Introduction
Gastric cancer (GC) is one of the most common
malignant tumors of the digestive system. It is the fifth leading
cause of cancer-related deaths and the fourth leading cause of
cancer-related morbidity (1). In
addition to chronic Helicobacter pylori (H. pylori)
infection, the carcinogenesis and progression of GC involve complex
genetic and epigenetic alterations, including chromosomal
instability, abnormalities in oncogenes, tumor suppressor genes,
several growth factors, DNA repair genes and metabolic
disturbances. Based on high throughput sequencing data in The
Cancer Genome Atlas (TCGA), GC has been categorized into four
molecular subtypes: i) Epstein-Barr virus (EBV)-positive; ii)
microsatellite-instable; iii) genomically stable; iv) and
chromosomally unstable (2).
Although the TCGA molecular subtypes provide a promising
application for stratified management and targeted therapy, the
prognosis for most patients remains poor. In contrast to the
EBV-positive subtype, which has an abundance of lymphoid stroma
that allows for possible immune checkpoint inhibition, most cases
of Epstein-Barr virus-negative GC (EBVnGC) lack targeted
therapeutic drugs due to tumor heterogeneity (2,3). Thus,
finding a potential biomarker for promising effective therapeutic
targets will become an essential strategy for GC, especially for
EBVnGC.
SH3 domain-binding glutamic acid-rich protein-like3
(SH3BGRL3), also named tumor necrosis factor inhibitory protein 1,
belongs to the thioredoxin superfamily, which maps to chromosome
1p34.3-p35 with 279 nucleotides (4). It encoding cytoplasmic protein is 27
kDa, showing a highly similar structure to that of the glutathione
peroxidase of Escherichia coli, an enzyme with
oxidoreductase activity (5).
SH3BGRL3 is a highly evolutionarily conserved gene in the early
stages of zebrafish embryonic development and diverse organogenesis
(6). Previous studies have reported
that SH3BGRL3 is upregulated and its expression is closely
associated with poor outcomes in certain human malignancies,
including urothelial carcinoma, clear cell renal carcinoma and
glioblastoma (7–13). In these studies, SH3BGRL3 was
reported to interact with the EGFR family, including ErbB1/EGFR and
ErbB2/human epidermal growth factor receptor 2 (HER-2) (10–12).
Despite this, the role of SH3BGRL3 expression and its clinical
significance in GC, particularly in EBVnGC, remains unclear.
The relationship between glucose metabolism,
diabetes and the development of GC has been reported in several
studies (14–17). Tumor cells rely on glucose for
energy supplements, and the status of hyperglycemia contributes to
tumor growth through the effects of genetic mutations, epigenetic
modification and proteomic alteration (14). Altered glucose metabolism, including
aerobic glycolysis and a dysfunctional mitochondrial oxidative
phosphorylation (OXPHOS) system, has been recognized as a critical
hallmark characteristic of cancer cells (18). In the process of a biochemical
reaction, many intermediate metabolic products are released, which
could promote tumor growth and induce alterations in immune
responses within the tumor microenvironment (TME) (19). It has been reported that
SH3BGRL3 potentiates the functions of several carcinogenic
pathways, including PI3K, Akt and Mammalian target of rapamycin
(mTOR), all of which are involved in glycolysis in tumor cells
(9–11). Nevertheless, whether SH3BGRL3 is
involved in glucose metabolism in patients with GC remains elusive.
Exploring the relationships between SH3BGRL3 expression and
preoperative blood glucose concentration and their possible
underlying mechanisms would be beneficial.
The expression of SH3BGRL3 and its associations with
clinicopathological parameters and patient outcomes were assessed
in the present study to determine its prognostic significance in
GC. To determine the messenger (m)RNA level of SH3BGRL3, RNA
sequencing data obtained from public databases and mRNA detection
by reverse transcription (RT)-quantitative (q)PCR on fresh tissues
were evaluated. Tissue microarrays (TMA) and immunohistochemistry
(IHC) were performed to assess the expression of SH3BGRL3 protein
in GC. In addition, a functional enrichment analysis of
differentially expressed genes (DEGs) and immune infiltrating cells
related to SH3BGRL3 were assessed and visualized.
Materials and methods
Data mining from public databases
Online bioinformatic tools, including the Tumor
Immune Estimation Resource 2.0 (TIMER 2.0; http://timer.cistrome.org), the Human Protein Atlas
(HPA; proteinatlas.org) and the Gene Expression Profiling
Interactive Analysis (GEPIA; http://gepia2.cancer-pku.cn/) databases were used to
assess the levels of SH3BGRL3 mRNA in GC tissues and their related
normal tissues. |log2 fold change|>1 and a q-value
<0.01 was considered significant. The Kaplan-Meier plotter
(https://kmplot.com/analysis/) was
employed for the analysis of the Gene Expression Omnibus (GEO)
database (https://www.ncbi.nlm.nih.gov/geo/; including GSE14210,
GSE15459, GSE22377, GSE29272, GSE51105 and GSE62254) to investigate
the association between SH3BGRL3 expression, overall survival (OS)
and post-progression survival (PPS).
To assess the potential functions of SH3BGRL3, the
related DEGs were identified from TCGA_STAD RNA-sequencing data
using LinkedOmics (http://www.linkedomics.org/), and functional
enrichment analysis was performed. Kyoto Encyclopedia of Genes and
Genomes (KEGG) pathway and Gene Ontology (GO) analyses, comprising
biological process (BP), cellular component (CC) and molecular
function (MF), were performed using the top 1,000 DEGs (|r|>0.3
and P<0.05) with the R package ‘cluster profiler’ (version
3.0.4; http://bioconductor.org/packages/clusterProfiler/).
Gene set enrichment analysis (GSEA) was performed using the
Molecular Signatures Database (MSigDB; http://www.gsea-msigdb.org/gsea/msigdb/collections.jsp),
according to the median levels of SH3BGRL3 expression, and an
enrichment with normalized enrichment score >1 or <-1, and
false discovery rate q-value <0.05 was considered significant.
In addition, GeneMANIA (http://genemania.org/) and Protein-Protein Interaction
Networks Functional Enrichment Analysis (STRING; version 11.5;
http://cn.string-db.org) were used to evaluate
the associated networks of SH3BGRL3. TIMER and Tumor and Immune
System Interaction Database (TISIDB; http://cis.hku.hk/), two online web servers, were used
to assess the relationships between SH3BGRL3 gene expression and
infiltrating immune cells in the TCGA_STAD database. In TIMER and
TISIDB, the Estimating the Proportion of Immune and Cancer cells
calculation was used to predict the tumor immune score.
Clinical populations
A total cohort of 607 consecutive patients from
Fujian Provincial Hospital (Fuzhou, China), who were pathologically
diagnosed as GC between January 2014 and December 2015, was
enrolled. The ages of the patients ranged from 27–89 years, with a
median age of 63 years. Two pathologists (HQL and LYC) reviewed
original hematoxylin and eosin (H&E) slides and recorded the
clinicopathological parameters of the patients from the hospital
medical systems. Preoperative blood glucose was also recorded. OS
was defined as the duration from the date of initial diagnosis to
the date of death. An additional cohort including seven pairs of
fresh EBVnGC tissues and their corresponding normal gastric tissues
were also collected at Fujian Provincial Hospital (Fuzhou, China)
between August 2021 and November 2021. In this cohort, the ratio of
men to women was 3:4, with a median age of 74 years (range, 54–75
years). The inclusion criteria of the aforementioned patient were
as follows: i) Pathologically diagnosed with gastric
adenocarcinoma; ii) available paired cancer tissues and
para-carcinoma tissues; and iii) radical gastrectomy performed.
Furthermore, the exclusion criteria were as follows: i) Recurrent
or metastatic disease; ii) neoadjuvant therapy administered; iii)
mixed adenocarcinoma and neuroendocrine tumors; iv) loss to
follow-up; and v) death within 1 month after surgery. The diagnosis
and staging were based on the 5th edition of the World Health
Organization (WHO) classification and staging of tumors of the
digestive system: Gastric tumors (20).
Assessment of tumor budding (BUD)
Similarly, two pathologists assessed the status of
BUD according to the guidelines of the International Tumor Budding
Consensus Conference (21).
Microscopically, a single tumor cell or a cell cluster of ≤4 tumor
cells in the invasive front area was identified as a BUD. BUD was
assessed in the ‘hot spot’ region at a ×200 high power field (Leica
light microscope DM3000; Leica Microsystems GmbH), which was
converted to standardized numbers every 0.785 mm2. BUD
was classified as the following: Bd1, 0–4 buds; Bd2, 5–9 buds; and
Bd3, ≥10 buds. Based on the results, cases were graded as budding
low (Bd1) and budding high (Bd2/3). A consensus review using a
multi-head microscope was performed when inconsistencies arose.
TMA construction
A TMA was constructed for IHC staining (Fig. S1A). For the donor formalin-fixed
paraffin-embedded (FFPE) tissues fixed with 10% neutral formalin
(PH 7.2–7.4; room temperature for 18 h), the representative regions
of each H&E-stained slide were labeled (Fig. S1B), including the GC tumor and the
paired adjacent normal gastric tissues. Cores with a 1.5-mm
diameter at each corresponding area were taken and transferred into
the ‘recipient’ paraffin blocks by punching tissue cylinders. The
resulting array had nine cores across (x-axis) and seven cores down
(y-axis). Unstained 4-µm sections of the TMA were prepared and then
adhered to SuperFrostPlus™ glass slides (Matsunami Glass
Ind., Ltd.). For every tissue array block, one slide was stained
with H&E to confirm the presence of representative tumors. The
TMA slides were stained with H&E using the HistoCore SPECTRA
Workstation (Leica Biosystems GmbH) according to a preset program.
The TMAs were placed on the burner at 70°C for 30 min and then
sequentially went through the steps of dewaxing, dehydration,
hematoxylin staining, differentiation, bluing, eosin staining,
dehydration, clearing and cover-slipping (Table SI).
IHC staining and scoring
For the IHC of SH3BGRL3, TMA sections were incubated
overnight at 4°C with a 1:500 dilution of rabbit SH3BGRL3
antibodies (cat. no. HPA030848; Sigma-Aldrich; Merck KGaA),
followed by secondary antibodies and DAB. IHC was performed as
described previously (13). The
SH3BGRL3 immunostaining was assessed microscopically, and ≥3 high
power fields (Leica light microscope DM3000; ×200) in hotspot areas
were imaged, respectively. According to the operating instructions
and the ImageJ software (https://imagej.nih.gov/ij/index.html; version 1.53 h),
the intensity of SH3BGRL3 staining was calculated and represented
with an average optical density (AOD). The optimal cut-off value of
the SH3BGRL3 expression was calculated by the R package
‘survminer’.
