Introduction
The incidence and mortality rates of patients with
breast cancer (BRCA) have emerged as a significant public health
concern among women, posing a threat to human well-being (1,2).
According to the latest GLOBOCAN 2021 data, there is an estimated
global burden of ~2.3 million new cases of BRCA, accompanied by a
mortality rate of 6.9% (3). The
lack of early symptoms at diagnosis leads to late detection and
even metastasis, resulting in poor prognosis (4). With the advancement of gene chips,
novel biomarkers that unravel the heterogeneity of BRCA have
gradually emerged (5). In recent
years, the utilization of immunotherapy and targeted therapy have
been extensively employed in the treatment of BRCA, with promising
outcomes (6). To enhance the early
diagnosis of BRCA, optimize the efficacy of immunotherapy and
prolong survival rates, it is imperative to identify more effective
biomarkers (7).
The UL16 binding protein (ULBP)1 is a gene that
belongs to the major histocompatibility complex class I-related
family (8). The protein encoded by
this gene acts as a ligand for the natural killer group 2, member D
(NKG2D), which is an immune system-activating receptor found on
natural killer cell (NK) cells and T-cells (9). Additionally, in cells infected with
cytomegalovirus, this ligand interacts with the UL16 glycoprotein
and is hindered from activating the immune system (10). This gene has been found to have four
transcript variants that encode different isoforms, including
ULBP1, ULBP2, ULBP3 and ULBP4 (11). The expression of ULBP1 is
upregulated in certain types of cancer, such as colon cancer
(12), hepatocellular carcinoma
(13) and cervical cancer (14); however, its expression in BRCA
remains unreported, at least to the best of our knowledge.
The present study evaluated the expression of the
ULBP1 gene in BRCA and assessed its association with the prognosis
of patients with BRCA and immune cell infiltration. Moreover, the
sequencing results were further validated through
immunohistochemistry conducted on both BRCA and adjacent
non-cancerous tissues. The findings presented in the current study
established ULBP1 as a potential biomarker for fundamental and
applied research on BRCA, providing crucial molecular evidence for
early diagnosis and immunotherapy.
Materials and methods
Data collection
Data collection was conducted from The Cancer Genome
Atlas (TCGA; http://portal.gdc.cancer.gov/) platform was utilized
to acquire clinical and RNA-sequencing data sets, including
GSE73540 (15), GSE3143 (16), GSE22820 (17), and GSE42568 (18) from a cohort of 1,226 BRCA patients.
Additionally, this dataset encompassed 113 corresponding non-tumor
samples. The data obtained from the TCGA database was collected and
analyzed by Xiantao Academic Online (https://www.xiantaozi.com). The Xiantao Academic tool
streamlines various analysis and visualization processes commonly
used in the R language. It presents these analyses and
visualizations as online pages, providing a comprehensive solution
for common statistical analysis and visualization tasks. A total of
1,098 clinical records were collected among the 1,226 patients with
BRCA. Furthermore, for the immunohistochemical analysis, the data
were obtained from specimens collected at Zibo Central Hospital
(Zibo, China).
Differentially expressed mRNA (DEmRNA)
analysis
The ggplot2 (version 3.3.6), stats (version 4.2.1)
and car (version 3.1–0) package were employed via Xiantao Academic
Online for conducting differential mRNA analysis to identify mRNAs
that exhibited a significant differential expression, characterized
by an absolute log2 fold change (|logFC|)>1.5 and a P.adj value
<0.05. The Xiantao Academic Online was utilized for the
identification of co-expressed mRNAs with target genes, while the
‘ggplot2’ package (version 3.3.6) in Xiantao Academic Online was
employed for visualizing mRNA volcano plots.
Survival analysis
The patients with BRCA were stratified into two
groups, namely the ULBP1 high expression group and ULBP1 low
expression group, based on the median level of ULBP1 mRNA
expression (cut-off level, 0.731). Survival analysis was performed
using the survival (version 3.3.1) package to investigate the
association between DEmRNA and the prognosis of patients with BRCA
(https://www.xiantaozi.com).
Identification of DEmRNAs associated
with the prognosis and immunity of patients with BRCA
In order to identify immune-related target genes,
the ImmPort database (https://www.immport.org/shared/home) was utilized to
obtain immunoregulatory genes. Subsequently, Venn overlap analysis
in Xiantao Academic Online was employed to demonstrate the
interaction between prognosis-associated DEmRNAs and immune-related
genes. Ultimately, ULBP1 was identified as the specific target
gene.
Functional enrichment analysis
The ‘cluster profiler’ package (version 4.4.4) in
Xiantao Academic Online was utilized to automate the process of
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes
(KEGG) term analysis. The patients with BRCA were stratified into
the low and high expression groups based on the median ULBP1
expression levels as aforementioned in survival analysis. The
protein-protein interaction analysis was performed utilizing the
STRING database (https://string-db.org).
Immune infiltration analysis
The single sample gene set enrichment analysis
(ssGSEA) algorithm in Xiantao Academic Online, which is further
normalized by the range of values across all gene sets and samples,
was employed to conduct the immune infiltration analysis of ULBP1
in BRCA tissue samples, evaluating 24 distinct immune cell types.
Spearman's correlation analysis was employed to evaluate the
correlation between ULBP1 expression and immune cells, as well as
immune checkpoint molecules, including programmed cell death
protein 1 (PDCD1), cytotoxic T lymphocyte-associated protein 4
(CTLA4) and programmed cell death ligand 1 (CD274). The Wilcoxon
rank-sum test was conducted to assess the enrichment of immune
infiltrating cells in patients with BRCA with a high expression of
ULBP1 compared with those with a low expression of ULBP1.
Immunohistochemical analysis
The expression levels of ULBP1 in biopsy samples
obtained from a total of 74 treatment-naive patients diagnosed with
primary BRCA at Zibo Central Hospital between May 2020 and July
2023 were assessed using immunohistochemical analysis. Patients
with a prior history of chemotherapy, radiotherapy or other
malignancies were also excluded from the current study. Written
consent was obtained from all patients. The sections (5 µm) were
dewaxed by heating at 55°C for 30 min and subjected to two 15 min
washes with xylene. Then, the sections were rehydrated by a series
of 5 min washes in ethanol. The sections were placed into an enamel
cylinder containing 10 mmol/l sodium citrate (pH 6.0), heated by
gas cooker at 95°C for 5 min for antigen unmasking, and then were
treated with 3% hydrogen peroxide for 30 min to inactivate
endogenous peroxidase activity. After being incubated with fetal
bovine serum at 37°C for 30 min, the sections were then incubated
at 4°C overnight with specific rabbit polyclonal antibodies
targeting human ULBP1 (cat. no. ab238331; Abcam), ULBP2 (cat. no.
ab275023, Abcam) and ULBP3 (cat. no. ab300102, Abcam), diluted at
1:1000. The sections were then washed with PBS and incubated for 30
min with biotinylated goat anti-rabbit secondary antibody (cat. no.
ab6721; Abcam) at 37°C. The substrate, 3′3-diaminobenzidin (DAB)
tetrachloride, dissolved in steamed water, was added to visualize
the positive expression. Negative control sections were
immune-stained as described above, but incubated with PBS instead
of a primary antibody. Negative control slides were treated with
isotype-matched antibodies at the same dilution (1:1,000) as the
primary anti-human ULBP1, ULBP2 or ULBP3 antibodies. The presence
of ULBP1, ULBP2 or ULBP3 was determined based on its positive
localization in cellular membrane regions. A fluorescence
microscope used to capture the images, and the protein expression
levels were quantitatively analyzed using Image-Pro Plus software
(Media Cybernetics, Inc.). To classify and score protein expression
levels, a semi-quantitative approach combining the percentage of
tumor cells stained positively, staining intensity and previous
descriptions of high and low expression groups based on ULBP1
molecule levels was employed (19).
Statistical analysis
SPSS 26.0 software (IBM Corp.) was utilized for data
processing purposes. Initially, the expression of ULBP1 in both
normal breast tissue and BRCA tissue was examined using the
Wilcoxon rank-sum test and Wilcoxon signed-rank test, respectively.
Receiver operating characteristic (ROC) curve analysis was
conducted to evaluate the diagnostic efficacy of ULBP1 expression
in BRCA. The association between ULBP1 expression and the
clinicopathological parameters of the patients was analyzed using
the χ2 test with counts (percentages). Univariate and
multivariate Cox regression analyses were performed to assess the
impact of ULBP1 expression and clinicopathological parameters on
the patient survival rate. Spearman's rank correlation coefficient
analysis was employed for correlation analysis. The Kaplan-Meier
method was applied to plot overall survival (OS) curves, with
differences in survival being assessed using log-rank tests. The
data presented are derived from the mean ± SD of a minimum of three
replicates. P<0.05 was considered to indicate a statistically
significant difference.
Results
Identification of DEmRNAs associated
with the prognosis and immunity of patients with BRCA
To identify DEmRNAs associated with BRCA, a total of
4,356 genes were identified as being significantly altered in BRCA
compared with normal breast tissue samples. The upregulated genes
are represented by red dots, while the downregulated genes are
represented by blue dots. Among these, ULBP1 exhibited a
significant upregulation, as depicted by the volcano plot
illustrated in Fig. 1A. By
employing the ‘survival’ package for the batch fitting of survival
regression, a comprehensive analysis was conducted to investigate
the association between multiple genes and the survival rate of
patients with BRCA using the Cox regression statistical method. As
a result, 1,078 genes were successfully identified that were
significantly associated with the prognosis of patients with BRCA.
Furthermore, the genes associated with the immune system were
classified as ‘immune-related genes’ and the gene list was
downloaded from the ImmPort database. A Venn overlap analysis was
conducted to identify shared target genes among the
prognosis-related genes, immune-related genes and DEmRNAs in
patients with BRCA. The analysis revealed a set of common target
DEmRNAs, namely TNF superfamily member 4, ULBP1, syndecan 1,
uromodulin-like 1, interleukin (IL)27, inhibin subunit α, thymosin
β 15A and IL36 receptor antagonist (Fig. 1B). The aforementioned genes
demonstrated substantial associations with both the prognosis and
immune infiltration of patients with BRCA. Through comprehensive
comparisons, ULBP1 was selected as the target gene. The heatmap
presented in Fig. 1C illustrates
the co-expression of ULBP1 and its associated mRNAs. The initial
five genes exhibit a positive correlation with ULBP1 expression,
whereas the latter five genes demonstrated a negative
correlation.
