259:1244752023.
View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Wu J, Lv J, Zheng X and Wu ZS:
Hybridization chain reaction and its applications in biosensing.
Talanta. 234:1226372021. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Kim J, Shim JS, Han BH, Kim HJ, Park J,
Cho IJ, Kang SG, Kang JY, Bong KW and Choi N: Hydrogel-based
hybridization chain reaction (HCR) for detection of urinary
exosomal miRNAs as a diagnostic tool of prostate cancer. Biosens
Bioelectron. 192:1135042021. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Papakonstantinou A, Gonzalez NS, Pimentel
I, Sunol A, Zamora E, Ortiz C, Espinosa-Bravo M, Peg V, Vivancos A,
Saura C, et al: Prognostic value of ctDNA detection in patients
with early breast cancer undergoing neoadjuvant therapy: A
systematic review and meta-analysis. Cancer Treat Rev.
104:1023622022. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Wang D, Zhao P, Lu T, Ren J, Zhu L, Han X,
Zhang G, Dong X, Ma H, Yu M and Cai H: ctDNA as a prognostic
biomarker in resectable CLM: Systematic review and meta-analysis.
Open Life Sci. 18:202206152023. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Wei J, Feng J, Weng Y, Xu Z, Jin Y, Wang
P, Cui X, Ruan P, Luo R, Li N and Peng M: The prognostic value of
ctDNA and bTMB on immune checkpoint inhibitors in human cancer.
Front Oncol. 11:7069102021. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Markou A, Tzanikou E and Lianidou E: The
potential of liquid biopsy in the management of cancer patients.
Semin Cancer Biol. 84:69–79. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Bratman SV, Yang S, Iafolla M, Liu Z,
Hansen AR, Bedard PL, Lheureux S, Spreafico A, Razak AA, Shchegrova
S, et al: Personalized circulating tumor DNA analysis as a
predictive biomarker in solid tumor patients treated with
pembrolizumab. Nat Cancer. 1:873–881. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Gale D, Heider K, Ruiz-Valdepenas A,
Hackinger S, Perry M, Marsico G, Rundell V, Wulff J, Sharma G,
Knock H, et al: Residual ctDNA after treatment predicts early
relapse in patients with early-stage non-small cell lung cancer.
Ann Oncol. 33:500–510. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Borcoman E, Kanjanapan Y, Champiat S, Kato
S, Servois V, Kurzrock R, Goel S, Bedard P and Le Tourneau C: Novel
patterns of response under immunotherapy. Ann Oncol. 30:385–396.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Young JS, Al-Adli N, Scotford K, Cha S and
Berger MS: Pseudoprogression versus true progression in
glioblastoma: What neurosurgeons need to know. J Neurosurg.
139:748–759. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Zheng J, Zhou X, Fu Y and Chen Q: Advances
in the study of hyperprogression of different tumors treated with
PD-1/PD-L1 antibody and the mechanisms of its occurrence. Cancers
(Basel). 15:13142023. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Vellanki PJ, Ghosh S, Pathak A, Fusco MJ,
Bloomquist EW, Tang S, Singh H, Philip R, Pazdur R and Beaver JA:
Regulatory implications of ctDNA in Immuno-oncology for solid
tumors. J Immunother Cancer. 11:e0053442023. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Mahuron KM and Fong Y: Applications of
liquid biopsy for surgical patients with cancer: A review. JAMA
Surg. 159:96–103. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Juarez-Avendano G, Mendez-Ramirez N,
Luna-Silva NC, Gomez-Almaguer D, Pelayo R and Balandran JC:
Molecular and cellular markers for measurable residual disease in
acute lymphoblastic leukemia. Bol Med Hosp Infant Mex. 78:159–170.
2021.PubMed/NCBI
|
|
143
|
Li Y, Solis-Ruiz J, Yang F, Long N, Tong
CH, Lacbawan FL, Racke FK and Press RD: NGS-defined measurable
residual disease (MRD) after initial chemotherapy as a prognostic
biomarker for acute myeloid leukemia. Blood Cancer J. 13:592023.
View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Gutman JA, Winters A, Kent A, Amaya M,
Mcmahon C, Smith C, Jordan CT, Stevens B, Minhajuddin M, Pei S, et
al: Higher-dose venetoclax with measurable residual disease-guided
azacitidine discontinuation in newly diagnosed acute myeloid
leukemia. Haematologica. 108:2616–2625. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Munir T, Cairns DA, Bloor A, Allsup D,
Cwynarski K, Pettitt A, Paneesha S, Fox CP, Eyre TA, Forconi F, et
al: Chronic lymphocytic leukemia therapy guided by measurable
residual disease. N Engl J Med. 390:326–337. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Zhang JT, Liu SY, Gao W, Liu SM, Yan HH,
Ji L, Chen Y, Gong Y, Lu HL, Lin JT, et al: Longitudinal
undetectable molecular residual disease defines potentially cured
population in localized non-small cell lung cancer. Cancer Discov.
