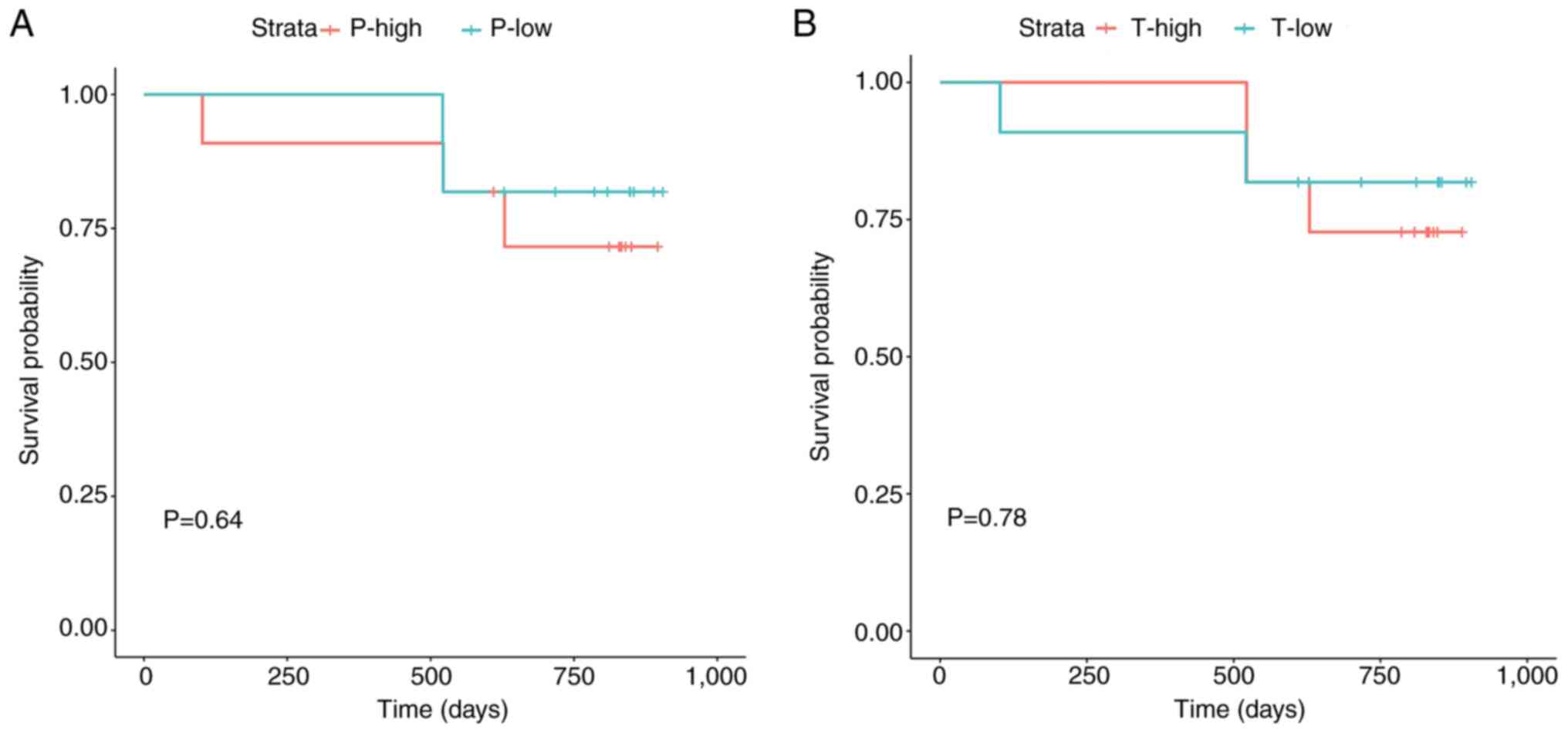
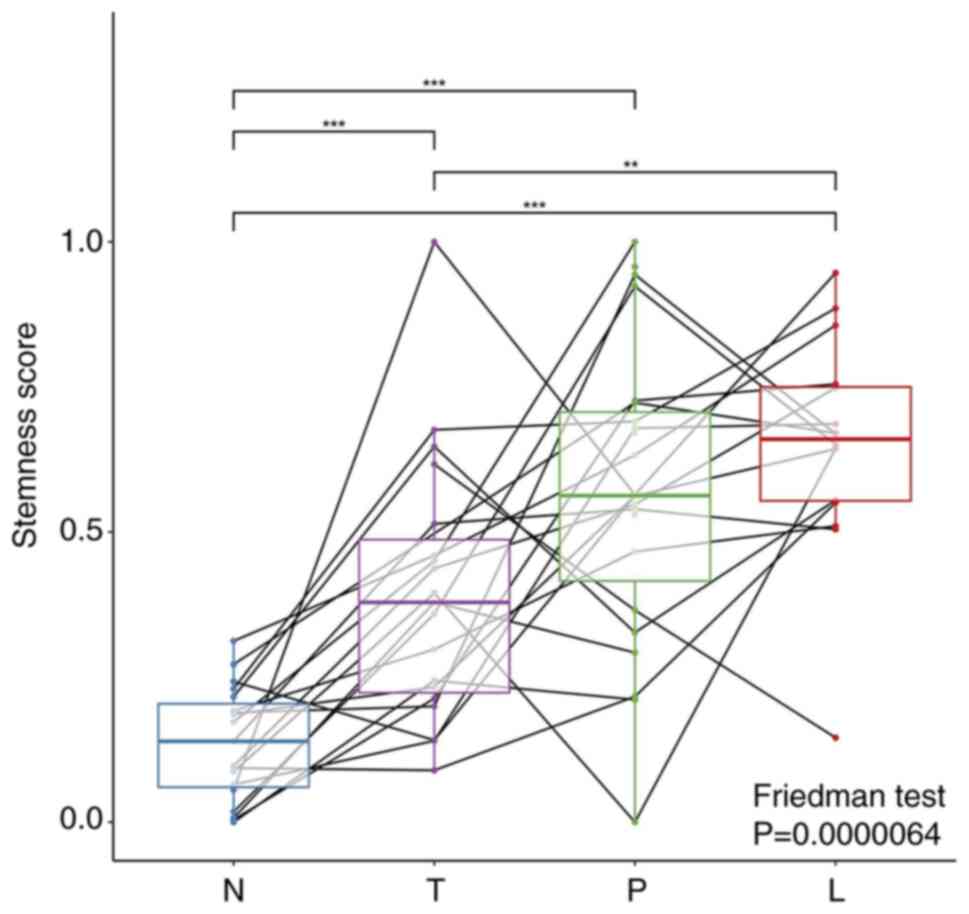
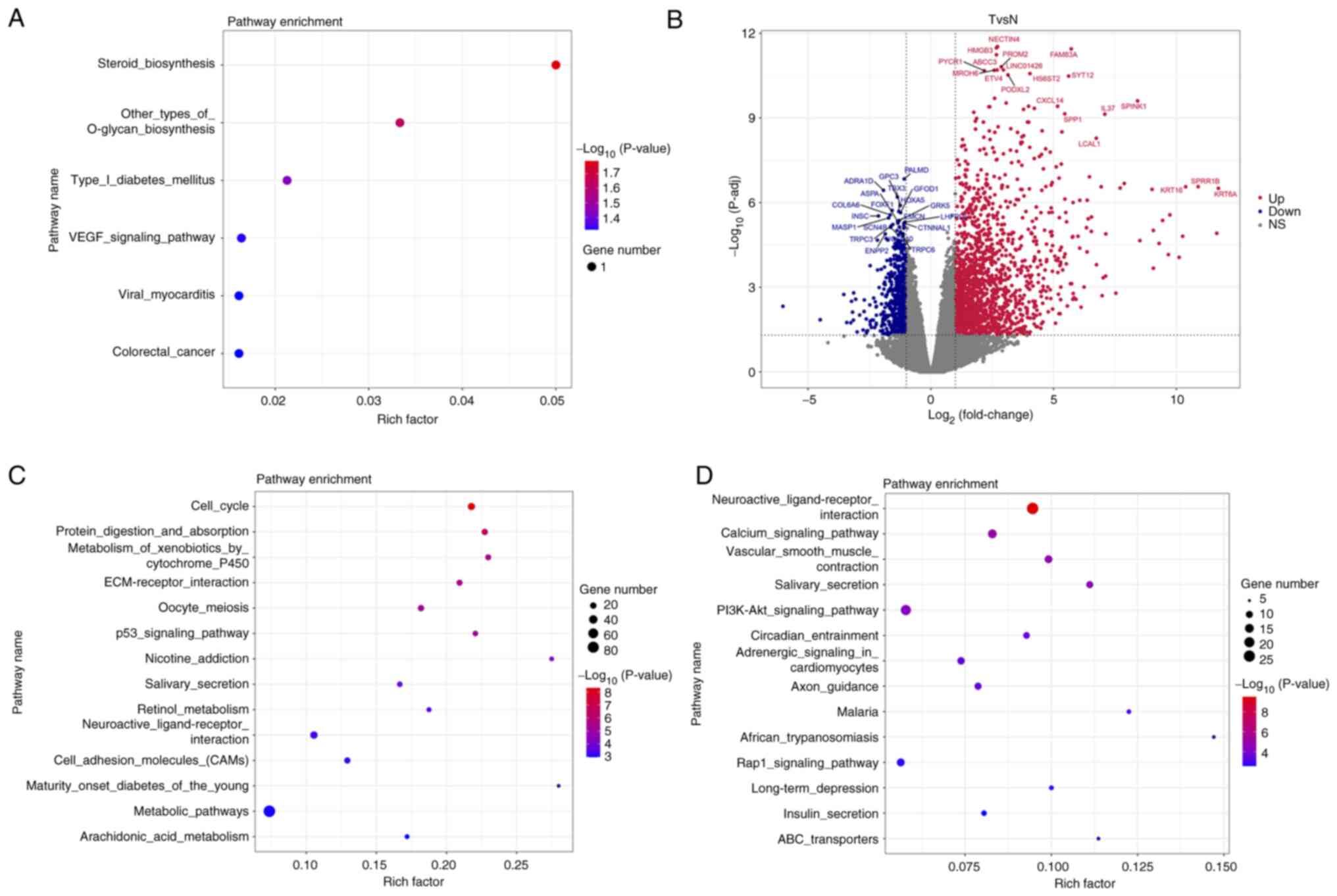