The TMA slides were also stained with the following
antibodies: MutL protein homolog 1 (MLH1) (cat. no. MX063),
postmeiotic segregation increased 2 (PMS2) (cat. no. EP51), MutS
homolog (MSH)2 (cat. no. MX061), MSH6 (cat. no. MX056) and EGFR
(cat. no. SP111), using the Lumatas platform (Fuzhou Maixin
Biotechnology Development Co., Ltd.). The IHC automated staining
protocol was performed according to the manufacturer's
instructions. In brief, after being deparaffinized in dewaxing
fluid at 50°C for 5 min, the 4-µm sections of the TMA were treated
with Max2inOneTM LP (Fuzhou Maixin Biotechnology Development Co.,
Ltd.) at 99°C for 20 min. The samples were cooled to room
temperature, washed three times with PBS, blocked with 3%
H2O2 at 32°C for 10 min and washed with PBS
twice. The primary antibodies were added to the section and the
section was incubated at 32°C for 30 min. After final washing with
PBS twice, pre-diluted HRP-Polymer Goat Anti-Rabbit or Mouse IgG
(cat. no. TT0801, Fuzhou Maixin Biotechnology Development Co.,
Ltd.) was dropped onto the slices and the slices were incubated for
10 min at 32°C, and then washed with PBS twice. The DAB detection
kit chromogenic liquid was added to the sections for 5 min and then
stopped with PBS twice. After washing with water twice, the section
was redyed with hematoxylin for 25 sec at 26°C, followed by PBS for
30 sec. The slices were dehydrated using a series of concentrations
of ethyl alcohol and treated with xylene for 3 min at room
temperature. Finally, the sections were sealed with neutral gum and
observed under a microscope. HER-2 (cat. no. 4B5) staining was
performed on 4-µm sections cut from the FFPE blocks using a
Benchmark ULTRA immunostainer (Roche Tissue Diagnostics). The HER-2
IHC automated staining protocol was performed as described
previously (22). The details of
antibodies, incubation conditions and antigen retrieval are listed
in Table SII. PBS was used as the
negative control.
EGFR staining was characterized as membranous and/or
cytoplasmic. EGFR staining was assessed as 0, 1, 2 or 3 according
to the recommendations in previous literature (23): 0, negative or weak staining in
<10% of tumor cells; 1+, weak staining in >10% of tumor
cells; 2+, moderate staining in >10% of tumor cells; and 3+,
intense staining in >10% of tumor cells (Fig. S2). Tumors classified as 3+ were
considered to have high EGFR expression. HER-2 immunostaining
(Fig. S1C) was scored according to
the American Society of Clinical Oncology (ASCO) guidelines
(24). The four mismatch repair
(MMR) proteins, including MLH1, PMS2, MSH2 and MSH6, showed nuclear
staining, which were classified as proficient (p)MMR (no loss of
MMR proteins) or defective (d)MMR (≥1 losses of MMR proteins)
(25). Immunohistochemical staining
was observed under a Leica light microscope DM3000 (Leica
Microsystems GmbH).
In situ hybridization (ISH) and
fluorescence ISH (FISH)
Unstained 4-µm sections of FFPE tissue fixed with
10% neutral formalin (pH 7.2–7.4; 26°C) were prepared for the ISH
and FISH assays. The ISH assay was performed with EBV-encoded small
RNA (EBER) probe (cat. no. PB0589; Leica Biosystems GmbH) using an
automatic staining device (Bond-III; Leica Biosystems GmbH). The
EBER-ISH automated staining protocol was performed according to the
manufacturer's instructions as previously described (26). In brief, slides were automatically
deparaffinized three times at 72°C for 1 min with BondDewax (cat.
no. AR9222, Leica Biosystems GmbH), rinsed with gradient alcohols
and four times with BOND Wash (cat. no. AR9590-CN; Leica Biosystems
GmbH) at room temperature. After incubation with enzyme proteinase
K (cat. no. RE7160-k; Leica Biosystems GmbH) for 15 min at 37°C.
The Ready-to-Use EBER probe (containing formamide) was applied for
the slides and incubated at 26°C for 2 h. After blocked with 3%
H2O2 at 26°C for 5 min, the slides were
incubated with BOND Ready-to-Use Anti-Fluorescein Antibody (cat.
no. AR0222; Leica Biosystems GmbH) for 15 min, post-primary reagent
for 8 min and polymer for 8 min (all at 26°C), with four times BOND
Wash rinses between steps, 3–5 sec each time. After the final
polymer incubation, slides were rinsed twice with BOND Wash and
once with distilled water. Staining was performed with DAB for 10
min at 26°C, followed by rinsing in distilled water, hematoxylin
counterstaining for 25 sec at 26°C, rinsing in BOND Wash and
distilled water at 26°C and cover-slipping. The results of the ISH
assay were observed under a Leica light microscope DM3000 (Leica
Microsystems GmbH). The brown nuclear staining was considered
positive. The known EBER-positive nasopharyngeal carcinoma tissues
were used as the positive control, and a sense probe for EBER was
used as the negative control.
The FISH assay was performed with PathVysion HER-2
DNA Probe Kit II (Abbott Pharmaceutical Co., Ltd.). All the
experimental operations were carried out according to the
manufacturer's instructions. In summary, prior to hybridization,
slices were deparaffinized with xylene twice for 10 min each and
hydration with ethyl alcohol (100, 95 and 75%) and distilled water,
all 5 min at room temperature. Following these, sections were
boiling in distilled water for 20 min and cooled to room
temperature. Subsequently, the sections were treated with enzyme
proteinase K (cat. no. 03L7860; Abbott Pharmaceutical Co., Ltd.) at
37°C for 10 min, and then washed with 2× SSC solution for 1 min at
room temperature. After that, the slices underwent dehydration with
gradient alcohols (100, 95 and 75%, all 3 min) at room temperature.
Then the PathVysion HER-2 DNA Probe Kit II was applied before
denaturation at 75°C for 5 min and hybridization at 37°C for After
14 h, the slides were washed with NP-40 (0.3%, pH 7.0–7.5) at 72°C
for two min to remove any unbound probes and counterstained with
DAPI at room temperature for 10 min. PathVysion HER-2 DNA Probe kit
II consists of two labeled DNA probes. The LSI HER-2/neu probe that
spans the HER2 gene (17q11.2–12) is labeled in SpectrumOrange,
while the CEP17 probe is labeled in SpectrumGreen and hybridizes to
the alpha satellite DNA located at the centromere of chromosome 17
(17p11.1-q11.1). Inclusion of the CEP17 probe allows detection of
the relative copy number of the HER2 gene. FISH signals were
assessed using an Olympus fluorescence microscope BX63 (Olympus
Corporation; objective lens, ×100), and the results were assessed
using the methods described in the ASCO guidelines (24). The total numbers of HER2 and CEP17
signals were counted in 20 adjacent interphase tumor cell nuclei,
using a fluorescence microscope and appropriate filters. The ratios
of HER2 signals to CEP17 signals were calculated regardless of IHC
status as follows: When the ratio was <1.8, the gene was
considered non-amplified, and when it was >2.2, the gene was
considered to be amplified. If the ratio was within the range of
1.8 to 2.2 at the initial count, an additional 20 tumor cells were
counted. If the final ratio for 40 nuclei was 2.0 or higher, the
case was deemed to have HER2 amplification (Fig. S1D).
RT-qPCR
Total RNA was extracted using TRIzol™
reagent (Takara Bio, Inc.). The cDNA was synthesized using the
PrimeScript™ RT Reagent Kit (cat. no. RR037; Takara Bio,
Inc.). Subsequently, cDNA was obtained following the completion of
reverse transcription at 37°C for 15 min and 85°C for 5 sec, during
which time the enzyme was inactivated. qPCR reactions were
performed using TB Green (cat. no. RR820; Takara Bio, Inc.) using
the LightCycler® 480 Real-Time PCR System (Roche
Diagnostics). The reaction conditions were as follows:
Pre-denaturation at 95°C for 30 sec, followed by 95°C for 5 sec and
60°C for 34 sec, 45 cycles; melting reaction at 95°C for 15 sec,
60°C for 1 min and 95°C for 15 sec, 1 cycle. The relative SH3BGRL3
mRNA expression was calculated using the 2−ΔΔCq method
(27). All the experimental
operations were carried out according to the manufacturer's
instructions. The sequences of the primers used are as follows:
SH3BGRL3 (forward), 5′-CCACCCCAGATTGTCAACGG-3′; SH3BGRL3 (reverse),
5′-TCAAGCCAGCTTCAGGAACTC-3′; GAPDH (forward),
5′-GGTGTGAACCATGAGAAGTATGA-3′; and GAPDH (reverse),
5′-GAGTCCTTCCACGATACCAAAG-3′.
Statistical analysis
Graphs were generated and statistical analysis was
performed using GraphPad Prism 9.0 (Dotmatics), RStudio (V1.4.110)
software (http://www.r-project.org/) and SPSS
Statistics 26.0 (IBM Corp.), respectively. An unpaired t-test was
employed for the analysis of SH3BGRL3 expression utilizing the
GEPIA database and TIMER database, and the AOD of SH3BGRL3
staining. The expression of SH3BGRL3 mRNA in the local datasets was
analyzed using a paired t-test. The correlation between SH3BGRL3
expression and clinicopathological parameters was evaluated using
Pearson's χ2 test. A log-rank test was performed to
perform survival analysis, and RStudio generated Kaplan-Meier
curves. Univariate and multivariate Cox regression analyses were
performed to analyze the prognostic variables associated with OS. A
nomogram was generated by RStudio based on the results of the
multivariate Cox regression analysis. Moreover, the predictive
ability was evaluated with the concordance index (C-index), and
calibration plots were generated to compare the predicted
probability of OS with the observed outcome. P<0.05 was
considered to indicate a statistically significant difference.
Results
SH3BGRL3 is upregulated and associated
with poor prognosis in GC in analysis of public databases
A comprehensive analysis of SH3BGRL3 expression in
normal human tissue and cancers was conducted using data from the
HPA and GEPIA. The results demonstrated that SH3BGRL3 was widely
distributed in human tissues and cancers (Fig. S3). The datasets from TIMER, a data
analysis platform, were analyzed with 415 TCGA_STAD and 35
corresponding normal samples. The results revealed that SH3BGRL3
was significantly upregulated in multiple malignancies compared
with the corresponding normal tissues, including GC (P<0.05;
Fig. 1A). Moreover, single-cell
analysis from the HPA revealed that SH3BGRL3 was weakly expressed
in normal gastric mucus-secreting cells, which was lower than that
in immune cells and muscle fiber cells (Fig. 1B).