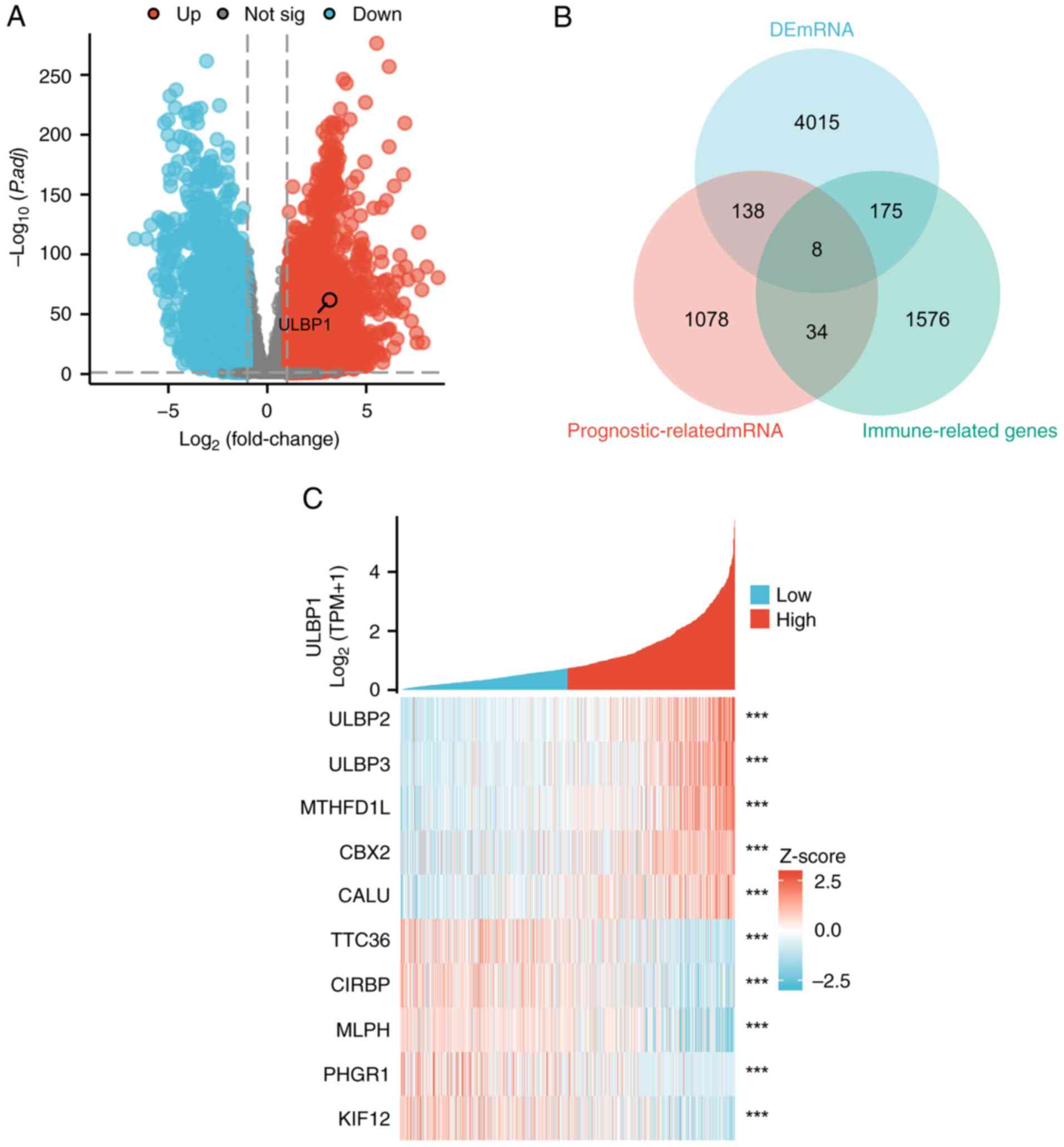 | Figure 1.Identification of DEmRNAs that are
associated with the prognosis and immunity of patients with BRCA.
(A) A volcanic map depicting the differential expression pattern of
mRNA, and (B) a Venn diagram highlighting the overlap between
target genes among prognosis-related genes, immune-related genes
and DEmRNAs in patients with BRCA. (C) Heatmap showcasing ULBP1 and
its co-expressed mRNA. ***P<0.001. DEmRNAs, differentially
expressed mRNAs; BRCA, breast cancer; ULBP, UL16 binding protein;
MTHFD1L, methylenetetrahydrofolate dehydrogenase (NADP+
dependent) 1; CBX2, chromobox protein homolog 2; CALU, calumenin;
TTC36, tetratricopeptide repeat domain 36; MLPH, melanophilin;
PHGR1, proline, histidine and glycine rich 1; KIF12, kinesin family
member 12. |
High expression of ULBP1 has a
specific predictive and diagnostic value for patients with
BRCA
The expression analysis of ULBP1 in pan-cancer using
the Xiantao database revealed that ULBP1 was highly expressed in
BRCA, bladder urothelial carcinoma, cervical squamous cell
carcinoma and endocervical adenocarcinoma, cholangiocarcinoma,
colon adenocarcinoma, esophageal carcinoma, head and neck squamous
cell carcinoma, kidney chromophobe, kidney renal papillary cell
carcinoma, lung squamous cell carcinoma, prostate adenocarcinoma,
rectal adenocarcinoma, THCA, stomach gastric adeno-cancers and
endometrial cancers compared with normal tissues (Fig. 2A) and the corresponding adjacent
normal tissue samples (Fig. 2B).
Because adjacent tissue may have different gene expression patterns
compared with normal tissue, this causes a slight difference in the
results of the two graphs (Fig. 2A and
B). By contrast, lung adenocarcinoma exhibited low levels of
ULBP1 expression in tumor tissues compared with normal tissues
(Fig. 2A). Furthermore, additional
visualization analysis of the expression pattern of ULBP1 was
performed by comparing both normal para-cancerous tissues and the
matched tumor-normal pairs obtained from TCGA database, with a
specific focus on patients with BRCA. The expression of ULBP1 was
significantly upregulated in BRCA tissues compared with normal
para-cancerous tissues (Fig. 2C)
and the matched tumor-normal paired tissue (Fig. 2D).
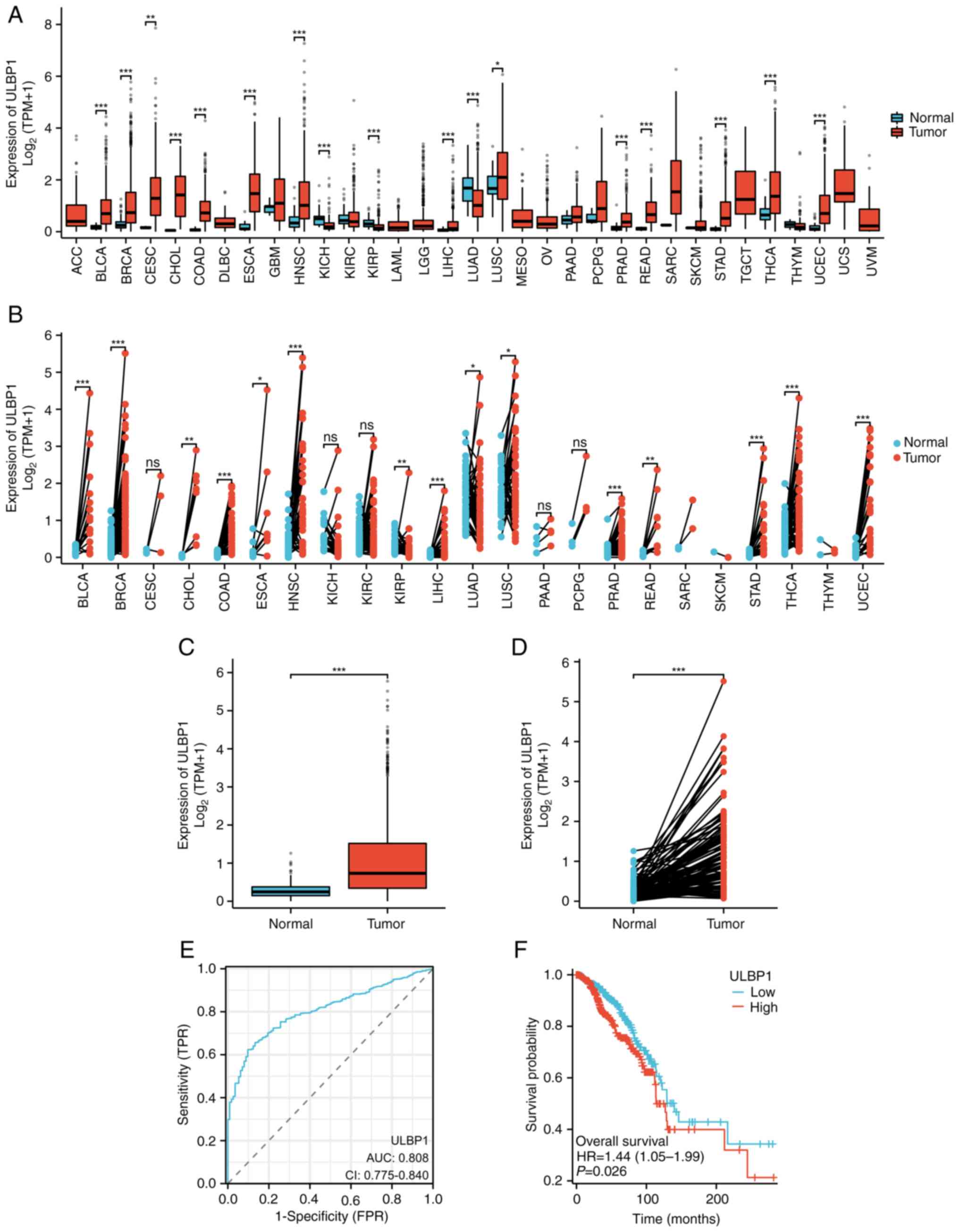 | Figure 2.Expression level of ULBP1 exhibits
good diagnostic predictive value for patients with BRCA. (A)
Comparative analysis of ULBP1 mRNA expression levels in tumor and
adjacent normal tissues across various malignancies. (B) ULBP1 mRNA
levels were quantified using RNA-sequencing data from tumor samples
and their matched normal tissues in The Cancer Genome Atlas
database. (C) Analysis of sequencing data revealed the differential
expression of ULBP1 mRNA between normal and BRCA tissues. (D)
Validation of ULBP1 mRNA expression through RNA-sequencing in
paired BRCA and normal breast tissue samples. (E) The diagnostic
predictive value of ULBP1 expression level was assessed using ROC
curve analysis. (F) Kaplan-Meier curve illustrating the comparison
of overall survival between subgroups of patients with BRCA with a
high/low ULBP1 mRNA expression. *P<0.05; **P<0.01;
***P<0.001. ULBP1, UL16 binding protein 1; BRCA, breast cancer;
BLCA, bladder urothelial carcinoma; CESC, cervical squamous cell
carcinoma and endocervical adenocarcinoma; CHOL,
cholangiocarcinoma; COAD, colon adenocarcinoma; ESCA, esophageal
carcinoma; HNSC, head and neck squamous cell carcinoma; KICH,
kidney chromophobe; KIRP, kidney renal papillary cell carcinoma;
LUSC, lung squamous cell carcinoma; PRAD, prostate adenocarcinoma;
READ, rectal adenocarcinoma; STAD, stomach gastric adeno-cancers;
UCEC, endometrial cancers; ns, not significant; AUC, area under the
curve; CI, confidence intervals; HR, hazard ratio; FPR, false
positive rate; TPR, true positive rate. |
Subsequently, ROC curve analysis demonstrated that
ULBP1 expression should be regarded as a discriminative factor. The
results revealed that compared with normal breast tissue, BRCA
tissue exhibited higher levels of ULBP1 expression, with an area
under the curve (AUC) of 0.808 and 95% confidence interval of
0.775–0.840 (Fig. 2E). In addition,
the AUC for the low expression of ULBP1 was found to be 0.498 (data
not shown). This further supports the notion that ULBP1 holds
potential as a diagnostic biomarker for BRCA. Furthermore, the
prognostic significance of ULBP1 in BRCA was evaluated by analyzing
data from TCGA database. Kaplan-Meier survival curves indicated
significantly higher overall survival rates in the low expression
group compared with the high expression group for ULBP1 (P=0.026),
as depicted in Fig. 2F. This
information revealed that the low expression of ULBP1 predicted a
higher overall survival rate.