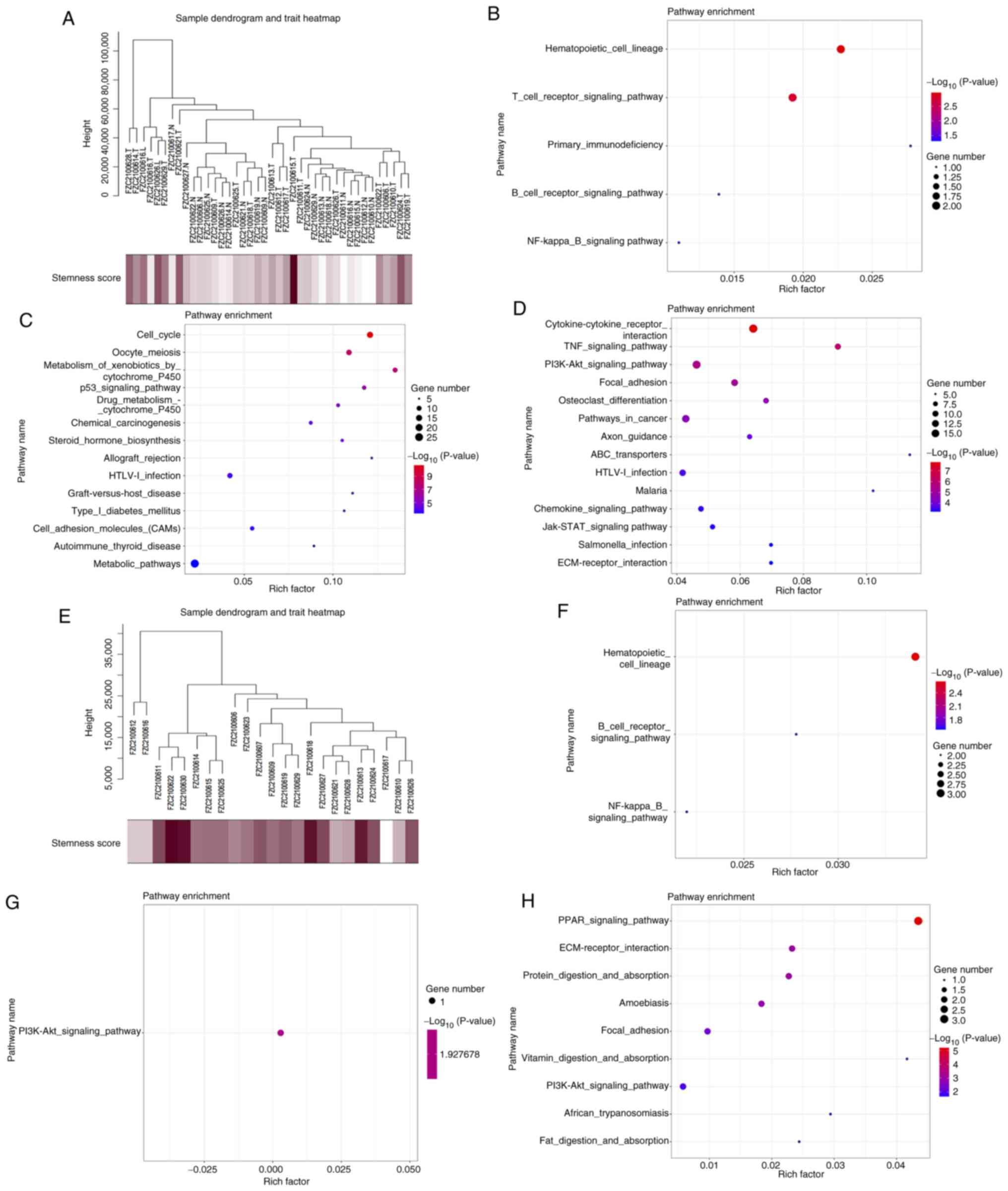
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
Globocan estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Samarth N, Gulhane P and Singh S:
Immunoregulatory framework and the role of miRNA in the
pathogenesis of NSCLC-A systematic review. Front Oncol.