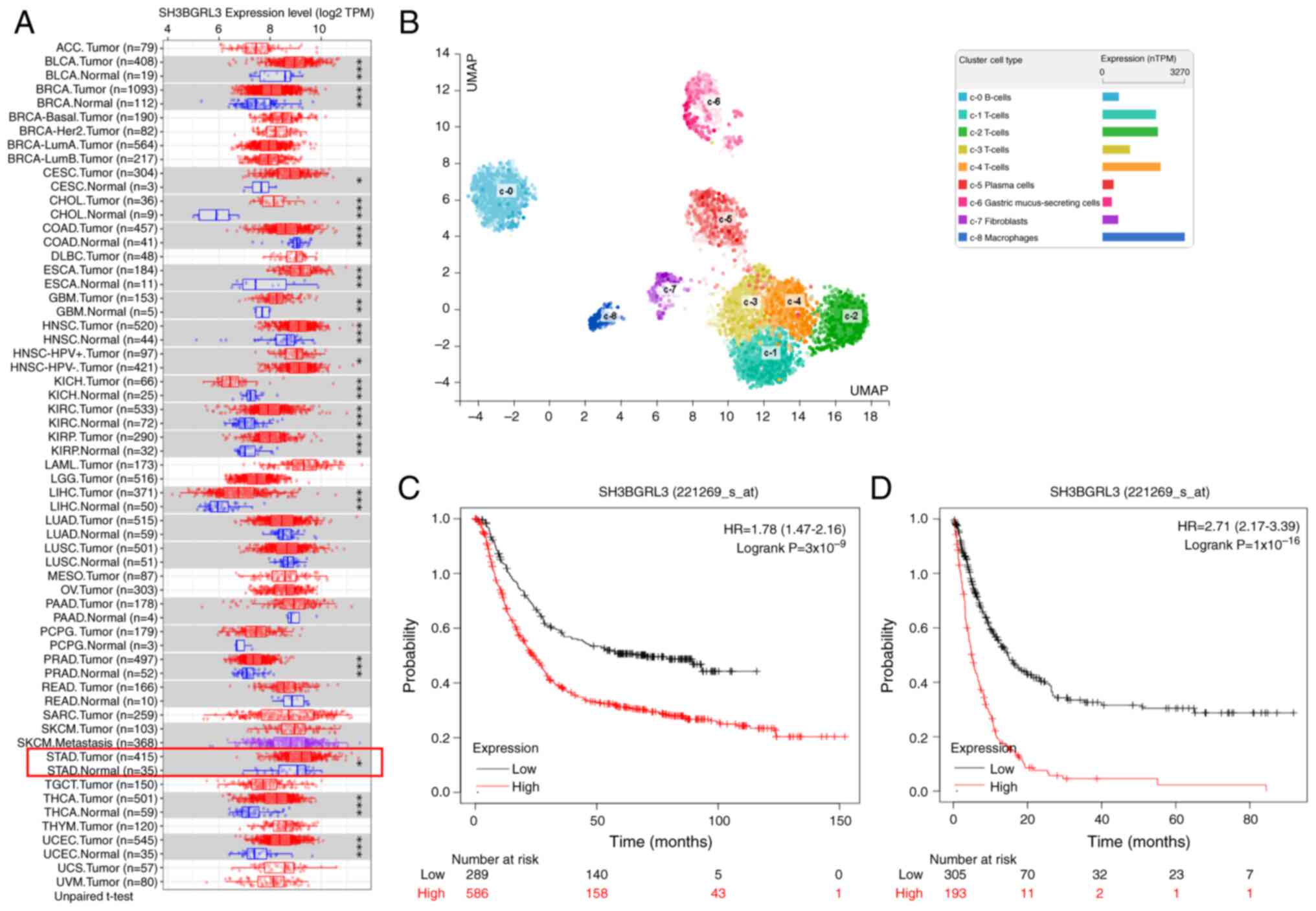 | Figure 1.Expression and prognosis analyses of
SH3BGRL3 in GC using different databases. (A) The Tumor Immune
Estimation Resource revealed that SH3BGRL3 was remarkably aberrant
and expressed in several human cancers, including GC. (B)
Single-cell analysis demonstrated that SH3BGRL3 was weakly
expressed in normal gastric epithelial cells. Kaplan-Meier plotters
revealed that SH3BGRL3 expression was significantly associated with
(C) overall survival and (D) post-progression survival in GC.
*P<0.05, **P<0.01, ***P<0.001, unpaired t-test. SH3BGRL3,
SH3 domain-binding glutamic acid-rich protein-like 3; GC, gastric
cancer; TPM, transcripts per million; nTPM, number of TPM; UMAP,
uniform manifold approximation and projection; HR, hazard ratio;
CI, confidence interval. |
Subsequently, the effect of SH3BGRL3 expression on
GC prognosis was assessed using the GEO database. A total of 875
and 498 GC samples were used to analyze the association between
SH3BGRL3 expression and OS and PPS, respectively. Kaplan-Meier
plots demonstrated that a high expression of SH3BGRL3 was
significantly associated with poor OS (P<0.0001; Fig. 1C) and PPS (P<0.0001; Fig. 1D) in patients with GC. Patients with
high SH3BGRL3 expression were 1.78 times more likely to die than
patients with low expression. The median time of OS was 23.6 months
for the high SH3BGRL3 group and 70.2 months for the low expression
group. Parallel to the OS results, patients with high SH3BGRL3
expression had a 2.71-fold higher risk of relapse than those with
low expression. In the high SH3BGRL3 group, the median time for PPS
was 4.9 months, whilst in the low expression group, the median time
was 14.8 months.
Furthermore, the stratified analysis revealed that
patients with GC with the characteristics of pathological
tumor-node-metastasis (pTNM) stage III, pTNM stage IV, lymph node
involvement, intestinal histological type, male sex and no HER-2
amplification, could be divided into groups with different outcomes
based on SH3BGRL3 expression. Cases with low SH3BGRL3 expression
had significantly longer OS compared with those with high
expression (Fig. S4). This
suggests that a high expression of SH3BGRL3 is an adverse factor
for the outcomes of patients with GC.
High SH3BGRL3 expression is associated
with high preoperative blood glucose concentration and aggressive
clinicopathological characteristics in patients with EBVnGC
As that the mRNA level of SH3BGRL3 was upregulated
in GC, the present study subsequently assessed SH3BGRL3 protein
expression in GC. First, TMA-based immunostaining was used to
detect SH3BGRL3 protein expression in nonselective GC and adjacent
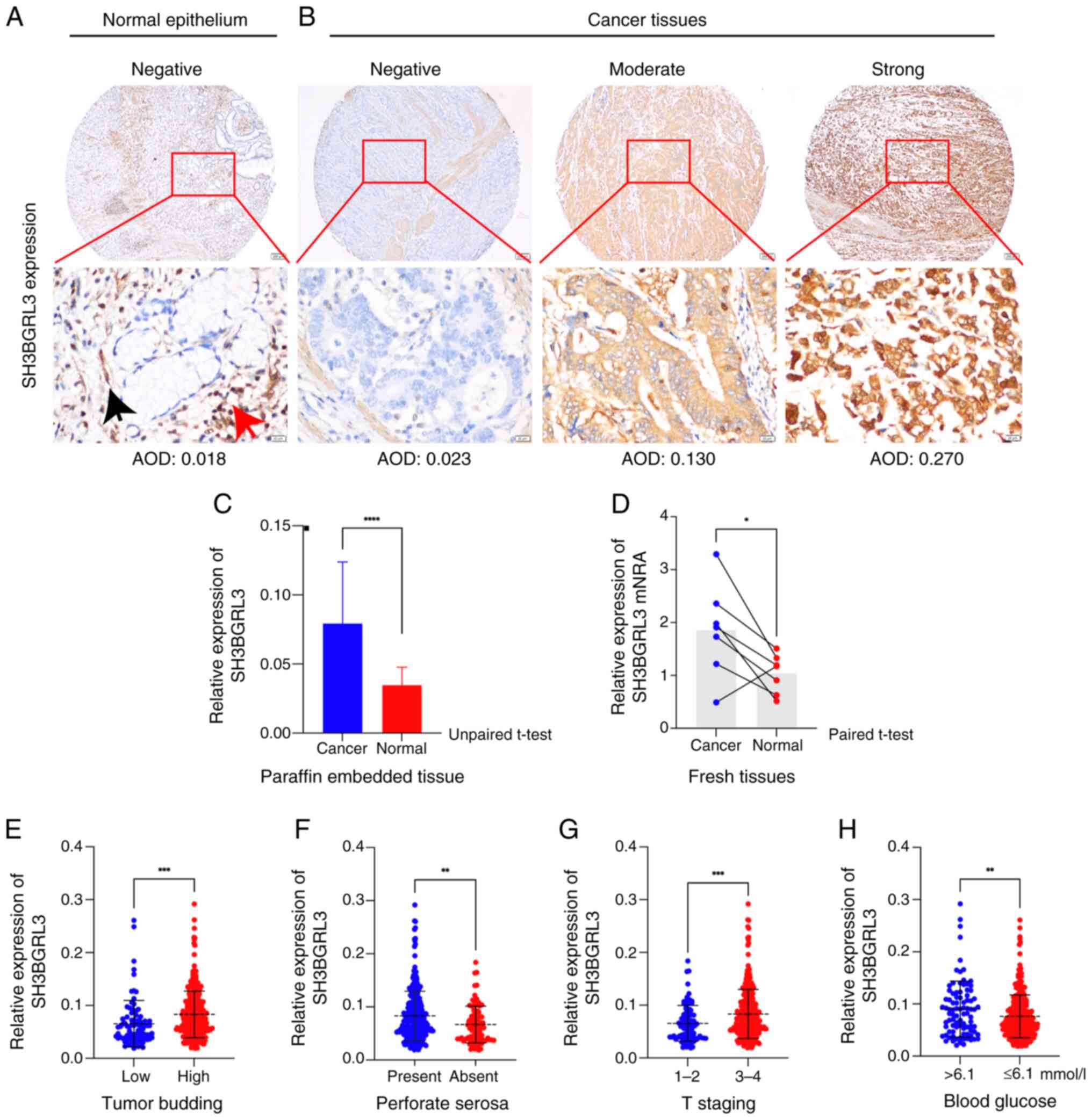
normal gastric tissues (Fig. 2).
There was weak or negative staining of SH3BGRL3 protein in normal
gastric epithelial cells, similar to the results of single-cell
sequencing (Fig. 2A). By contrast,
there was stronger staining in the cytoplasm of the lymphocytes and
muscle fibrocytes (Figs. 2A and
S5C) and GC cells (Figs. 2B and S5C). Subsequently, due to the abundant
lymphoid stroma in EBV-positive gastric cancers (Fig. S5A and B), the present study
evaluated the role of SH3BGRL3 in EBVnGC. An ISH assay of EBER was
performed, and the EBV-negative cases were selected for further
analysis. Finally, 398 patients with EBVnGC with complete
clinicopathological and follow-up data were enrolled, including 287
(72.1%) males and 111 (27.9%) females (Table I). The male-to-female ratio was
2.5:1, and the age ranged from 27–89 years, with a median age of
63. Of the 398 patients, 201 died, and the average follow-up time
was 45.58 months (ranging from 1–87 months).
 | Table I.Association between SH3
domain-binding glutamic acid-rich protein-like 3 expression and
clinicopathological features in patients with Epstein-Barr
virus-negative gastric cancer. |
Table I.