Association between ULBP1 expression
and clinicopathological features of patients with BRCA
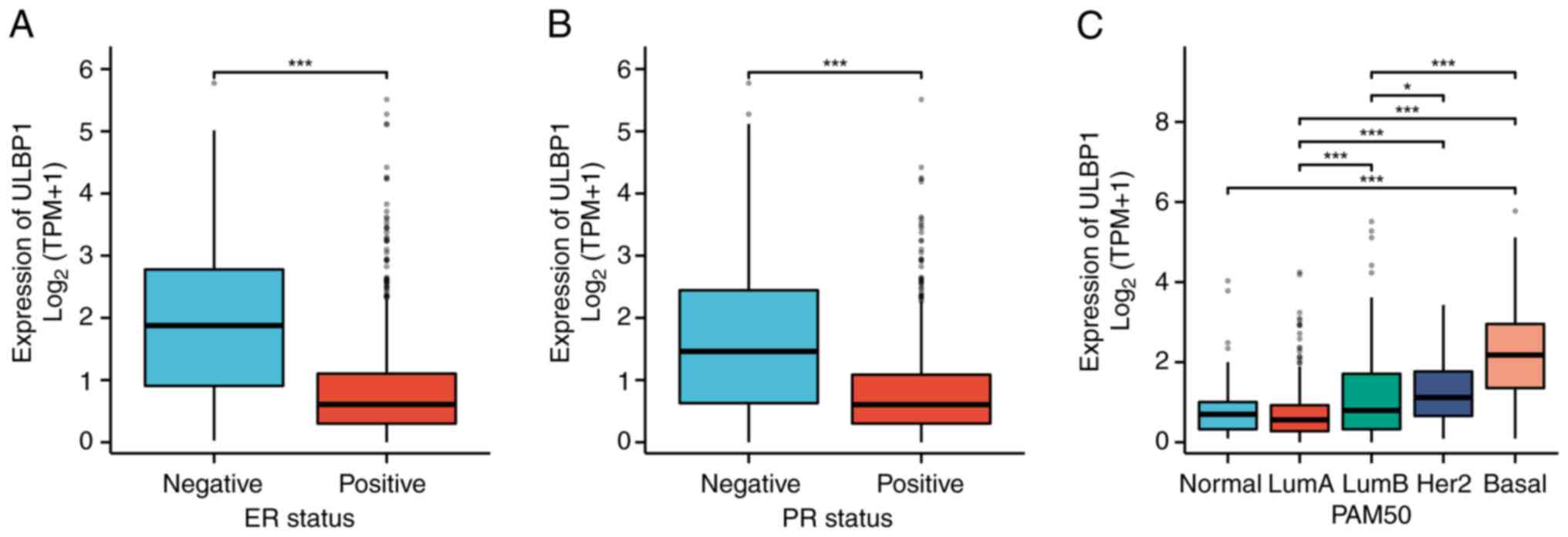
The associations between ULBP1 expression and the
clinicopathological features of patients with BRCA were assessed
using the χ2 test and logistic regression models
(Fig. 3). The expression of ULBP1
exhibited significant associations with estrogen receptor (ER)
expression (P<0.001), progesterone receptor (PR) expression
(P<0.001) and the prediction analysis of microarray 50 (PAM50)
(P<0.05). As presented in Table
I, the χ2 test revealed significant associations
between ULBP1 expression and the age of patients with BRCA
(P=0.007), race (P=0.012), PR expression (P<0.001), ER
expression (P<0.001), PAM50 (P<0.001) and histological type
(P<0.001). However, no significant associations were observed
with other clinicopathological features, such as pathological T
stage (P=0.216), pathological N stage (P=0.580), pathological M
stage (P=0.771), pathological stage (P=0.605), human epidermal
growth factor receptor 2 (HER2) status (P=0.529) and menopause
status (P=0.382).
 | Table I.Association between ULBP1 and
clinicopathological features of patients with BRCA. |
Table I.
Association between ULBP1 and
clinicopathological features of patients with BRCA.
| Characteristics | Low expression of
ULBP1 | High expression of
ULBP1 | P-value |
|---|
| n | 543 | 544 |
|
| Pathological T stage,
n (%) |
|
| 0.216 |
| T1 +
T2 | 447 (41.2%) | 462 (42.6%) |
|
| T3 +
T4 | 95 (8.8%) | 80 (7.4%) |
|
| Pathological N
stage, n (%) |
|
| 0.580 |
| N0 | 253 (23.7%) | 263 (24.6%) |
|
| N1 + N2
+ N3 | 280 (26.2%) | 272 (25.5%) |
|
| Pathological M
stage, n (%) |
|
| 0.771 |
| M0 | 437 (47.2%) | 468 (50.6%) |
|
| M1 | 9 (1%) | 11 (1.2%) |
|
| Pathological stage,
n (%) |
|
| 0.605 |
| Stage I
+ stage II | 398 (37.4%) | 403 (37.9%) |
|
| Stage
III + stage IV | 135 (12.7%) | 127 (11.9%) |
|
| Race, n (%) |
|
| 0.012 |
|
White | 401 (40.2%) | 354 (35.5%) |
|
| Asian +
Black or African American | 106 (10.6%) | 136 (13.6%) |
|
| Age, n (%) |
|
| 0.007 |
|
≤60 | 279 (25.7%) | 324 (29.8%) |
|
|
>60 | 264 (24.3%) | 220 (20.2%) |
|
| Histological type,
n (%) |
|
| <0.001 |
|
Infiltrating ductal
cancer | 345 (35.2%) | 431 (43.9%) |
|
|
Infiltrating lobular
cancer | 141 (14.4%) | 64 (6.5%) |
|
| ER status, n
(%) |
|
| <0.001 |
|
Negative | 43 (4.1%) | 197 (19%) |
|
|
Positive | 472 (45.5%) | 325 (31.3%) |
|
| PR status, n
(%) |
|
| <0.001 |
|
Negative | 104 (10.1%) | 238 (23%) |
|
|
Positive | 410 (39.7%) | 282 (27.3%) |
|
| HER2 status, n
(%) |
|
| 0.529 |
|
Negative | 262 (36.5%) | 298 (41.6%) |
|
|
Positive | 69 (9.6%) | 88 (12.3%) |
|
| PAM50, n (%) |
|
| <0.001 |
|
LumA | 373 (35.6%) | 191 (18.2%) |
|
|
LumB | 99 (9.5%) | 107 (10.2%) |
|
|
HER2 | 27 (2.6%) | 55 (5.3%) |
|
|
Basal | 23 (2.2%) | 172 (16.4%) |
|
| Menopause status, n
(%) |
|
| 0.382 |
|
Pre | 109 (11.6%) | 121 (12.9%) |
|
|
Post | 358 (38.2%) | 348 (37.2%) |
|
| OS event, n
(%) |
|
| 0.056 |
|
Alive | 478 (44%) | 457 (42%) |
|
|
Dead | 65 (6%) | 87 (8%) |
|
| DSS event, n
(%) |
|
| 0.082 |
|
Yes | 35 (3.3%) | 50 (4.7%) |
|
| No | 501 (47%) | 481 (45.1%) |
|
| PFI event, n
(%) |
|
| 0.135 |
|
Yes | 65 (6%) | 82 (7.5%) |
|
| No | 478 (44%) | 462 (42.5%) |
|
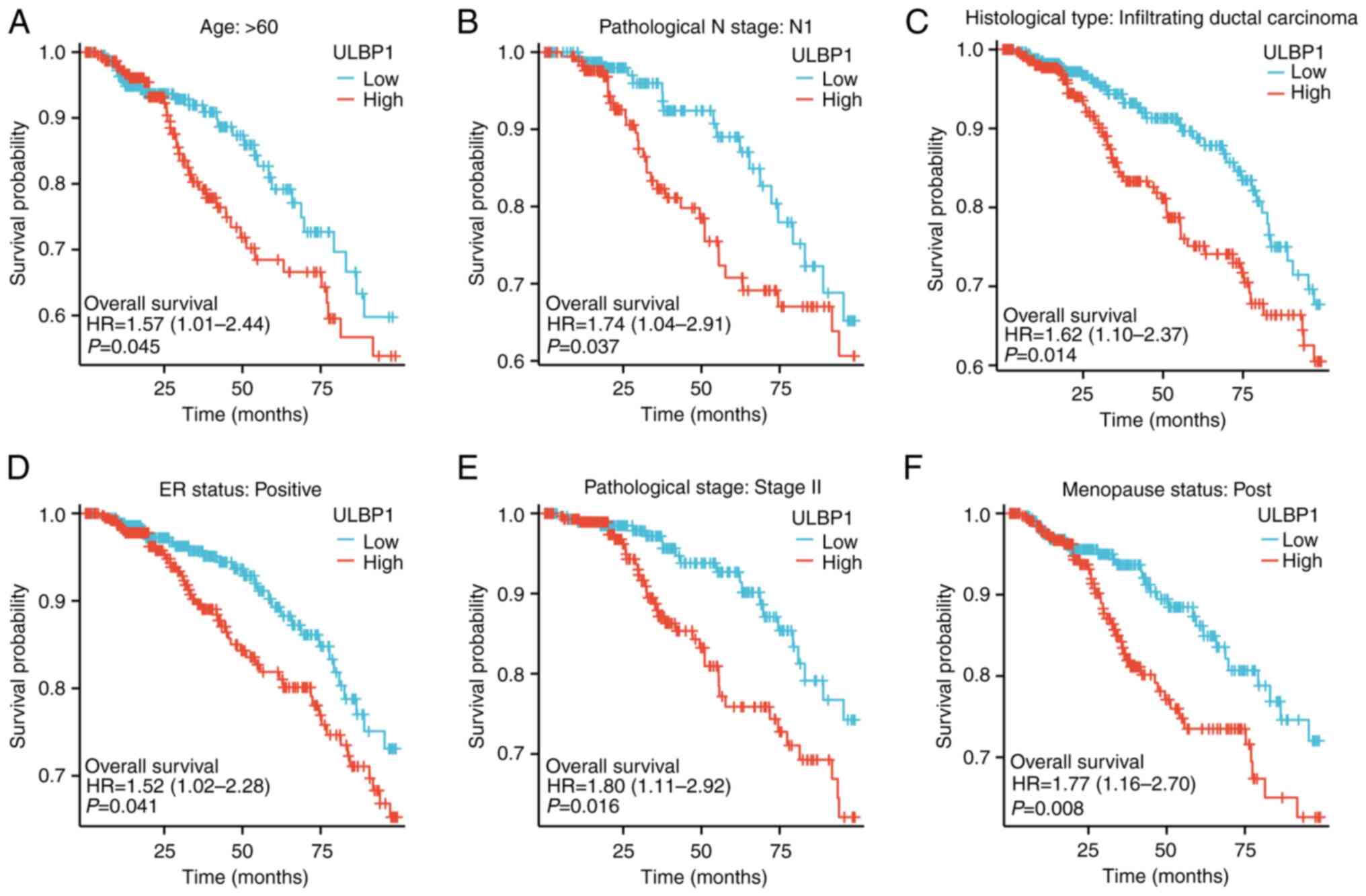