12:10893202022. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shi JF, Wang L, Wu N, Li JL, Hui ZG, Liu
SM, Yang BY, Gao SG, Ren JS, Huang HY, et al: Clinical
characteristics and medical service utilization of lung cancer in
China, 2005–2014: Overall design and results from a multicenter
retrospective epidemiologic survey. Lung Cancer. 128:91–100. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shi X, Liu Y, Cheng S, Hu H, Zhang J, Wei
M, Zhao L and Xin S: Cancer stemness associated with prognosis and
the efficacy of immunotherapy in adrenocortical carcinoma. Front
Oncol. 11:6516222021. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bhuvaneswari MS, Priyadharsini S,
Balaganesh N, Theenathayalan R and Hailu TA: Investigating the lung
adenocarcinoma stem cell biomarker expressions using machine
learning approaches. Biomed Res Int. 2022:35181902022. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wan R, Liao H, Liu J, Zhou L, Yin Y, Mu T
and Wei J: Development of a 5-gene signature to evaluate lung
adenocarcinoma prognosis based on the features of cancer stem
cells. Biomed Res Int. 2022:44044062022. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Su C, Zheng J, Chen S, Tuo J, Su J, Ou X,
Chen S and Wang C: Identification of key genes associated with
cancer stem cell characteristics in Wilms' tumor based on
bioinformatics analysis. Ann Transl Med. 10:12042022. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Malta TM, Sokolov A, Gentles AJ,
Burzykowski T, Poisson L, Weinstein JN, Kamińska B, Huelsken J,
Omberg L, Gevaert O, et al: Machine learning identifies stemness
features associated with oncogenic dedifferentiation. Cell.
173:338–354.e15. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liao Y, Xiao H, Cheng M and Fan X:
Bioinformatics analysis reveals biomarkers with cancer stem cell
characteristics in lung squamous cell carcinoma. Front Genet.
11:4272020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hou S, Xu H, Liu S, Yang B, Li L, Zhao H
and Jiang C: Integrated bioinformatics analysis identifies a new
stemness index-related survival model for prognostic prediction in
lung adenocarcinoma. Front Genet. 13:8602682022. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li N, Li Y, Zheng P and Zhan X: Cancer
stemness-based prognostic immune-related gene signatures in lung
adenocarcinoma and lung squamous cell carcinoma. Front Endocrinol
(Lausanne). 12:7558052021. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li RY and Liang ZY: Circulating tumor DNA
in lung cancer: Real-time monitoring of disease evolution and
treatment response. Chin Med J (Engl). 133:2476–2485. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Heitzer E, Haque IS, Roberts CES and
Speicher MR: Current and future perspectives of liquid biopsies in
genomics-driven oncology. Nat Rev Genet. 20:71–88. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Roskams-Hieter B, Kim HJ, Anur P, Wagner
JT, Callahan R, Spiliotopoulos E, Kirschbaum CW, Civitci F,
Spellman PT, Thompson RF, et al: Plasma cell-free RNA profiling
distinguishes cancers from pre-malignant conditions in solid and
hematologic malignancies. NPJ Precis Oncol. 6:282022. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Larson MH, Pan W, Kim HJ, Mauntz RE,
Stuart SM, Pimentel M, Zhou Y, Knudsgaard P, Demas V, Aravanis AM
and Jamshidi A: A comprehensive characterization of the cell-free
transcriptome reveals tissue- and subtype-specific biomarkers for
cancer detection. Nat Commun. 12:23572021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sorber L, Zwaenepoel K, Jacobs J, De Winne
K, Goethals S, Reclusa P, Van Casteren K, Augustus E, Lardon F,
Roeyen G, et al: Circulating cell-free DNA and RNA analysis as
liquid biopsy: Optimal centrifugation protocol. Cancers (Basel).
11:4582019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Seneviratne C, Shetty AC, Geng X,
McCracken C, Cornell J, Mullins K, Jiang F and Stass S: A pilot
analysis of circulating cfRNA transcripts for the detection of lung
cancer. Diagnostics (Basel). 12:28972022. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Goldstraw P, Chansky K, Crowley J,
Rami-Porta R, Asamura H, Eberhardt WE, Nicholson AG, Groome P,
Mitchell A, Bolejack V, et al: The IASLC lung cancer staging
project: Proposals for revision of the TNM stage groupings in the
forthcoming (Eighth) edition of the TNM classification for lung
cancer. J Thorac Oncol. 11:39–51. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jacobsen N, Nielsen PS, Jeffares DC,
Eriksen J, Ohlsson H, Arctander P and Kauppinen S: Direct isolation
of poly(A)+ RNA from 4 M guanidine thiocyanate-lysed cell extracts
using locked nucleic acid-oligo(T) capture. Nucleic Acids Res.