Association between SH3
domain-binding glutamic acid-rich protein-like 3 expression and
clinicopathological features in patients with Epstein-Barr
virus-negative gastric cancer.
|
|
| SH3BGRL3
expression |
|
|---|
|
|
|
|
|
|---|
| Parameter | Total (n=398) | High (n=313) | Low (n=85) | P-value |
|---|
| Sex |
|
|
| 0.3700 |
|
Male | 287 | 229 | 58 |
|
|
Female | 111 | 84 | 27 |
|
| Age |
|
|
| 0.5600 |
| >65
years | 152 | 118 | 34 |
|
| ≤65
years | 246 | 195 | 51 |
|
| Tumor location |
|
|
| 0.8690 |
| Body
and cardia | 231 | 181 | 50 |
|
|
Antrum | 167 | 132 | 35 |
|
| Perforate
serosa |
|
|
| <0.0001 |
|
Presence | 304 | 255 | 49 |
|
|
Absence | 94 | 58 | 36 |
|
| WHO grading |
|
|
| 0.8620 |
|
G1-2 | 201 | 158 | 43 |
|
| G3 | 197 | 155 | 42 |
|
| T staging |
|
|
| <0.0001 |
|
T1-T2 | 92 | 56 | 36 |
|
|
T3-T4 | 306 | 257 | 49 |
|
| N staging |
|
|
| 0.0080 |
| N0 | 117 | 79 | 38 |
|
|
N1-3 | 291 | 244 | 47 |
|
| pTNM staging |
|
|
| 0.0010 |
|
I–II | 153 | 107 | 46 |
|
|
III–IV | 245 | 206 | 39 |
|
| Cancer embolus |
|
|
| 0.0030 |
|
Presence | 105 | 72 | 33 |
|
|
Absence | 293 | 241 | 52 |
|
| Perineural
invasion |
|
|
| <0.001 |
|
Presence | 182 | 129 | 53 |
|
|
Absence | 216 | 184 | 32 |
|
| BUD |
|
|
| <0.0001 |
|
BUD-L | 88 | 53 | 35 |
|
|
BUD-H | 310 | 260 | 50 |
|
| Lauren type |
|
|
| 0.4610 |
|
IGC | 235 | 187 | 48 |
|
|
nIGC | 163 | 126 | 37 |
|
| MMR |
|
|
| 0.3970 |
|
dMMR | 87 | 67 | 20 |
|
|
pMMR | 311 | 246 | 65 |
|
| HER-2a amplification |
|
|
| 0.2800 |
|
Positive | 44 | 37 | 7 |
|
|
Negative | 334 | 260 | 74 |
|
| EGFR
expression |
|
|
| 0.0130 |
|
High | 37 | 35 | 2 |
|
|
Low | 361 | 278 | 83 |
|
For SH3BGRL3 expression, the mean AOD of normal
gastric epithelial cells was 0.0348±0.0129 (range, 0.017–0.071;
Fig. 2A). Furthermore, the mean AOD
of SH3BGRL3 staining in GC tissues was 0.0792±0.0445 (range,
0.019–0.292; Fig. 2B), which was
significantly higher than that in normal gastric epithelial cells
(P<0.0001; Fig. 2C). The mRNA
level of SH3BGRL3 was also demonstrated to be significantly
upregulated in the EBVnGC tissues compared with that in its
corresponding normal tissues (P=0.039; Fig. 2D and Table SIII). Moreover, high SH3BGRL3
expression was strongly significantly associated with higher BUD
(P=0.001; Fig. 2E), the presence of
perforate serosa (P=0.0021; Fig.
2F), a higher tumor (T) stage (P=0.0008; Fig. 2G) and a blood glucose level of
>6.1 mmol/l (P=0.0096; Fig. 2H)
compared with low SH3BGRL3 expression.
High SH3BGRL3 expression is an
independent adverse prognostic factor in patients with EBVnGC
To further assess the role of SH3BGRL3 in EBVnGC,
the present study dichotomized the SH3BGRL3 expression into high or
low groups (cut-off=0.0467, calculated using RStudio software). The
results demonstrated that 313 cases had high expression and 85
cases had low expression. In comparison with low expression, high
SH3BGRL3 expression was significantly associated with the presence
of tumor perforate serosa (P<0.0001), perineural invasion
(P<0.001), lymphovascular embolus (P<0.01), pathological T
stage (P<0.0001), pathological node stage (P<0.01), pTNM
stage (P<0.001) and high BUD (P<0.0001). In addition, high
SH3BGRL3 expression was significantly associated with higher EGFR
expression, in comparison with low SH3BGRL3 expression (P=0.013;
Table I).
Subsequently, the associations between SH3BGRL3
protein expression and outcomes in patients with EBVnGC were
analyzed. Kaplan Meier analysis indicated that SH3BGRL3 expression
was an influential prognostic factor in patients with EBVnGC.
Patients with EBVnGC with high SH3BGRL3 expression demonstrated
worse outcomes for OS compared with those with low expression
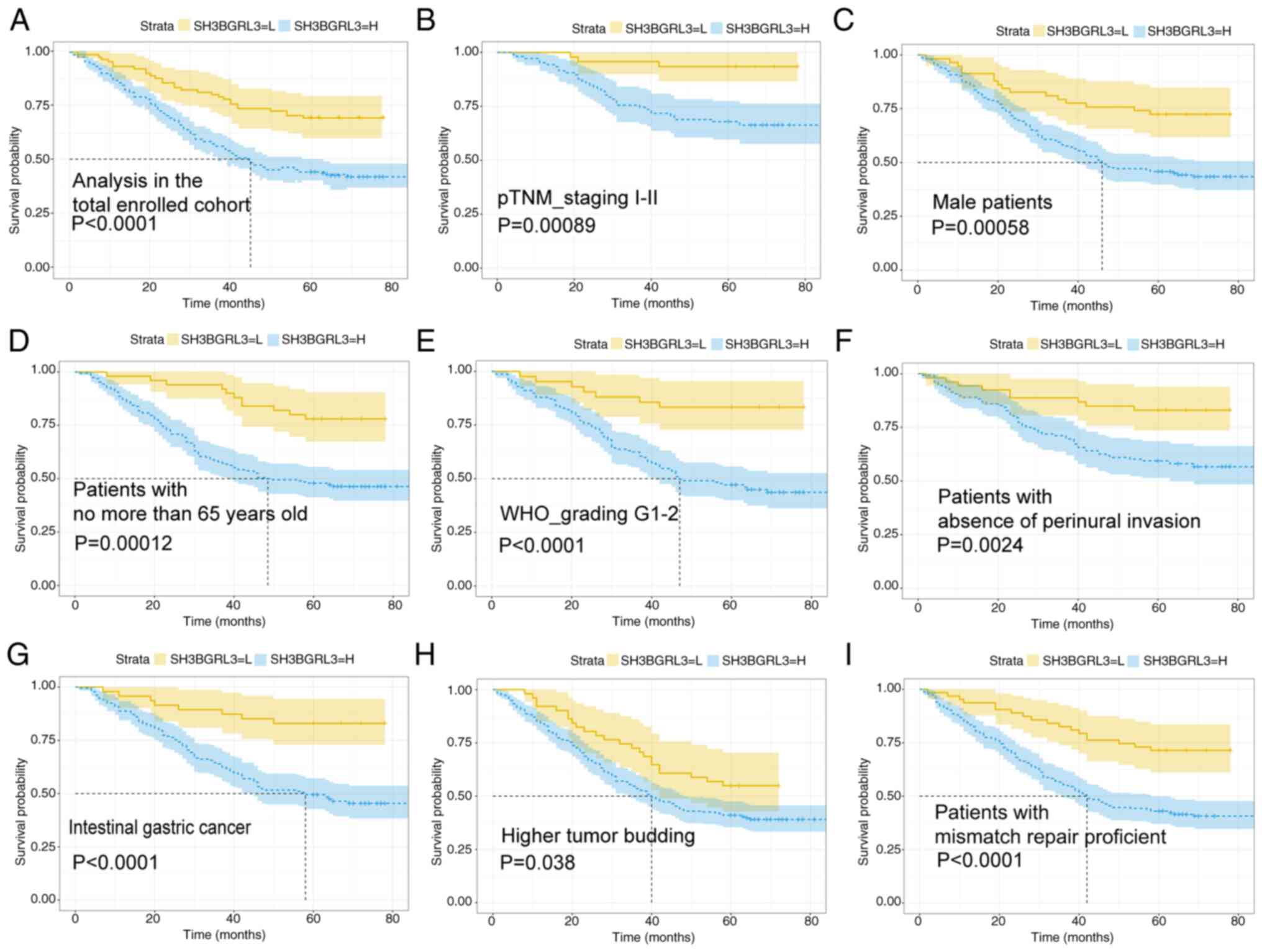
(Logrank=15.085; P<0.0001; Fig. 3A). Furthermore, subgroup analysis
suggested that, compared with patients with EBVnGC with low
expression, patients with high SH3BGRL3 expression had a worse OS
rate in the following subgroups: TNM I–II stage (Logrank=11.037;
P<0.001; Fig. 3B), male sex
(Logrank=11.459; P<0.001; Fig.
3C), aged ≤65 years (Logrank=14.352; P<0.001; Fig. 3D), WHO grade 1–2 (Logrank=15.222;
P<0.0001; Fig. 3E), absence of
perineural invasion (Logrank=9.231; P<0.01; Fig. 3F), Lauren intestinal type (20) (Logrank=15.517; P<0.0001; Fig. 3G), high BUD (Logrank=4.099; P=0.038;
Fig. 3H) and proficient mismatch
repair status (Logrank=15.240; P<0.0001; Fig. 3I). Additionally, multivariate Cox
analysis demonstrated that SH3BGRL3 expression was an independent
prognostic factor in patients with EBVnGC. Patients with high
SH3BGRL3 expression had an increased risk of death compared with
those with low SH3BGRL3 phenotype [hazard ratio (HR), 1.666; 95%
confidence interval (CI), 1.093–2.541; P=0.018; Table II].
 | Table II.Univariate and multivariate analysis
of prognostic factors in patients with Epstein-Barr virus-negative
gastric cancer. |
Table II.
Univariate and multivariate analysis
of prognostic factors in patients with Epstein-Barr virus-negative
gastric cancer.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Parameter | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age (>65 vs. ≤65
years) | 1.363
(1.031–1.802) | 0.0300 | 1.269
(0.950–1.697) | 0.107 |
| Sex (Male vs.
female) | 0.923
(0.680–1.252) | 0.6060 | NA | NA |
| Tumor location
(Body and cardia vs. antrum) | 1.171
(0.882–1.554) | 0.2740 | NA | NA |
| WHO grading (G3 vs.