OS rates of patients with BRCA with
varying ULBP1 expression levels in the different subgroups
The OS rates of patients with BRCA with a high or
low expression of ULBP1 in the different subgroups are presented in
Fig. 4A-F. The results indicated
that a poor OS rate was associated with an increased ULBP1
expression in the subgroups aged >60 years (P=0.045), N1
subgroup (P=0.037), infiltrating ductal cancer subgroup (P=0.014),
ER-positive subgroup (P=0.041), subgroup of stage II (P=0.016) and
post-menopause subgroup (P=0.008).
ULBP1 is an independent risk factor
significantly affecting survival
In order to assess the impact of ULBP1 expression
and clinicopathological parameters on survival, univariate and
multivariate Cox regression analyses were conducted. In the
univariate Cox regression model, T4 stage (P<0.001), N1 stage
(P<0.001), N2 stage (P<0.001), N3 stage (P<0.001), M1
stage (P<0.001) and the ULBP1 expression level (P=0.026) were
all statistically significant variables. Subsequently, the
multivariate Cox regression analysis revealed that T4 stage
(P=0.032), M1 stage (P=0.034), N1 stage (P=0.020), N2 stage
(P=0.006), N3 stage (P=0.004) and the ULBP1 expression level
(P=0.046) were independent risk factors significantly affecting
survival, as presented in Table
II.
 | Table II.Prognostic value of ULBP1 in patients
with BRCA determined through both univariate and multivariate Cox
regression analyses. |
Table II.
Prognostic value of ULBP1 in patients
with BRCA determined through both univariate and multivariate Cox
regression analyses.
|
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|
|---|
|
Characteristics | Total (n) | Hazard ratio (95%
CI) | P-value | Hazard ratio (95%
CI) | P-value |
|---|
| Pathologic T
stage | 1,083 |
|
|
|
|
| T1 | 277 | Reference |
| Reference |
|
| T2 | 631 | 1.336
(0.890–2.006) | 0.162 | 1.115
(0.702–1.770) | 0.646 |
| T3 | 140 | 1.551
(0.921–2.612) | 0.099 | 1.110
(0.599–2.054) | 0.740 |
| T4 | 35 | 3.759
(1.959–7.213) | <0.001 | 2.336
(1.077–5.066) | 0.032 |
| Pathological N
stage | 1,067 |
|
|
|
|
| N0 | 516 | Reference |
| Reference |
|
| N1 | 358 | 1.947
(1.322–2.865) | <0.001 | 1.646
(1.080–2.508) | 0.020 |
| N2 | 116 | 2.522
(1.484–4.287) | <0.001 | 2.223
(1.258–3.931) | 0.006 |
| N3 | 77 | 4.191
(2.318–7.580) | <0.001 | 2.973
(1.413–6.255) | 0.004 |
| ULBP1 | 1,086 |
|
|
|
|
|
Low | 542 | Reference |
| Reference |
|
|
High | 544 | 1.443
(1.046–1.990) | 0.026 | 1.431
(1.007–2.033) | 0.046 |
| Pathological M
stage | 925 |
|
|
|
|
| M0 | 905 | Reference |
| Reference |
|
| M1 | 20 | 4.266
(2.474–7.354) | <0.001 | 2.018
(1.053–3.870) | 0.034 |
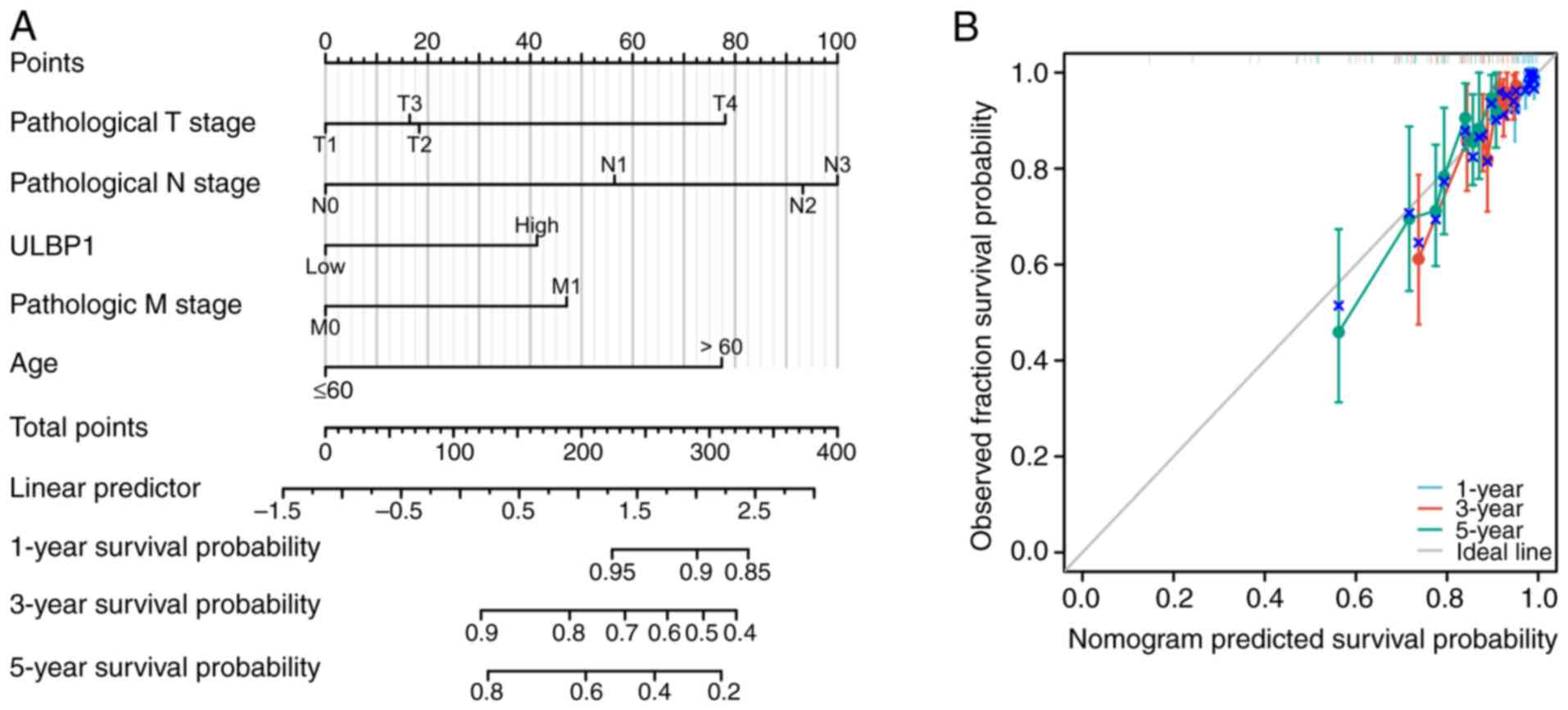
Furthermore, a nomogram was developed to predict the
OS rates of patients with BRCA at 1, 3 and 5 years based on the
pathological tumor node metastasis (TNM) stage, age and ULBP1
expression (Fig. 5A). The
combination of pathological TNM stage and ULBP1 expression was
utilized to forecast the OS of patients with BRCA at different
intervals. A bias-corrected line was constructed to approximate the
ideal curve that demonstrated agreement between predicted outcomes
and observed results at all time points (Fig. 5B). These findings highlight the
significant clinical relevance of ULBP1 in the assessment of the
prognosis of individuals diagnosed with BRCA.