32:e642004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Colaprico A, Silva TC, Olsen C, Garofano
L, Cava C, Garolini D, Sabedot TS, Malta TM, Pagnotta SM,
Castiglioni I, et al: TCGAbiolinks: An R/Bioconductor package for
integrative analysis of TCGA data. Nucleic Acids Res. 44:e712016.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen D, Liu J, Zang L, Xiao T, Zhang X, Li
Z, Zhu H, Gao W and Yu X: Integrated machine learning and
bioinformatic analyses constructed a novel stemness-related
classifier to predict prognosis and immunotherapy responses for
hepatocellular carcinoma patients. Int J Biol Sci. 18:360–373.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Langfelder P and Horvath S: WGCNA: An R
package for weighted correlation network analysis. BMC
Bioinformatics. 9:5592008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Robinson MD, McCarthy DJ and Smyth GK:
edgeR: A Bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang X, Wang F, Yan F, Huang D, Wang H,
Gao B, Gao Y, Hou Z, Lou J, Li W and Yan J: Identification of a
novel HOOK3-FGFR1 fusion gene involved in activation of the
NF-kappaB pathway. Cancer Cell Int. 22:402022. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Trapnell C, Roberts A, Goff L, Pertea G,
Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL and Pachter L:
Differential gene and transcript expression analysis of RNA-seq
experiments with TopHat and Cufflinks. Nat Protoc. 7:562–578. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhao Y, Jia S, Zhang K and Zhang L: Serum
cytokine levels and other associated factors as possible
immunotherapeutic targets and prognostic indicators for lung
cancer. Front Oncol. 13:10646162023. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shai S, Patolsky F, Drori H, Scheinman EJ,
Davidovits E, Davidovits G, Tirman S, Arber N, Katz A and Adir Y: A
novel, accurate, and non-invasive liquid biopsy test to measure
cellular immune responses as a tool to diagnose early-stage lung
cancer: A clinical trials study. Respir Res. 24:522023. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mathios D, Johansen JS, Cristiano S,
Medina JE, Phallen J, Larsen KR, Bruhm DC, Niknafs N, Ferreira L,
Adleff V, et al: Detection and characterization of lung cancer
using cell-free DNA fragmentomes. Nat Commun. 12:50602021.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sugimoto A, Matsumoto S, Udagawa H,
Itotani R, Usui Y, Umemura S, Nishino K, Nakachi I, Kuyama S, Daga
H, et al: A large-scale prospective concordance study of plasma-
and tissue-based next-generation targeted sequencing for advanced
non-small cell lung cancer (LC-SCRUM-Liquid). Clin Cancer Res.
29:1506–1514. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang S, Meng F, Li M, Bao H, Chen X, Zhu
M, Liu R, Xu X, Yang S, Wu X, et al: Multidimensional cell-free DNA
fragmentomic assay for detection of early-stage lung cancer. Am J
Respir Crit Care Med. 207:1203–1213. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen M, Wang X, Wang W, Gui X and Li Z:
Immune- and stemness-related genes revealed by comprehensive
analysis and validation for cancer immunity and prognosis and its
nomogram in lung adenocarcinoma. Front Immunol. 13:8290572022.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang H, Wang Y, Luo W, Zhang X, Cao R,
Yang Z, Duan J and Wang K: Integrative stemness characteristics
associated with prognosis and the immune microenvironment in lung
adenocarcinoma. BMC Pulm Med. 22:4632022. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Müller S, Janke F, Dietz S and Sültmann H:
Circulating MicroRNAs as potential biomarkers for lung cancer.