G1-2) | 1.285
(0.973–1.692) | 0.0780 | 1.480
(0.987–2.219) | 0.058 |
| Lauren types (IGC
vs. nIGC) | 0.674
(0.511–0.889) | 0.0050 | 0.662
(0.442–0.991) | 0.038 |
| Perineural invasion
(Presence vs. absence) | 2.233
(1.730–3.146) | <0.0001 | 1.534
(1.118–2.106) | 0.008 |
| Cancer embolus
(Presence vs. absence) | 3.676
(2.398–5.637) | <0.0001 | 1.741
(1.030–2.943) | 0.038 |
| Perforate serosa
(Presence vs. absence) | 3.519
(2.238–5.533) | <0.0001 | 1.387
(0.394–4.878) | 0.610 |
| T staging (T3-4 vs.
T1-2) | 3.632
(2.287–5.767) | <0.0001 | 1.019
(0.291–3.832) | 0.977 |
| N staging (N1-3 vs.
N0) | 2.923
(2.054–4.164) | <0.0001 | 1.088
(0.660–1.794) | 0.742 |
| pTNM staging
(III–IV vs. I–II) | 3.629
(2.555–5.154) | <0.0001 | 2.953
(1.326–3.179) | <0.001 |
| BUD (BUD-H vs.
BUD-L) | 2.897
(1.859–4.513) | <0.0001 | 1.840
(1.145–2.958) | 0.012 |
| SH3BGRL3 expression
(High vs. low) | 2.228
(1.475–3.366) | <0.0001 | 1.666
(1.093–2.541) | 0.018 |
| Blood glucose
(>6.1 vs. ≤6.1 mmol/l) | 1.572
(1.155–2.141) | 0.0040 | 1.515
(1.109–2.069) | 0.009 |
Subsequently, a prognostic nomogram was constructed
to predict the 3- and 5-year survival probabilities in patients
with EBVnGC, including the parameters where P<0.1 in the results
of multivariate cox regression analysis using the backward method.
Finally, a nomogram integrated eight factors, including WHO grading
(HR, 1.480; P=0.058), Lauren types (HR, 0.662; P=0.038), perineural
invasion (HR=1.534; P=0.008), cancer embolus (HR, 1.741; P=0.038),
pTNM stage (HR, 2.953; P<0.001), BUD (HR, 1.840; P=0.012), blood
preoperative glucose level (HR, 1.515; P=0.009) and SH3BGRL3
expression (HR, 1.666; P=0.018; Table
II and Fig. 4A). The results
revealed that the predictive accuracy of the nomogram was good,
with a C-index of 0.740 (95% CI, 0.706–0.773). The calibration
curves for the 3- and 5-year survival prediction indicated a good
accuracy and consistency between predicted probabilities and actual
observations (Fig. 4B and C). These
results suggest that the nomogram has good discriminative ability
and reliability for predicting prognosis.
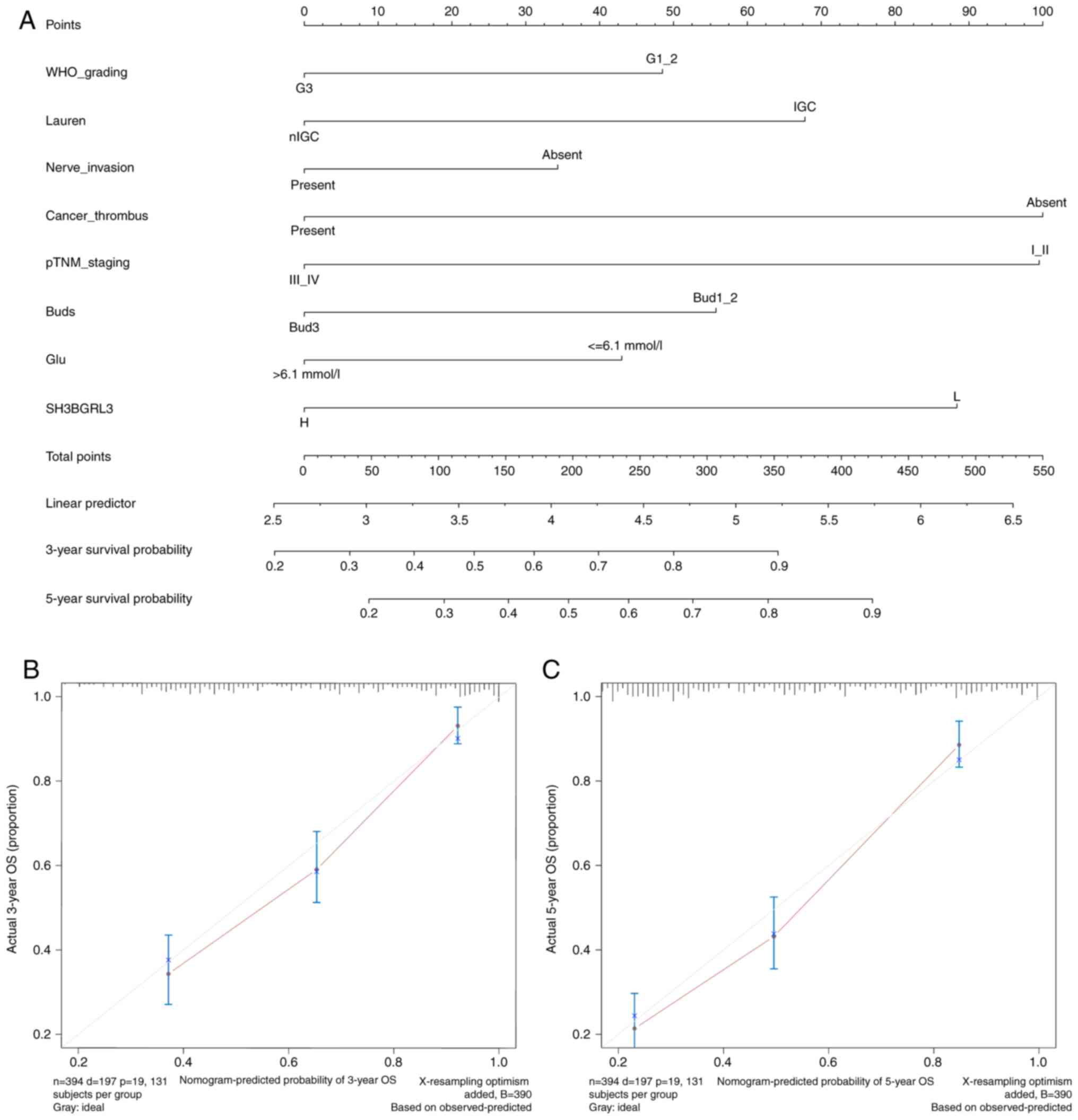 | Figure 4.Nomogram for predicting the prognosis
of patients with EBVnGC. (A) Nomogram predicting the 3-year and
5-year OS of patients with EBVnGC, including the parameters of WHO
grading, Lauren types, perineural invasion, tumor embolus, pTNM
staging, tumor budding, blood glucose level and SH3BGRL3
expression. The total score was calculated by adding the scores
corresponding to each variable, and the total scores predicted the
3-year or 5-year survival probability of a patient to the lowest
survival rate scale. Nomogram calibration for (B) 3-year and (C)
5-year OS. EBVnGC, Epstein-Barr virus-negative gastric cancer; OS,
overall survival; WHO, World Health Organization; pTNM,
pathological tumor-node-metastasis; SH3BGRL3, SH3 domain-binding
glutamic acid-rich protein-like 3; IGC, intestinal gastric cancer;
nIGC, non-IGC; Bud, tumor budding; Glu, glucose level. |
DEGs of SH3BGRL3 and associated
pathways in GC
The present study used the DEGs of SH3BGRL3 to
analyze and visualize the possible underlying mechanisms of
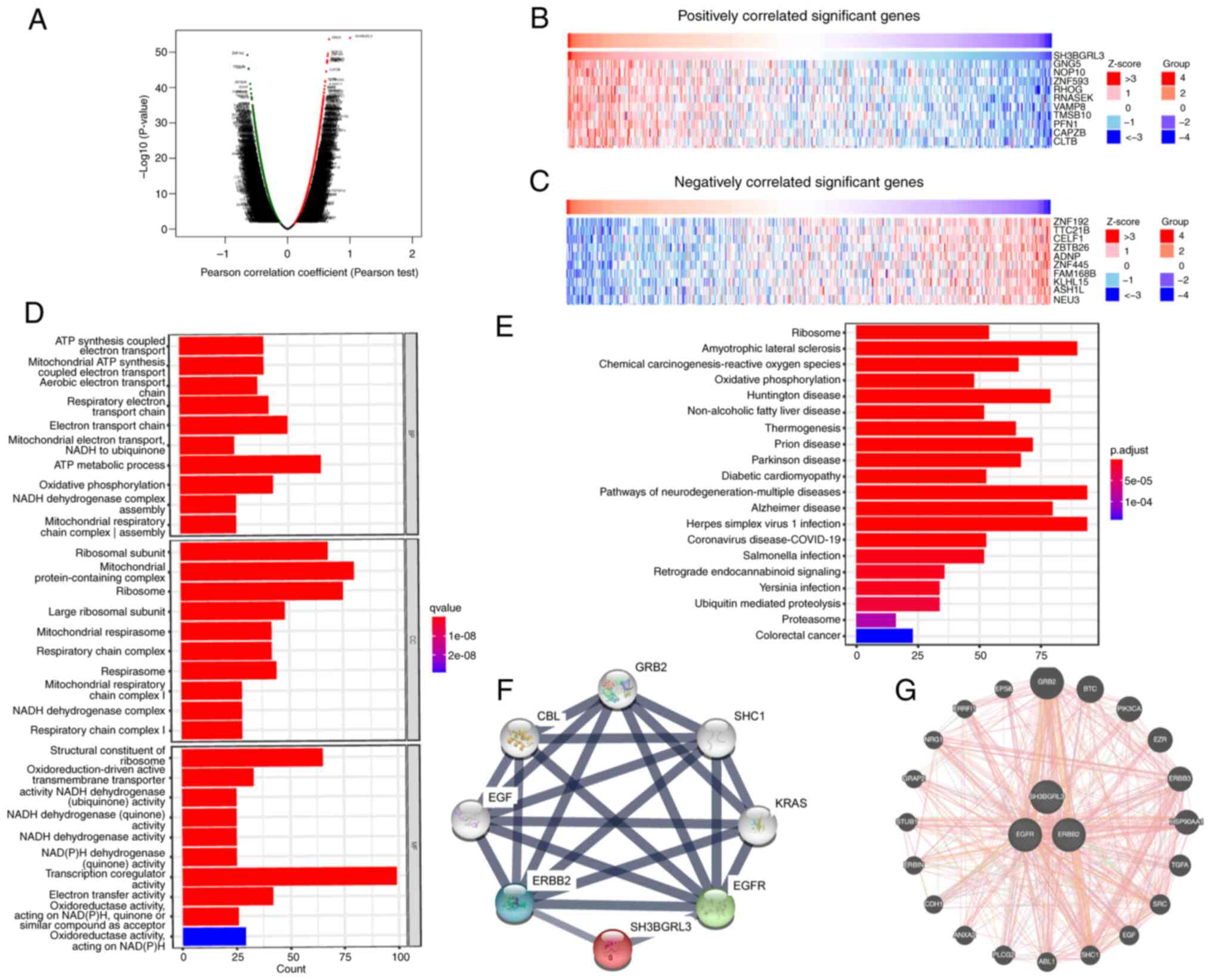
SH3BGRL3 in GC. LinkedOmics revealed that a total of 4,403 genes
(dark red dots) were significantly positively correlated with
SH3BGRL3, whilst 7,101 genes (dark green dots) were significantly
negatively correlated with SH3BGRL3 (Fig. 5A). The top 10 positively correlated
genes identified were GNG5, NOP10, ZNF593, RHOG, RNASEK, VAMP8,
TMSB0, PFN1, CAPZB and CLTB (Fig. 5B). Meanwhile, the top 10 negatively
correlated genes identified were ZNF192, TTC21B, CELF1, ZBTB26,
ADNP, ZNF445, FAM168B, KLHL15, ASH1L and NEU3 (Fig. 5C).