Role of ULBP1 in BRCA investigated
through gene enrichment analysis
To delineate potential gene regulatory networks
associated with ULBP1, the RNA-sequencing data of corresponding
molecules were extracted from TCGA public database and divided into
high and low expression groups, based on ULBP1 molecule expression
levels; gene enrichment analysis was performed, and the ‘ggplot2’
package was utilized to visualize the results of this analysis.
Moreover, GO/KEGG analysis was employed to classify the gene list.
The gene set enrichment analysis indicated the significant
enrichment of several gene functional clusters, including
‘intermediate filament organization’ (P<0.001), ‘intermediate
filament cytoskeleton organization’ (P<0.001) and ‘intermediate
filament-based process’ (P<0.001) in patients with BRCA with a
high expression of ULBP1 (Fig. 6A).
In addition, genes were arranged in a descending order according to
their average resemblance to other genes. The gene at the top of
the list signified the greatest level of similarity with other
genes, suggesting its significant correlation and potential pivotal
function. The data analysis suggested that ULBP3, neuron
proteoglycan (‘NCAN’), SRY-box transcription factor 8 (‘SOX8’),
keratin 81 (‘KRT81’), retinaldehyde-binding protein 1 (‘RLBP1’),
claudin-6 (‘CLDN6’), collagen type IX alpha 1 Chain (‘COL9A1’),
ring finger protein 182 (‘RNF182’), nuclear factor erythroid 4
(‘NFE4’), cholecystokinin B receptor (‘CCKBR’), CaM kinase-like
vesicle-associated protein (‘CAMKV’), transient receptor potential
cation channel subfamily M member 8 (‘TRPM8’) and ADP-ribosyl
transferase C2 and C3 toxin-like 3 (‘ART3’) may play a crucial role
in the functioning of ULBP1 (Fig.
6B). The protein-protein interaction analysis revealed that
ULBP1 exhibits interactions with both ULBP2 and ULBP3, while ULBP2
interacts with retinoic acid early transcript 1K (RAET1K) (Fig. 6C). The expression levels of ULBP2,
ULBP3 and RAET1K in patients with BRCA were analyzed using heatmaps
to demonstrate associations. The data revealed significant
associations among the expression levels of ULBP1, ULBP2, ULBP3 and
RAET1K in BRCA (Fig. 6D).
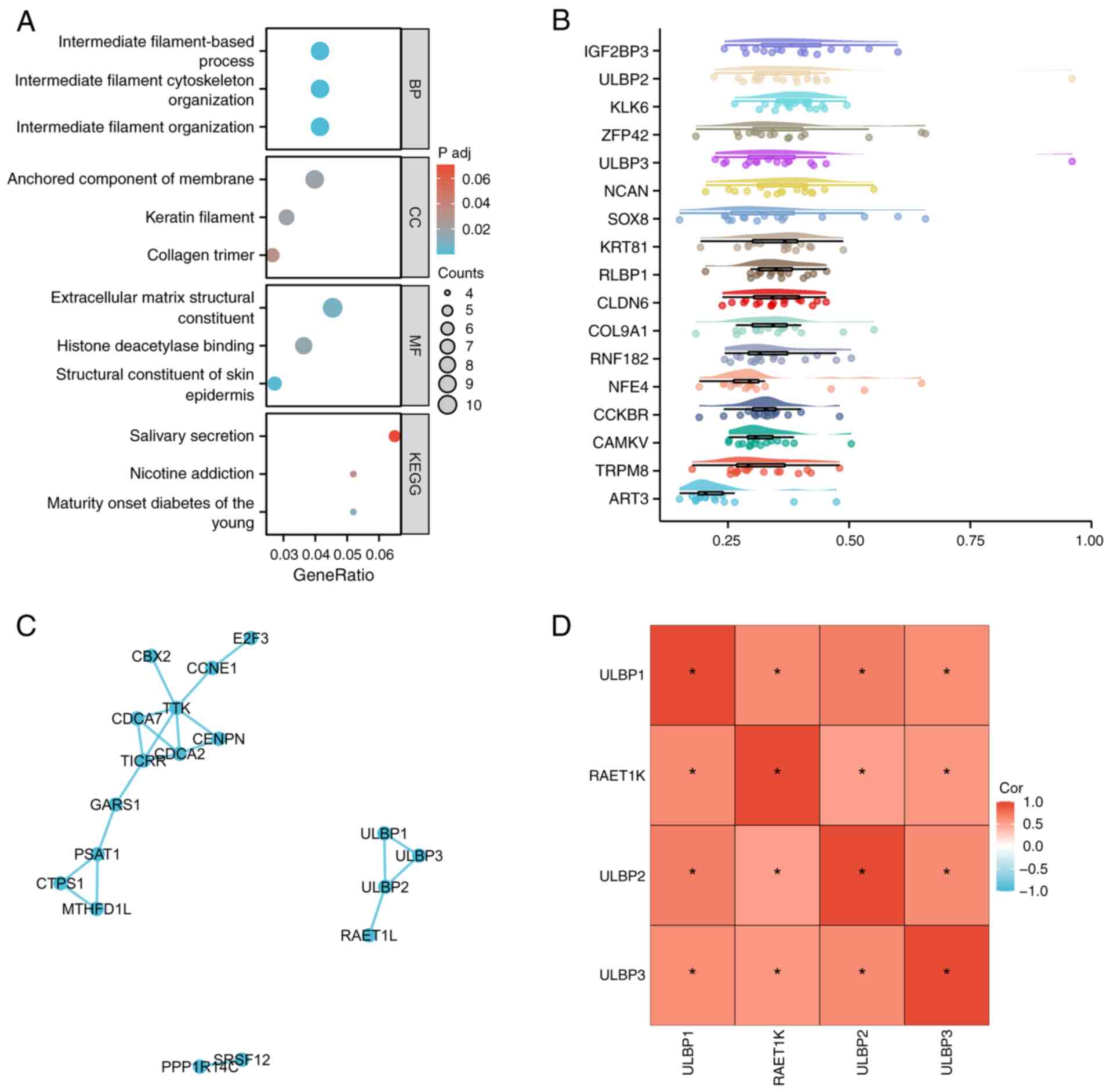 | Figure 6.Role of ULBP1 in BRCA. The role of
ULBP1 in BRCA was investigated using gene enrichment analysis. (A)
The differentially expressed genes were ranked based on the
correlation factor, and subsequently, the gene list underwent Gene
Ontology/Kyoto Encyclopedia of Genes and Genomes analysis for
clustering purposes. (B) The importance of each gene was calculated
using network topology, followed by screening key genes from the
gene list. (C) Protein-protein interaction analysis was performed
utilizing the STRING database (https://string-db.org). (D) Heatmaps were used to
analyze the expression levels of ULBP2, ULBP3 and RAET1K in
patients with BRCA to demonstrate correlations. *P<0.05. BRCA,
breast cancer; ULBP, UL16 binding protein; RAET1K, retinoic acid
early transcript 1K; NCAN, neuron proteoglycan; SOX8, SRY-box
transcription factor 8; KRT81, keratin 81; RLBP1,
retinaldehyde-binding protein 1; CLDN6, claudin-6; COL9A1, collagen
type IX alpha 1 chain; RNF182, ring finger protein 182; NFE4,
nuclear factor erythroid 4; CCKBR, cholecystokinin B receptor;
CAMKV, CaM kinase-like vesicle-associated protein; TRPM8, transient
receptor potential cation channel subfamily M member 8; ART3,
ADP-ribosyl transferase C2 and C3 toxin-like 3. |
Correlation between ULBP1 expression
and tumor immune-infiltration in BRCA
The correlation between ULBP1 expression and tumor
immunity is demonstrated in Fig. 7,
where the ssGSEA algorithm was utilized to evaluate the association
between the relative abundance of 24 immune cell types and ULBP1
expression in BRCA (Fig. 7A). As
illustrated in Fig. 7B-I, various
subsets of immune cells exhibited significant associations with
ULBP1 expression, including CD8+ T-cells (P<0.001;
R=−0.119), NK cells (P<0.001; R=−0.200), dendritic cells (DC)
cells (P<0.001; R=0.135), T-cells (P=0.004; R=0.086),
macrophages (P<0.001; R=0.316), type 2 T helper (Th2) cells
(P<000l; R=0.282), regulatory T (Treg) cells (P<0.00l;
R=0.238) and type 1 T helper (Th1) cells (P<0.00l; R=0.341).