Recent Results Cancer Res. 215:299–318. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Narayanan S, Wu ZX, Wang JQ, Ma H,
Acharekar N, Koya J, Yoganathan S, Fang S, Chen ZS and Pan Y: The
spleen tyrosine kinase inhibitor, entospletinib (GS-9973) restores
chemosensitivity in lung cancer cells by modulating ABCG2-mediated
multidrug resistance. Int J Biol Sci. 17:2652–2665. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Marchetti P, Antonov A, Anemona L,
Vangapandou C, Montanaro M, Botticelli A, Mauriello A, Melino G and
Catani MV: New immunological potential markers for triple negative
breast cancer: IL18R1, CD53, TRIM, Jaw1, LTB, PTPRCAP. Discov
Oncol. 12:62021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Park H, Kang S, Kim I, Kim S, Kim HJ, Shin
DY, Kim DY, Lee KH, Ahn JS, Sohn SK, et al: The association of
genetic alterations with response rate in newly diagnosed chronic
myeloid leukemia patients. Leuk Res. 114:1067912022. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wu R, Liu S, Lv G, Deng C, Wang R, Zhang
S, Zhu D, Wang L, Lei Y and Luo Z: First-line crizotinib therapy is
effective for a novel SEC31A-anaplastic lymphoma kinase fusion in a
patient with stage IV lung adenocarcinoma: A case report and
literature reviews. Anticancer Drugs. 34:294–301. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zou Z, Gu Y, Liang L, Hao X, Fan C, Xin T,
Zhao S, Liu Z, Guo Y, Ma K, et al: Alectinib as first-line
treatment for advanced ALK-positive non-small cell lung cancer in
the real-world setting: Preliminary analysis in a Chinese cohort.
Transl Lung Cancer Res. 11:2495–2506. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Shaw AT, Solomon BJ, Chiari R, Riely GJ,
Besse B, Soo RA, Kao S, Lin CC, Bauer TM, Clancy JS, et al:
Lorlatinib in advanced ROS1-positive non-small-cell lung cancer: A
multicentre, open-label, single-arm, phase 1–2 trial. Lancet Oncol.
20:1691–1701. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Guo W, Liang J, Zhang D, Huang X and Lv Y:
Lung adenocarcinoma harboring complex EML4-ALK fusion and BRAF
V600E co-mutation responded to alectinib. Medicine (Baltimore).
101:e309132022. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ettinger DS, Wood DE, Aisner DL, Akerley
W, Bauman JR, Bharat A, Bruno DS, Chang JY, Chirieac LR, D'Amico
TA, et al: NCCN guidelines insights: Non-small cell lung cancer,
version 2.2021. J Natl Compr Canc Netw. 19:254–266. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Steven A, Fisher SA and Robinson BW:
Immunotherapy for lung cancer. Respirology. 21:821–833. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Liu P, Liu Y, Chen L, Fan Z, Luo Y and Cui
Y: Anemoside A3 inhibits macrophage M2-like polarization to prevent
triple-negative breast cancer metastasis. Molecules. 28:16112023.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Najafi M, Goradel NH, Farhood B, Salehi E,
Nashtaei MS, Khanlarkhani N, Khezri Z, Majidpoor J, Abouzaripour M,
Habibi M, et al: Macrophage polarity in cancer: A review. J Cell
Biochem. 120:2756–2765. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Chen H, Lin R, Lin W, Chen Q, Ye D, Li J,
Feng J, Cheng W, Zhang M and Qi Y: An immune gene signature to
predict prognosis and immunotherapeutic response in lung
adenocarcinoma. Sci Rep. 12:82302022. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Fridman WH, Zitvogel L, Sautès-Fridman C
and Kroemer G: The immune contexture in cancer prognosis and
treatment. Nat Rev Clin Oncol. 14:717–734. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Müller L, Tunger A, Plesca I, Wehner R,
Temme A, Westphal D, Meier F, Bachmann M and Schmitz M:
Bidirectional crosstalk between cancer stem cells and immune cell
subsets. Front Immunol. 11:1402020. View Article : Google Scholar : PubMed/NCBI
|