A GO and KEGG pathway analysis was performed using
RStudio software to evaluate the biological function and molecular
mechanism of SH3BGRL3-associated genes. The top 1,000 positively
and negatively correlated DEGs were selected for GO and KEGG
pathway analysis. In the GO analysis, BP terms were implicated in
ATP metabolism processes, the electron transport chain and OXPHOS.
CC terms were implicated in mitochondrial protein-containing
complexes, ribosomes and ribosomal subunits. Moreover, it was
demonstrated that MF terms serves a significant role in
transcription coregulator activities, ribosome structural
components and electron transfer activities (Fig. 5D). The KEGG pathway analysis
indicated the SH3BGRL3-related signaling pathways were enriched in
multiple neurodegenerative diseases, ribosome, diabetic
cardiomyopathy, OXPHOS, coronavirus disease 2019, colorectal cancer
and the proteasome (Fig. 5E).
Furthermore, the related interactive networks of SH3BGRL3 were
predicted by the online servers of STRING (Fig. 5F) and GeneMANIA (Fig. 5G). It was demonstrated that SH3BGRL3
protein and mRNA were co-expressed and physically interacted with
ErbB1/EGFR and ErbB2/HER-2.
To further assess the possible biological pathways
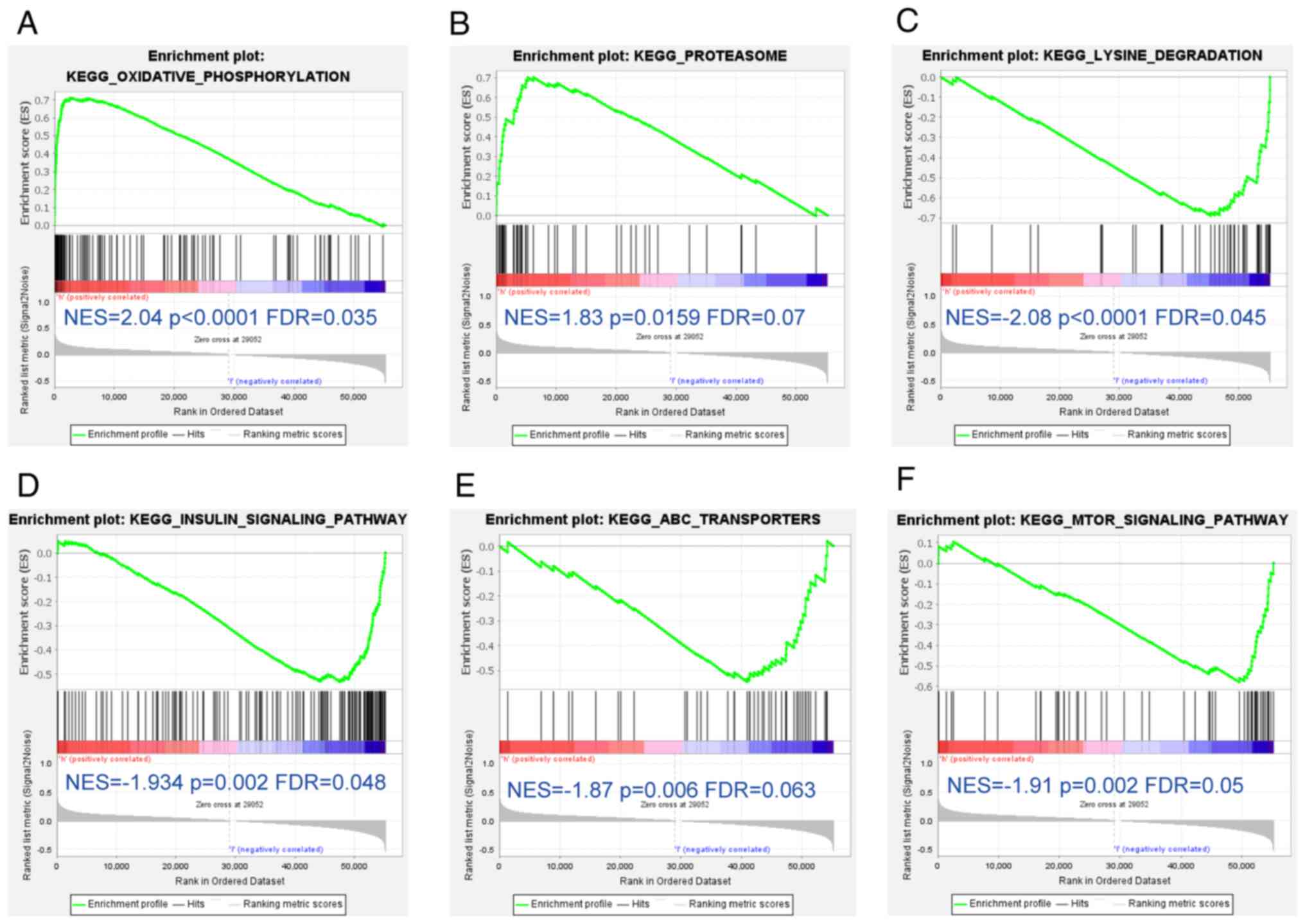
enriched by SH3BGRL3 expression, the present study performed GSEA
between the SH3BGRL3 low-expression group and high-expression group
based on the TCGA_STAD and MSigDB datasets. Based on the main
results, the high-expression group had significantly more positive
regulatory gene sets for OXPHOS and proteasome than the
low-expression group (Fig. 6A and
B). In contrast, the negative regulatory gene sets of lysine
degradation (Fig. 6C), the insulin
signaling pathway (Fig. 6D), ATP
synthase (ATP)-binding cassette transporters (Fig. 6E) and the mTOR signaling pathway
(Fig. 6F) were significantly
enriched in the low-expression group, in comparison with the
high-expression group.
Correlation of SH3BGRL3 expression
with immune cell infiltration
The results of TIMER analysis revealed that high
SH3BGRL3 expression was positively correlated with more macrophages
(P<0.001) and natural killer (NK; P<0.0001) infiltrating
cells but negatively correlated with CD4+ T (P<0.001)
and CD8+ T (P<0.001) infiltrating cells
(Fig. 7A). The results of TISIDB
analysis demonstrated that the expression of SH3BGRL3 widely
affected the infiltrating immune cells within TME in 28 types of
malignant tumors (Fig. 7B). Further
analysis revealed significant positive correlations between
SH3BGRL3 expression and infiltrating macrophages, activated
dendritic cells, NK-CD56dim subgroups and monocytes
(P<0.0001; Fig. 7C).
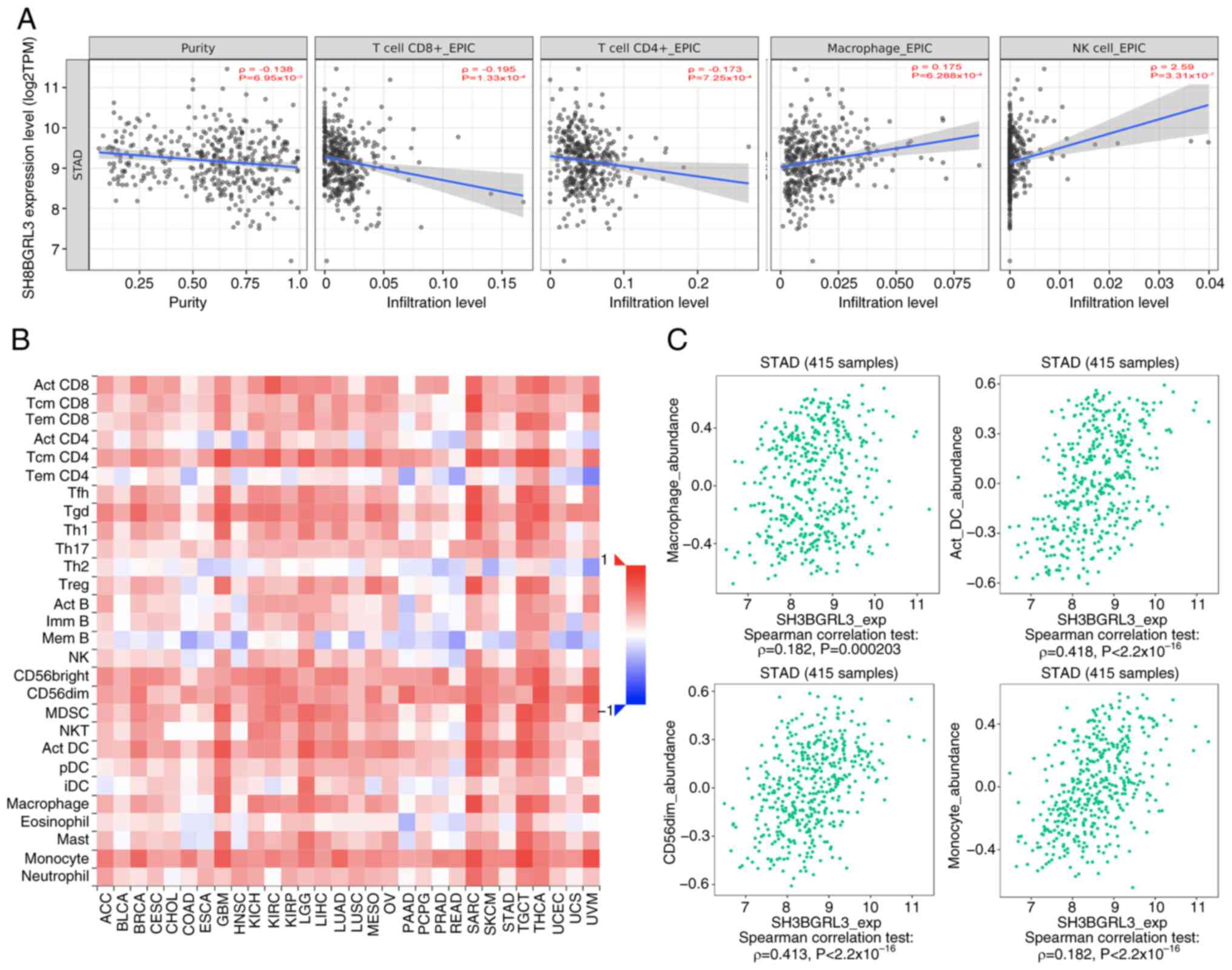 | Figure 7.Association between SH3BGRL3
expression level and infiltrating immune cells in gastric cancer
from the TIMER and TISIDB databases. (A) TIMER revealed that
SH3BGRL3 expression was negatively correlated with CD4+
and CD8+ T, and positively correlated with macrophages
and NK cells. TISIDB demonstrated that (B) SH3BGRL3 was associated
with infiltrating immune cells within the tumor microenvironment in
malignant tumors, including in gastric cancer, and (C) there was a
positive correlation between SH3BGRL3 expression and macrophages,
dendritic cells, NK CD56dim cells and monocytes.