12:1690–1701. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Jung HA, Ku BM, Kim YJ, Park S, Sun JM,
Lee SH, Ahn JS, Cho JH, Kim HK, Choi YS, et al: Longitudinal
monitoring of circulating tumor DNA from plasma in patients with
curative resected Stages I to IIIA EGFR-Mutant Non-Small cell lung
cancer. J Thorac Oncol. 18:1199–1208. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Costa LJ, Chhabra S, Medvedova E, Dholaria
BR, Schmidt TM, Godby KN, Silbermann R, Dhakal B, Bal S, Giri S, et
al: Daratumumab, Carfilzomib, Lenalidomide, and dexamethasone with
minimal residual disease Response-Adapted therapy in newly
diagnosed multiple myeloma. J Clin Oncol. 40:2901–2912. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
149
|
San-Miguel J, Avet-Loiseau H, Paiva B,
Kumar S, Dimopoulos MA, Facon T, Mateos MV, Touzeau C, Jakubowiak
A, Usmani SZ, et al: Sustained minimal residual disease negativity
in newly diagnosed multiple myeloma and the impact of daratumumab
in MAIA and ALCYONE. Blood. 139:492–501. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Costa LJ, Chhabra S, Medvedova E, Dholaria
BR, Schmidt TM, Godby KN, Silbermann R, Dhakal B, Bal S, Giri S, et
al: Minimal residual disease response-adapted therapy in newly
diagnosed multiple myeloma (MASTER): Final report of the
multicentre, single-arm, phase 2 trial. Lancet Haematol.
10:e890–e901. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
D'Agostino M, Bertuglia G, Rota-Scalabrini
D, Belotti A, More S, Corradini P, Oliva S, Ledda A, Grasso M,
Pavone V, et al: Predictors of unsustained minimal residual disease
negativity in multiple myeloma (MM) Patients. Blood.
1432023.doi:10.1182/blood.2023022080.
|
|
152
|
Medford AJ, Moy B, Spring LM, Hurvitz SA,
Turner NC and Bardia A: Molecular residual disease in breast
cancer: Detection and therapeutic interception. Clin Cancer Res.
29:4540–4548. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Patel RP, Somasundram PM, Smith LK,
Sheppard KE and Mcarthur GA: The therapeutic potential of targeting
minimal residual disease in melanoma. Clin Transl Med.
13:e11972023. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Honore N, van Marcke C, Galot R, Helaers
R, Ambroise J, van Maanen A, Mendola A, Dahou H, Marbaix E, Van
Eeckhout P, et al: Tumor-agnostic plasma assay for circulating
tumor DNA detects minimal residual disease and predicts outcome in
locally advanced squamous cell carcinoma of the head and neck. Ann
Oncol. 34:1175–1186. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Pott C, Jurinovic V, Trotman J, Kehden B,
Unterhalt M, Herold M, Jagt RV, Janssens A, Kneba M, Mayer J, et
al: Minimal residual disease status predicts outcome in patients
with previously untreated follicular lymphoma: A prospective
analysis of the Phase III GALLIUM study. J Clin Oncol. 42:550–561.
2024. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Yang K, Hu H, Wu J, Wang H, Guo Z, Yu W,
Yao L, Ding F, Zhou T, Wang W, et al: Letter to the Editor:
Clinical utility of urine DNA for noninvasive detection and minimal
residual disease monitoring in urothelial carcinoma. Mol Cancer.
22:252023. View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Mo S, Ye L, Wang D, Han L, Zhou S, Wang H,
Dai W, Wang Y, Luo W, Wang R, et al: Early detection of molecular
residual disease and risk stratification for stage I to III
colorectal cancer via circulating tumor DNA Methylation. JAMA
Oncol. 9:770–778. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Slater S, Bryant A, Chen HC, Begum R, Rana
I, Aresu M, Peckitt C, Zhitkov O, Lazaro-Alcausi R, Borja V, et al:
ctDNA guided adjuvant chemotherapy versus standard of care adjuvant
chemotherapy after curative surgery in patients with high risk
stage II or stage III colorectal cancer: A multi-centre,
prospective, randomised control trial (TRACC Part C). BMC Cancer.
23:2572023. View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Armakolas A, Kotsari M and Koskinas J:
Liquid biopsies, novel approaches and future directions. Cancers
(Basel). 15:15792023. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Xie J, Yao W, Chen L, Zhu W, Liu Q, Geng
G, Fang J, Zhao Y, Xiao L, Huang Z and Zhao J: Plasma ctDNA
increases tissue NGS-based detection of therapeutically targetable
mutations in lung cancers. BMC Cancer. 23:2942023. View Article : Google Scholar : PubMed/NCBI
|