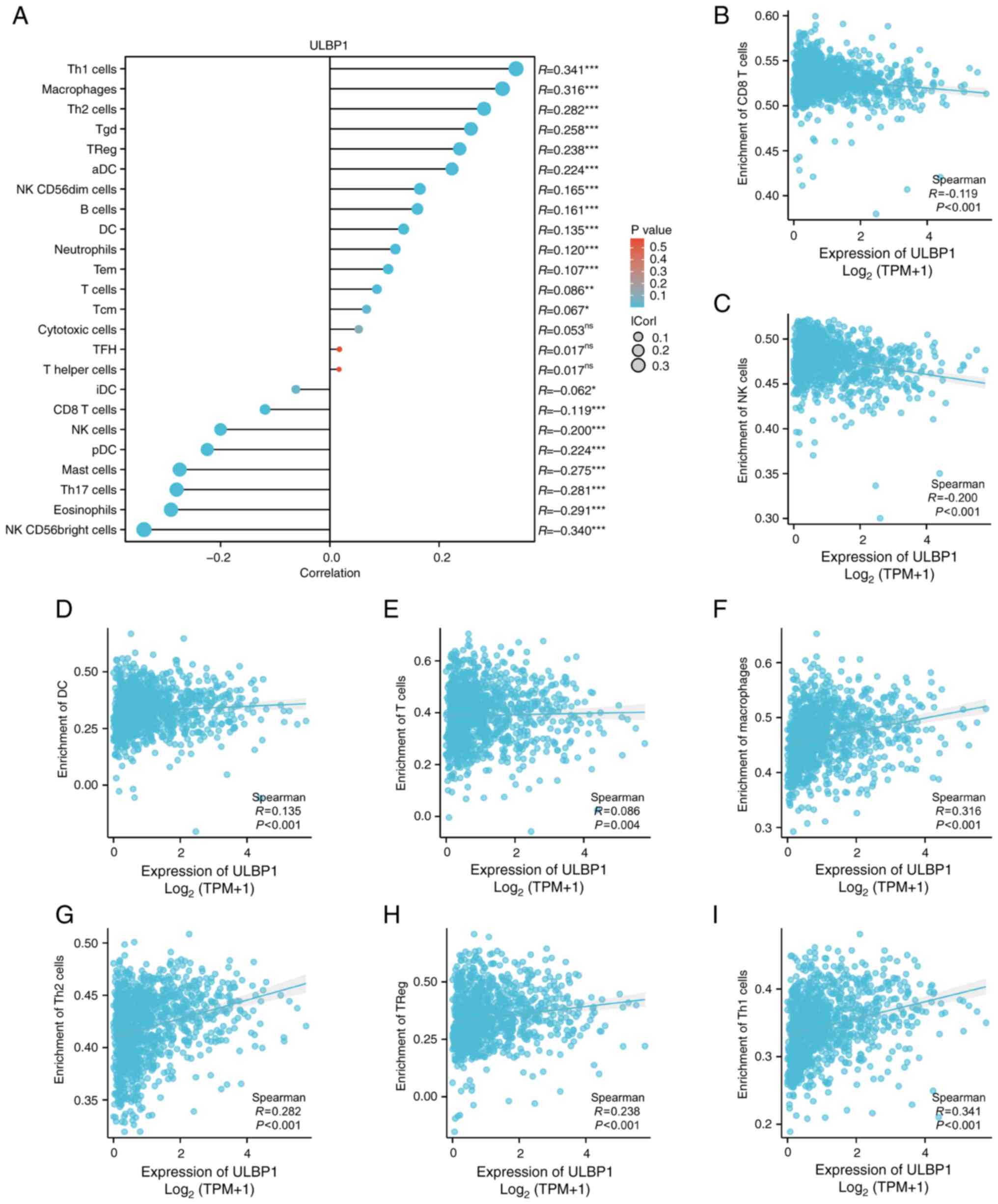 | Figure 7.Correlations between immune cells and
ULBP1. (A) The correlation between infiltrating immune cells and
the expression levels of ULBP1 was investigated in clinical samples
from patients with breast cancer, revealing significant
associations with various subsets of immune cells. The immune cells
included (B) CD8+ T cells, (C) NK cells, (D) DC cells,
(E) T cells, (F) macrophages, (G) Th2 cells, (H) Tregs, and (I) Th1
cells. *P<0.05; **P<0.01; ***P<0.001. ULBP1, UL16 binding
protein 1; DC, dendritic cells; Th2, type 2 T helper cell; Treg,
regulatory T cell; Th1, type 1 T helper cell; NK, natural
killer. |
Enrichment of immune cells in the
ULBP1 high and low expression groups
The Wilcoxon rank-sum test was employed to assess
the enrichment of immune cells in the ULBP1 high and low expression
groups. The findings revealed that, compared with the low
expression group, the high ULBP1 expression group exhibited
significantly reduced levels of CD8+ T-cells (Fig. 8A) and NK cells (Fig. 8B). Conversely, when compared to the
low expression group, the ULBP1 high expression group demonstrated
significantly elevated levels of DC cells, T-cells, Th1 cells, Th2
cells and macrophages (Fig.
8C-G).
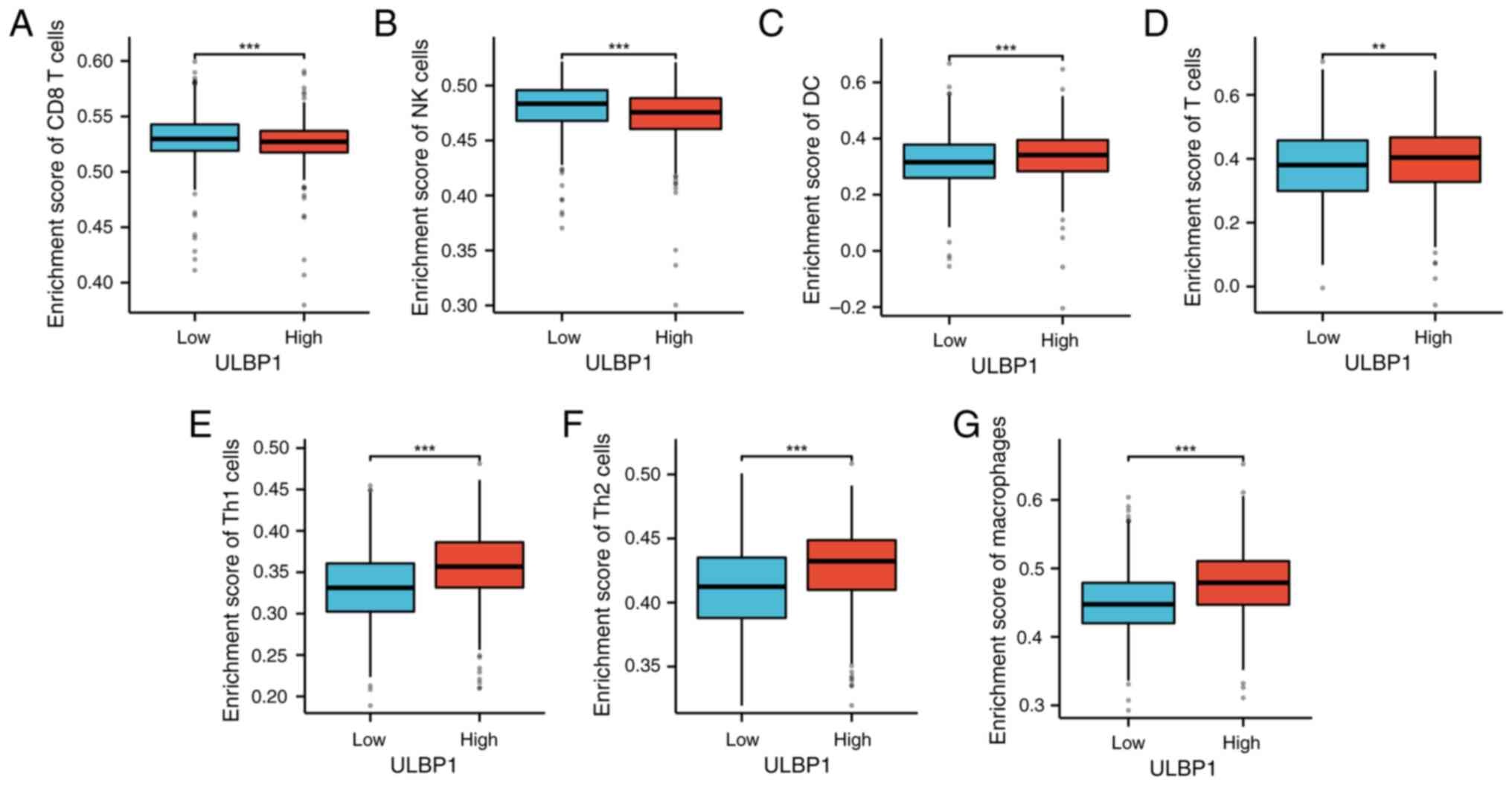 | Figure 8.ULBP1 expression and immune cells. The
high and low expression group of ULBP1 exhibited differential
enrichment of immune cells. Specifically, the high expression group
exhibited a significant decrease in (A) CD8+ T cells and
(B) NK cells. By contrast, compared with the low expression group,
the high expression group demonstrated a notable increase in (C) DC
cells, (D) T cells, (E) Th1 cells, (F) Th2 cells and (G)
macrophages. **P<0.01; ***P<0.001. ULBP1, UL16 binding
protein 1; DC, dendritic cells; Th2, type 2 T helper cell; Th1,
type 1 T helper cell; NK, natural killer. |
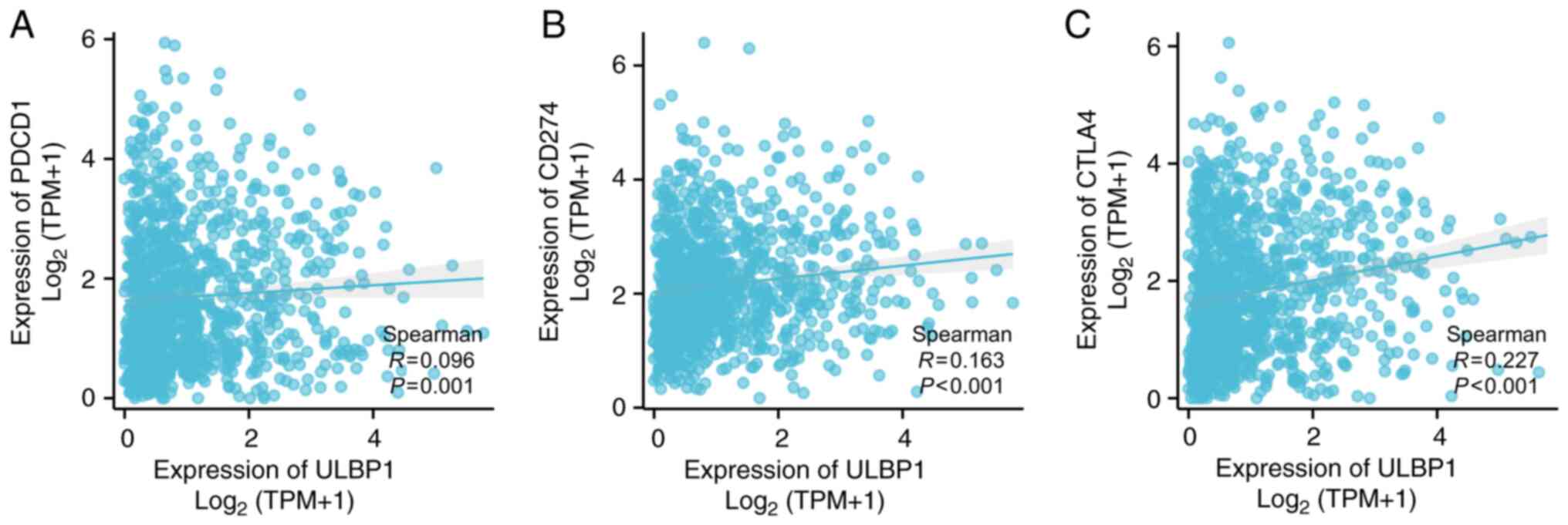
Correlation between tumor immune
checkpoints and ULBP1 expression in BRCA
The correlation between tumor immune checkpoints and
ULBP1 expression in BRCA was also examined. A positive correlation
was observed between the expression of ULBP1 and PDCD1 (P=0.001;
R=0.096), CD274 (P<0.001; R=0.163), as well as CTLA4 expression
(P<0.001; R=0.227) (Fig. 9).