SH3BGRL3, SH3 domain-binding glutamic acid-rich protein-like 3;
TIMER, Tumor Immune Estimation Resource; TISIDB, Tumor and Immune
System Interaction Database; NK, natural killer; TPM, transcripts
per million. |
Discussion
Although targeted therapies, including anti-HER-2,
anti-VEGF and immune checkpoint therapy, hold promise for GC, there
is still a lack of effective treatment for most patients in
clinical practice. Thus, searching for an alternative prognostic
marker and a potential target agent in GC is urgent. The results of
the present study indicate that SH3BGRL3 may be an attractive
predictive molecule for GC, particularly in EBVnGC. The present
study demonstrated that SH3BGRL3 mRNA and protein expression was
increased in tumor tissues using data from multiple public datasets
and clinical specimens. In GC, especially in EBVnGC, high SH3BGRL3
expression was significantly associated with more aggressive
clinicopathological features, such as the presence of perforate
serosa, lymph node involvements, cancer embolus and perineural
invasion, together with a higher TNM stage. Patients with high
SH3BGRL3 expression demonstrated worse outcomes than those with low
expression. The findings of the present study indicate that
SH3BGRL3 expression could further refine prognosis in patients with
GC with the same status of systems such as TNM staging and WHO
grading, which could significantly contribute to hierarchical
clinical management. Moreover, the nomogram based on SH3BGRL3,
preoperative blood glucose and other prognostic variables in EBVnGC
in the present study, demonstrated good accuracy and consistency,
indicating its predictive value for predicting the prognosis in
EBVnGC.
The present study confirmed an association between
SH3BGRL3 expression and BUD formation. BUD is usually considered a
morphological marker for epithelial-mesenchymal transformation
(EMT), which results in cancer invasion and metastasis and has been
indicated as an independent adverse prognostic factor for
colorectal cancer and other solid tumors, including GC (21). As in previous studies (28,29),
the present study demonstrated that BUD was associated with a poor
prognosis in patients with EBVnGC. Furthermore, the present study
revealed that high BUD was associated with SH3BGRL3 expression. To
the best of our knowledge, this is the first study to assess the
relationship between SH3BGRL3 expression and BUD. SH3BGRL3 has
proline-rich motifs, which could interact with phosphorylated-EGFR
through growth factor receptor-bound protein 2 in Akt-related
signaling pathways to promote EMT in tumor cells (11). The two online tools, GeneMANIA and
STRING, respectively, predicted that the mRNA and protein
expression levels of SH3BGRL3 were associated with ErbB1/EGFR and
ErbB2/ERBB2. There was a positive correlation between the
expression of ErbB1/EGFR in EBVnGC but not ErbB2/ERBB2, which
indicates the potential role of ErbB1/EGFR in cancer
progression.
Notably, the results of the present study revealed
that SH3BGRL3 expression was significantly associated with
preoperative blood glucose levels in EBVnGC. The relationship
between blood glucose and GC has been established in previous
studies (16,30–32).
Hyperglycemia is emerging as a vital factor in the progression of
GC, which is considered a cofactor by increasing the risk posed by
H. pylori-mediated gastric carcinogenesis (30). The present study demonstrated that
patients with high blood glucose levels had more adverse outcomes
than those without aberrant blood glucose levels in patients with
EBVnGC. Furthermore, using KEGG and GO analysis, it was
demonstrated that DEGs of SH3BGRL3 were mainly enriched in the
regulation of ATP metabolism and synthesis, OXPHOS and the electron
transport chain. However, when the data were subdivided into high
or low SH3BGRL3 expression, GSEA analysis revealed that the
positive regulatory gene sets of OXPHOS were significantly enriched
in the high expression group only. In contrast, the insulin and
mTOR signaling pathways were significantly enriched in the low
expression group, which are classic pathways that involve aerobic
glycolysis (33). Aberrant energy
metabolism, also called metabolic reprogramming, has been
demonstrated in cancer cells for several decades. The Warburg
effect indicates that tumor cells could utilize oxygen for aerobic
glycolysis to provide energy and meet the requirements for
proliferation. It has been previously reported that
mitochondrion-mediated OXPHOS is another essential energy source
and metabolic precursor for tumor cells by driving ATP synthesis
and governing energy metabolism to maintain redox balance. In
general, glycolysis and OXPHOS maintain the energy balance in tumor
cells (34). However, tumor energy
metabolism exhibits more flexibility and extensive heterogeneity.
Cancer cells can switch their metabolism from glycolysis to
restored suppressed mitochondrial OXPHOS to gain energy, causing
hyperglycemia and nutrient shortage (33,35).
Recently, studies have reported that enhanced OXPHOS serves
essential roles in the malignant development of tumors, including
promoting EMT, invasiveness, metastasis, maintaining stem cell
properties, and inducing TME remodeling, which may be a potent
therapeutic target for advanced solid tumors (34,36,37).
The close relationship between SH3BGRL3 and hyperglycemia suggests
that SH3BGRL3 may promote glucose metabolism in tumor cells via
enhanced OXPHOS, resulting in aggressive GC phenotypes. The
association between SH3BGRL3, glycolysis and OXPHOS may lead to
studies on the influence of other metabolic reprogramming processes
on EBVnGC transformation and cancer development. Further functional
analyses, including the identification of SH3BGRL3 associated
glycolysis-associated genes, the investigation of the interaction
mechanism between SH3BGRL3 and glycolysis, and the elucidation of
the molecular mechanism of SH3BGRL3 involved in regulating the
biological behaviors of EBVnGC, are essential areas of future
research.
Using two online servers, TIMER and TISIDB, the
present study demonstrated that SH3BGRL3 expression was primarily
associated with macrophage and NK cell infiltration. In contrast,
it was negatively correlated with T lymphocytes within the TME.
Macrophages infiltrating malignant tumors are called
tumor-associated macrophages (TAMs), which serve a critical role in
promoting a suppressive tumor immune microenvironment and escape.
TAMs are heterogeneous cells with two dominant populations of
classically (M1) and alternatively (M2) activated phenotypes that
differ depending on the microenvironmental stimulus (38). Previous studies have reported that
TAMs can promote malignant progression involving tumor growth,
prognosis and therapeutic resistance in GC (38,39).
Furthermore, a meta-analysis demonstrated that the higher
quantities of M2 subtypes and total TAMs were both adverse
prognostic factors for patients with GC (40). SH3BGRL3 was reported to have a
positive association with macrophage aggregation, which may have a
positive impact on tumor growth. However, the results of the
present study also revealed that SH3BGRL3 expression was positively
correlated with NK-CD56dim cells infiltrating in GC,
which are innate immune cells and exert an antitumor effect in
solid tumors (41). The TME of a
solid tumor is complex and highly heterogeneous. The
immunosuppressive microenvironment of the tumor could destroy NK
cell-mediated immune surveillance, even if NK cells are enriched in
abundance (42). Regarding the
possible role in regulating metabolic reprogramming, SH3BGRL3 could
induce TME remodeling, which may be linked with complex
infiltrating immune cells within the GC.
There are certain limitations to the present study.
First, although the association between SH3BGRL3 expression and OS
in GC and EBVnGC were demonstrated both using public datasets and
single-center clinical specimens, further external validation
should be performed by multi-center prospective studies. Second,
although the relationship between SH3BGRL3 expression and
preoperative blood glucose was established and the possible
molecular mechanism was explored, further in vitro and in
vivo functional analyses are required to verify the present
results in the future.
In summary, the present study demonstrated that
SH3BGRL3 is significantly upregulated in GC tissues. Furthermore,
SH3BGRL3 is an adverse prognostic factor for GC, particularly in
EBVnGC, which could serve as a potential biomarker for patients
with GC.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Startup Fund for
Scientific Research, Fujian Medical University (grant no.
2020QH1168), the Natural Science Foundation of Fujian Province
(grant no. 2024J011006). the Open Fund from Fujian Key Laboratory
of Translational Research in Cancer and Neurodegenerative Diseases
(grant no. FKLTR-202101) and Scientific Research Project of the
National Key Clinical Specialty Construction Project (grant no.
2022YBL-ZD-05).
Availability of data and materials
The data generated in the present study may be from
the corresponding author.
Authors' contributions
HQL, LYC, XYC and XC conceived and designed the
experiments. HQL, LQZ, XBY and XZ performed the experiments. HQL,
LQZ and GDZ analyzed the data. HQL, LQZ, XBY, XZ, XYC, LYC and XC
provided the reagents, materials and/or the analysis tools. HQL and
LYC wrote the paper. HQL, XYC, LYC and XC confirm the authenticity
of all the raw data. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
The Ethics Committee of Fujian Provincial Hospital
(Fuzhou, China) approved the present study with a waiver of
informed consent (approval no. K2021-04-094). The present study was
performed under the principles of the Declaration of Helsinki.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar
|
|
2
|
Cancer Genome Atlas Research Network, .
Comprehensive molecular characterization of gastric adenocarcinoma.
Nature. 513:202–209. 2014. View Article : Google Scholar
|
|
3
|
Setia N, Agoston AT, Han HS, Mullen JT,
Duda DG, Clark JW, Deshpande V, Mino-Kenudson M, Srivastava A,
Lennerz JK, et al: A protein and mRNA expression-based
classification of gastric cancer. Mod Pathol. 29:772–784. 2016.
View Article : Google Scholar
|
|
4
|
Mazzocco M, Maffei M, Egeo A, Vergano A,
Arrigo P, Di Lisi R, Ghiotto F and Scartezzini P: The
identification of a novel human homologue of the SH3 binding
glutamic acid-rich (SH3BGR) gene establishes a new family of highly
conserved small proteins related to thioredoxin superfamily. Gene.
291:233–239. 2002. View Article : Google Scholar
|
|
5
|
Berleth ES, Masso-Welch PA, Kazim LA, Ip
MM, Mihich E and Ehrke MJ: Expression, tissue distribution, and
cellular localization of the antiapoptotic TIP-B1 protein. J Leukoc
Biol. 69:995–1005. 2001. View Article : Google Scholar
|
|
6
|
Tong F, Zhang M, Guo X, Shi H, Li L, Guan
W, Wang H and Yang S: Expression patterns of SH3BGR family members
in zebrafish development. Dev Genes Evol. 226:287–295. 2016.
View Article : Google Scholar
|
|
7
|
Song J and Shen SH: Effects of SH3BGRL3
overexpression on the proliferation and differentiation of human
acute promyelocytic leukemia cells. J China Med Univ. 48:338–341.
2019.
|
|
8
|
Lee MJ, Kim J, Kim MY, Bae YS, Ryu SH, Lee
TG and Kim JH: Proteomic analysis of tumor necrosis
factor-alpha-induced secretome of human adipose tissue-derived
mesenchymal stem cells. J Proteome Res. 9:1754–1762. 2010.
View Article : Google Scholar
|
|
9
|
Jiang M, Lash GE, Zeng S, Liu F, Han M,
Long Y, Cai M, Hou H, Ning F, Hu Y and Yang H: Differential
expression of serum proteins before 20 weeks gestation in women
with hypertensive disorders of pregnancy: A potential role for
SH3BGRL3. Placenta. 104:20–30. 2021. View Article : Google Scholar
|
|
10
|
Chiang CY, Pan CC, Chang HY, Lai MD, Tzai
TS, Tsai YS, Ling P, Liu HS, Lee BF, Cheng HL, et al: SH3BGRL3
protein as a potential prognostic biomarker for urothelial
carcinoma: A novel binding partner of epidermal growth factor
receptor. Clin Cancer Res. 21:5601–5611. 2015. View Article : Google Scholar
|
|
11
|
Yin L, Gao S, Shi H, Wang K, Yang H and
Peng B: TIP-B1 promotes kidney clear cell carcinoma growth and
metastasis via EGFR/AKT signaling. Aging (Albany NY). 11:7914–7937.
2019. View Article : Google Scholar
|
|
12
|
Xiang Z, Huang X, Wang J, Zhang J, Ji J,
Yan R, Zhu Z, Cai W and Yu Y: Cross-database analysis reveals
sensitive biomarkers for combined therapy for ERBB2+ gastric
cancer. Front Pharmacol. 9:8612018. View Article : Google Scholar
|
|
13
|
Nie Z, Cheng D, Pan C, Wei Z and Wang C
and Wang C: SH3BGRL3, transcribed by STAT3, facilitates
glioblastoma tumorigenesis by activating STAT3 signaling. Biochem
Biophys Res Commun. 556:114–120. 2021. View Article : Google Scholar
|
|
14
|
Lega IC and Lipscombe LL: Review:
Diabetes, obesity, and cancer-pathophysiology and clinical
implications. Endocr Rev. 41:bnz0142020. View Article : Google Scholar
|
|
15
|
Shimoyama S: Diabetes mellitus carries a
risk of gastric cancer: A meta-analysis. World J Gastroenterol.
19:6902–6910. 2013. View Article : Google Scholar
|
|
16
|
Sheng L, Peng H, Pan Y, Wang C and Zhu Y:
Evaluating the effect of diabetes on the prognosis of gastric
cancer using a propensity score matching method. J Gastrointest
Oncol. 11:999–1008. 2020. View Article : Google Scholar
|
|
17
|
Miao ZF, Xu H, Xu YY, Wang ZN, Zhao TT,
Song YX and Xu HM: Diabetes mellitus and the risk of gastric
cancer: A meta-analysis of cohort studies. Oncotarget.
8:44881–44892. 2017. View Article : Google Scholar
|
|
18
|
Faubert B, Solmonson A and DeBerardinis
RJ: Metabolic reprogramming and cancer progression. Science.
368:eaaw54732020. View Article : Google Scholar
|
|
19
|
Zhao L, Liu Y, Zhang S, Wei L, Cheng H and
Wang J and Wang J: Impacts and mechanisms of metabolic
reprogramming of tumor microenvironment for immunotherapy in
gastric cancer. Cell Death Dis. 13:3782022. View Article : Google Scholar
|
|
20
|
WHO Classification of Tumours Editorial
Board, . Digestive system tumours: WHO classification of tumours.
5th edition. volume 1. Lyon: IARC; 2019
|
|
21
|
Lugli A, Kirsch R, Ajioka Y, Bosman F,
Cathomas G, Dawson H, El Zimaity H, Fléjou JF, Hansen TP, Hartmann
A, et al: Recommendations for reporting tumor budding in colorectal
cancer based on the international tumor budding consensus
conference (ITBCC) 2016. Mod Pathol. 30:1299–1311. 2017. View Article : Google Scholar
|
|
22
|
Package Insert and PATHWAY anti-HER-2/NEU
(4B5) rabbit monoclonal primary antibody. German Created.
17.03.2020. 01–12. 2021
|
|
23
|
Kim CH, Kim SH, Park SY, Yoo J, Kim SK and
Kim HK: Identification of EGFR mutations by immunohistochemistry
with EGFR mutation-specific antibodies in biopsy and resection
specimens from pulmonary adenocarcinoma. Cancer Res Treat.
47:653–660. 2015. View Article : Google Scholar
|
|
24
|
Wolff AC, Hammond MEH, Allison KH, Harvey
BE, Mangu PB, Bartlett JMS, Bilous M, Ellis IO, Fitzgibbons P,
Hanna W, et al: Human epidermal growth factor receptor 2 testing in
breast cancer: American society of clinical oncology/college of
American pathologists clinical practice guideline focused update. J
Clin Oncol. 36:2105–2122. 2018. View Article : Google Scholar
|
|
25
|
Wang HL, Kim CJ, Koo J, Zhou W, Choi EK,
Arcega R, Chen ZE, Wang H, Zhang L and Lin F: Practical
immunohistochemistry in neoplastic pathology of the
gastrointestinal tract, liver, biliary tract, and pancreas. Arch
Pathol Lab Med. 141:1155–1180. 2017. View Article : Google Scholar
|
|
26
|
Ikeda T, Gion Y, Sakamoto M, Tachibana T,
Nishikori A, Nishimura MF, Yoshino T and Sato Y:
Clinicopathological analysis of 34 Japanese patients with
EBV-positive mucocutaneous ulcer. Mod Pathol. 33:2437–2448. 2020.
View Article : Google Scholar
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
28
|
Kemi N, Eskuri M, Ikäläinen J, Karttunen
TJ and Kauppila JH: Tumor budding and prognosis in gastric
adenocarcinoma. Am J Surg Pathol. 43:229–234. 2019. View Article : Google Scholar
|
|
29
|
Ulase D, Heckl S, Behrens HM, Krüger S and
Röcken C: Prognostic significance of tumour budding assessed in
gastric carcinoma according to the criteria of the international
tumour budding consensus conference. Histopathology. 76:433–446.
2020. View Article : Google Scholar
|
|
30
|
Yang HJ, Kang D, Chang Y, Ahn J, Ryu S,
Cho J, Guallar E and Sohn CI: Diabetes mellitus is associated with
an increased risk of gastric cancer: A cohort study. Gastric
Cancer. 23:382–390. 2020. View Article : Google Scholar
|
|
31
|
Kim YM, Kim JH, Park JS, Baik SJ, Chun J,
Youn YH and Park H: Association between triglyceride-glucose index
and gastric carcinogenesis: A health checkup cohort study. Gastric
Cancer. 25:33–41. 2022. View Article : Google Scholar
|
|
32
|
Karlin NJ, Buras MR, Kosiorek HE, Verona
PM and Cook CB: Glycemic control and survival of patients with
coexisting diabetes mellitus and gastric or esophageal cancer.
Future Sci OA. 5:FSO3972019. View Article : Google Scholar
|
|
33
|
Yuan LW, Yamashita H and Seto Y: Glucose
metabolism in gastric cancer: The cutting-edge. World J
Gastroenterol. 22:2046–2059. 2016. View Article : Google Scholar
|
|
34
|
Hu Y, Xu W, Zeng H, He Z, Lu X, Zuo D, Qin
G and Chen W: OXPHOS-dependent metabolic reprogramming prompts
metastatic potential of breast cancer cells under osteogenic
differentiation. Br J Cancer. 123:1644–1655. 2020. View Article : Google Scholar
|
|
35
|
Thankamony AP, Saxena K, Murali R, Jolly
MK and Nair R: Cancer stem cell plasticity-a deadly deal. Front Mol
Biosci. 7:792020. View Article : Google Scholar
|
|
36
|
Kalyanaraman B, Cheng G and Hardy M:
Therapeutic targeting of tumor cells and tumor immune
microenvironment vulnerabilities. Front Oncol. 12:8165042022.
View Article : Google Scholar
|
|
37
|
Janku F, Beom SH, Moon YW, Kim TW, Shin
YG, Yim DS, Kim GM, Kim HS, Kim SY, Cheong JH, et al:
First-in-human study of IM156, a novel potent biguanide oxidative
phosphorylation (OXPHOS) inhibitor, in patients with advanced solid
tumors. Invest New Drugs. 40:1001–1010. 2022. View Article : Google Scholar
|
|
38
|
Ruffell B and Coussens LM: Macrophages and
therapeutic resistance in cancer. Cancer Cell. 27:462–472. 2015.
View Article : Google Scholar
|
|
39
|
Gambardella V, Castillo J, Tarazona N,
Gimeno-Valiente F, Martínez-Ciarpaglini C, Cabeza-Segura M, Roselló
S, Roda D, Huerta M, Cervantes A and Fleitas T: The role of
tumor-associated macrophages in gastric cancer development and
their potential as a therapeutic target. Cancer Treat Rev.
86:1020152020. View Article : Google Scholar
|
|
40
|
Wang XL, Jiang JT and Wu CP: Prognostic
significance of tumor-associated macrophage infiltration in gastric
cancer: A meta-analysis. Genet Mol Res. 15:gmr150490402016.
View Article : Google Scholar
|
|
41
|
Oya Y, Hayakawa Y and Koike K: Tumor
microenvironment in gastric cancers. Cancer Sci. 111:2696–2707.
2020. View Article : Google Scholar
|
|
42
|
Mylod E, Lysaght J and Conroy MJ: Natural
killer cell therapy: A new frontier for obesity-associated cancer.
Cancer Lett. 535:2156202022. View Article : Google Scholar
|