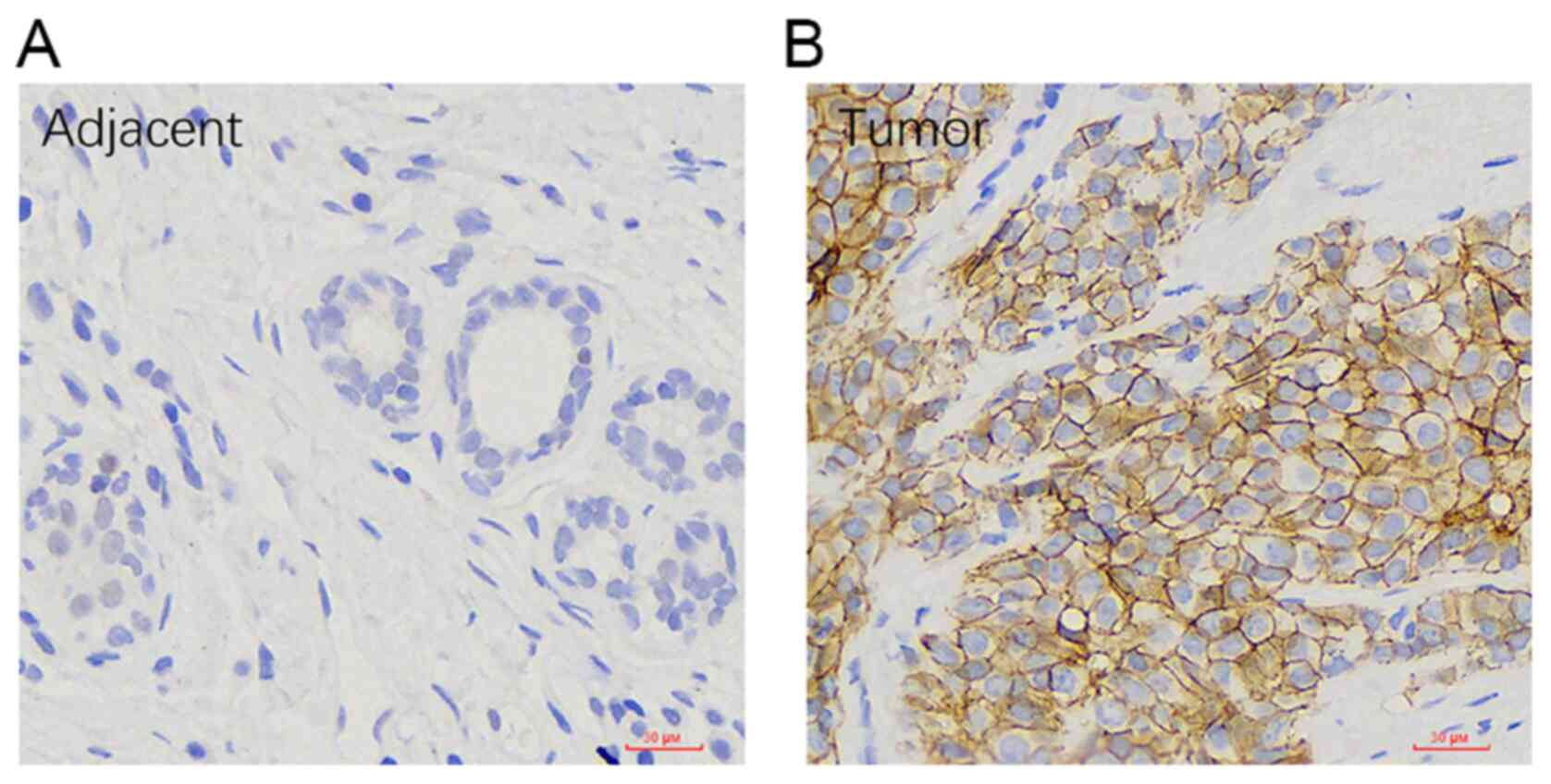
Assessment of ULBP1 expression in BRCA
using immunohistochemistry
The expression of ULBP1 in BRCA and its
corresponding para-cancerous tissue was confirmed using
immunohistochemistry. The predominant localization of ULBP protein
expression was observed on the cell membrane of BRCA tissues. The
findings demonstrated an increase in ULBP1 expression in 37 out of
74 cases of BRCA, as well as in 13 out of 74 paired para-cancer
normal tissue (Fig. 10).
Additionally, the results indicated an association between ULBP1
expression and clinical indicators among patients with BRCA,
including ER status (P<0.001) and PR status (P<0.001);
however, no association was observed with pathological T stage
(P=0.355), pathological N stage (P=0.831), pathological M stage
(P=0.792), pathological stage (P=0.916) or HER2 status (P=0.239).
Furthermore, there was a significant association between ULBP1
expression, and both ULBP2 expression (P<0.001) and ULBP3
expression (P<0.001) in patients with BRCA (Table III).
 | Table III.Association between ULBP1 and
clinical features of patients with BRCA. |
Table III.
Association between ULBP1 and
clinical features of patients with BRCA.
|
Characteristics | Low expression of
ULBP1 | High expression of
ULBP1 | P-value |
|---|
| n | 37 | 37 |
|
| Pathological T
stage |
|
| 0.355 |
| T1 +
T2 | 27 | 29 |
|
| T3 +
T4 | 10 | 8 |
|
| Pathological N
stage |
|
| 0.831 |
| N0 | 21 | 20 |
|
| N1 + N2
+ N3 | 16 | 17 |
|
| Pathological M
stage |
|
| 0.792 |
| M0 | 34 | 33 |
|
| M1 | 3 | 4 |
|
| Pathological
stage |
|
| 0.916 |
| Stage I
+ stage II | 28 | 27 |
|
| Stage
III + stage IV | 9 | 10 |
|
| ULBP2 |
|
| <0.001 |
| + | 24 | 14 |
|
| - | 13 | 23 |
|
| ULBP3 |
|
| <0.001 |
| + | 22 | 17 |
|
| - | 15 | 20 |
|
| ER status |
|
| <0.001 |
|
Negative | 5 | 17 |
|
|
Positive | 32 | 20 |
|
| HER2 status |
|
| 0.239 |
|
Negative | 26 | 29 |
|
|
Positive | 11 | 8 |
|
| PR status |
|
| <0.001 |
|
Negative | 7 | 19 |
|
|
Positive | 30 | 18 |
|
Discussion
In both experimental animals and patients with
cancer, the presence of tumor NKG2D ligands has been positively
associated with tumor eradication and improved patient survival
rates (20,21). These ligands are recognized by NKG2D
receptors at levels typically higher on tumor cells compared with
surrounding normal tissues, and can be further induced through
gastric cancer therapies (22).
Therefore, effective cancer treatment may directly induce damage to
tumor cells in order to stimulate the expression of NKG2D ligands
followed by subsequent attack from cytotoxic lymphocytes (23).
ULBP1 is a ligand that activates the NKG2D receptor,
playing a crucial role in immune regulation. NK cells express NKG2D
as an activating receptor (24).
Associated with major histocompatibility complex class I genes,
ULBP1 can be independently expressed in human cell lines and
primary tumors (25,26). The previous years have witnessed the
emergence of molecular targeted therapy and immunotherapy as
significant treatment modalities for BRCA (27). Notably, the clinical efficacy of
immune checkpoint inhibitors has been demonstrated (28); however, their utilization is limited
due to associated adverse events (29). Consequently, there is a need for the
further investigation of immune-related genes in order to enhance
the prognosis of patients with BRCA (30).
In the present study, based on TCGA database, it was
found that the expression level of ULBP1 was higher in BRCA tissues
compared with normal breast tissue samples. The upregulation of
ULBP1 expression in BRCA was associated with advanced clinical
pathological parameters such as age, race, PR expression, ER
expression, molecular subtypes and histological types. Patients
with a positive ER/PR status exhibit a higher cure rate and lower
recurrence rate compared with those with a negative status
(31). Subsequently, the results
obtained using bioinformatics were validated through the collection
of clinical patient data at the Zibo Central hospital. The evidence
obtained from immunohistochemical staining also indicated an
association between ULBP1 expression and clinical indicators among
patients with BRCA. These findings suggest that ULBP1 plays a
pivotal role in the management of BRCA and can serve as a molecular
marker for assessing the efficacy of endocrine therapy.
Additionally, ULBP1 exhibited a high diagnostic rate
and was identified as an independent prognostic factor through
multivariate regression analysis in BRCA. Furthermore, through GO
and KEGG pathway enrichment analysis, it was discovered that both
ULBP1 and its co-expressed mRNAs were enriched in certain signaling
pathways within tumors, such as ‘intermediate filament
organization’, ‘intermediate filament cytoskeleton organization’
and ‘intermediate filament-based process’. Moreover, the data
revealed significant associations among the expression levels of
ULBP1, ULBP2 and ULBP3 in BRCA. Moreover, the evidence obtained
from immunohistochemical staining also confirmed that there was a
significant association between ULBP1 expression, and both ULBP2
expression and ULBP3 expression in patients with BRCA. The
presented evidence suggests that ULBPs, namely ULBP1, ULBP2 and
ULBP3, exhibit co-expression in BRCA, and possess the ability to
interact synergistically with each other.
Immunotherapy utilizing immune checkpoint inhibitors
has demonstrated notable efficacy in tumor treatment and has
enhanced the prognosis of patients with cancer, particularly with
CD274/PD-1 inhibitors (32). In the
present study, correlation analysis revealed significant
associations between ULBP1 and CD274, CTLA4, PD-1 (PDCD1), as well
as various subsets of immune cells including DC cells, T-cells,
macrophages, Th2 cells, Treg cells and Th1 cells. However, a
negative association was observed with CD8+ T-cells and
NK cells. These findings suggest a correlation between ULBP1 and
immune infiltration, as well as immunosuppression within the
microenvironment of BRCA tumors.
In conclusion, the present study suggests an
association between ULBP1 and tumor immune response, highlighting
its potential as an attractive candidate for immunotherapeutic
interventions in BRCA. This biomarker holds promise in the
prediction and diagnosis of BRCA. However, future research
endeavors are required to validate the cellular functionality of
ULBP1.
Acknowledgements
Not applicable.
Funding
The present study received financial support from The Natural
Science Foundation of Shandong Province (grant no. ZR2021QH032) and
The Medical and Health Science and Technology Project of Shandong
Province (grant no. 202304070941).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
XZ, MD and PW were involved in conducting a portion
of the experiments and played a role in shaping the design of the
study. LL and SD provided support during the experimental
procedures and made valuable contributions to refining the
manuscript. All authors have read and approved the final version of
the manuscript. XZ and PW confirm the authenticity of all the raw
data. Each author significantly contributed to conceptualizing and
designing the study, acquiring, analyzing and interpreting data;
drafting or critically revising important intellectual content
within the article; as well as granting final approval for
publication.
Ethics approval and consent to
participate
The study has been approved by the Ethics Committee
of Zibo Central Hospital. The research program strictly follows the
scientific and ethical guidelines stated in the Declaration of
Helsinki, and written consent has been acquired from all
patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Wilkinson L and Gathani T: Understanding
breast cancer as a global health concern. The Br J Radiol.
95:202110332022. View Article : Google Scholar
|
|
2
|
Kawiak A: Molecular research and treatment
of breast cancer. Int J Mol Sci. 23:96172022. View Article : Google Scholar
|
|
3
|
Trapani D, Ginsburg O, Fadelu T, Lin NU,
Hassett M, Ilbawi AM, Anderson BO and Curigliano G: Global
challenges and policy solutions in breast cancer control. Cancer
Treat Rev. 104:1023392022. View Article : Google Scholar
|
|
4
|
Ben-Dror J, Shalamov M and Sonnenblick A:
The history of early breast cancer treatment. Genes (Basel).
13:9602022. View Article : Google Scholar
|
|
5
|
Katsura C, Ogunmwonyi I, Kankam HK and
Saha S: Breast cancer: Presentation, investigation and management.
Br J Hosp Med (Lond). 83:1–7. 2022. View Article : Google Scholar
|
|
6
|
Sarhangi N, Hajjari S, Heydari SF,
Ganjizadeh M, Rouhollah F and Hasanzad M: Breast cancer in the era
of precision medicine. Mol Biol Rep. 49:10023–10037. 2022.
View Article : Google Scholar
|
|
7
|
Goel S and Chandarlapaty S: Emerging
therapies for breast cancer. Cold Spring Harb Perspect Med.
13:a0413332023. View Article : Google Scholar
|
|
8
|
Lança T, Correia DV, Moita CF, Raquel H,
Neves-Costa A, Ferreira C, Ramalho JS, Barata JT, Moita LF, Gomes
AQ and Silva-Santos B: The MHC class Ib protein ULBP1 is a
nonredundant determinant of leukemia/lymphoma susceptibility to
gammadelta T-cell cytotoxicity. Blood. 115:2407–2411. 2010.
View Article : Google Scholar
|
|
9
|
López-Soto A, Quiñones-Lombraña A,
López-Arbesú R, López-Larrea C and González S: Transcriptional
regulation of ULBP1, a human ligand of the NKG2D receptor. J Biol
Chem. 281:30419–30430. 2006. View Article : Google Scholar
|
|
10
|
Hu B, Tian X and Li Y, Liu Y, Yang T, Han
Z, An J, Kong L and Li Y: Epithelial-mesenchymal transition may be
involved in the immune evasion of circulating gastric tumor cells
via downregulation of ULBP1. Cancer Med. 9:2686–2697. 2020.
View Article : Google Scholar
|
|
11
|
Zuo J, Willcox BE and Moss P: ULBPs:
Regulators of human lymphocyte stress recognition. Oncotarget.
8:106157–106158. 2017. View Article : Google Scholar
|
|
12
|
Ruan GT, Xie HL, Zhu LC, Ge YZ, Yan L,
Liao C, Gong YZ and Shi HP: Immune ULBP1 is elevated in colon
adenocarcinoma and predicts prognosis. Front Genet. 13:7625142022.
View Article : Google Scholar
|
|
13
|
Easom NJW, Marks M, Jobe D, Gillmore R,
Meyer T, Maini MK and Njie R: ULBP1 is elevated in human
hepatocellular carcinoma and predicts outcome. Front Oncol.
10:9712020. View Article : Google Scholar
|
|
14
|
Cho H, Chung JY, Kim S, Braunschweig T,
Kang TH, Kim J, Chung EJ, Hewitt SM and Kim JH: MICA/B and ULBP1
NKG2D ligands are independent predictors of good prognosis in
cervical cancer. BMC Cancer. 14:9572014. View Article : Google Scholar
|
|
15
|
Wen WX, Soo JS, Kwan PY, Hong E, Khang TF,
Mariapun S, Lee CS, Hasan SN, Rajadurai P, Yip CH, et al: Germline
APOBEC3B deletion is associated with breast cancer risk in an Asian
multi-ethnic cohort and with immune cell presentation. Breast
Cancer Res. 18:562016. View Article : Google Scholar
|
|
16
|
Bild AH, Yao G, Chang JT, Wang Q, Potti A,
Chasse D, Joshi MB, Harpole D, Lancaster JM, Berchuck A, et al:
Oncogenic pathway signatures in human cancers as a guide to
targeted therapies. Nature. 439:353–357. 2006. View Article : Google Scholar
|
|
17
|
Liu RZ, Graham K, Glubrecht DD, Germain
DR, Mackey JR and Godbout R: Association of FABP5 expression with
poor survival in triple-negative breast cancer: Implication for
retinoic acid therapy. Am J Pathol. 178:997–1008. 2011. View Article : Google Scholar
|
|
18
|
Clarke C, Madden SF, Doolan P, Aherne ST,
Joyce H, O'Driscoll L, Gallagher WM, Hennessy BT, Moriarty M, Crown
J, et al: Correlating transcriptional networks to breast cancer
survival: A large-scale coexpression analysis. Carcinogenesis.
34:2300–2308. 2013. View Article : Google Scholar
|
|
19
|
Zhang X, Ruan Y, Li Y, Lin D and Quan C:
Tight junction protein claudin-6 inhibits growth and induces the
apoptosis of cervical carcinoma cells in vitro and in
vivo. Med Oncol. 32:1482015. View Article : Google Scholar
|
|
20
|
Kamimura H, Yamagiwa S, Tsuchiya A,
Takamura M, Matsuda Y, Ohkoshi S, Inoue M, Wakai T, Shirai Y,
Nomoto M, et al: Reduced NKG2D ligand expression in hepatocellular
carcinoma correlates with early recurrence. J Hepatol. 56:381–388.
2012. View Article : Google Scholar
|
|
21
|
Lee GH, An HJ, Kim TH, Kim G, Park KS,
Park H, Lee TH and Kwon AY: Clinical impact of natural killer group
2D receptor expression and that of its ligand in ovarian
carcinomas: A retrospective study. Yonsei Med J. 62:288–297. 2021.
View Article : Google Scholar
|
|
22
|
Liu X, Sun M, Yu S, Liu K, Li X and Shi H:
Potential therapeutic strategy for gastric cancer peritoneal
metastasis by NKG2D ligands-specific T cells. Onco Targets Ther.
8:3095–3104. 2015.
|
|
23
|
Bae JH, Kim SJ, Kim MJ, Oh SO, Chung JS,
Kim SH and Kang CD: Susceptibility to natural killer cell-mediated
lysis of colon cancer cells is enhanced by treatment with epidermal
growth factor receptor inhibitors through UL16-binding protein-1
induction. Cancer Sci. 103:7–16. 2012. View Article : Google Scholar
|
|
24
|
Himmelreich H, Mathys A, Wodnar-Filipowicz
A and Kalberer CP: Post-transcriptional regulation of ULBP1 ligand
for the activating immunoreceptor NKG2D involves 3′ untranslated
region. Hum Immunol. 72:470–478. 2011. View Article : Google Scholar
|
|
25
|
Nanbakhsh A, Pochon C, Mallavialle A,
Amsellem S, Bourhis JH and Chouaib S: c-Myc regulates expression of
NKG2D ligands ULBP1/2/3 in AML and modulates their susceptibility
to NK-mediated lysis. Blood. 123:3585–3595. 2014. View Article : Google Scholar
|
|
26
|
Textor S, Fiegler N, Arnold A, Porgador A,
Hofmann TG and Cerwenka A: Human NK cells are alerted to induction
of p53 in cancer cells by upregulation of the NKG2D ligands ULBP1
and ULBP2. Cancer Res. 71:5998–6009. 2011. View Article : Google Scholar
|
|
27
|
Odle TG: Precision medicine in breast
cancer. Radiol Technol. 88:401M–421M. 2017.
|
|
28
|
Postow MA, Sidlow R and Hellmann MD:
Immune-related adverse events associated with immune checkpoint
blockade. N Engl J Med. 378:158–168. 2018. View Article : Google Scholar
|
|
29
|
Zhang Y and Zheng J: Functions of immune
checkpoint molecules beyond immune evasion. Adv Exp Med Biol.
1248:201–226. 2020. View Article : Google Scholar
|
|
30
|
Onkar SS, Carleton NM, Lucas PC, Bruno TC,
Lee AV, Vignali DAA and Oesterreich S: The great immune escape:
understanding the divergent immune response in breast cancer
subtypes. Cancer discov. 13:23–40. 2023. View Article : Google Scholar
|
|
31
|
da Silva JL, Cardoso Nunes NC, Izetti P,
de Mesquita GG and de Melo AC: Triple negative breast cancer: A
thorough review of biomarkers. Crit Rev Oncol Hematol. 145:102855.
2020. View Article : Google Scholar
|
|
32
|
Dermani FK, Samadi P, Rahmani G, Kohlan AK
and Najafi R: PD-1/PD-L1 immune checkpoint: Potential target for
cancer therapy. J Cell Physiol. 234:1313–1325. 2019. View Article : Google Scholar
|