Introduction
Breast cancer is the most prevalent form of cancer
among women worldwide, and notably, breast cancer has the highest
incidence rate among all cancer types. Furthermore, ~2.26 million
[95% uncertainty interval, 2.24–2.79 million] new cases of breast
cancer were reported in 2020 worldwide (1). Human epidermal growth factor receptor
2 (HER2), a tyrosine kinase receptor, is intricately linked with
cellular proliferation (2–4), metastasis (5–8),
invasion (9–12) and angiogenesis (13,14),
and is thus often considered a negative prognostic factor for
breast cancer. In total, 30–40% of breast cancer cases lack HER2
expression (HER2-0) (15), 15–20%
are HER2-positive [including HER2 immunohistochemistry (IHC) 2+ and
fluorescence in situ hybridization (FISH) positive or IHC
3+] and 45–55% have low HER2 expression [including HER2 IHC 1+ or
IHC 2+ and FISH negative] (16).
Thus, breast cancer cases with low HER2 expression represent
approximately half of all types, underscoring their growing
significance.
Although targeted therapies can improve the quality
of life of patients with cancer, anti-HER2 therapies such as
trastuzumab, are limited to patients with HER2 + cancer, and thus
do not improve the prognoses of patients with low HER2 expression
(HER2 IHC 1+ or IHC 2+ and FISH negative) (17,18).
Antibody-drug conjugates (ADCs), including trastuzumab deruxtecan
(a compound containing an anti-HER2 antibody and a cytotoxic
topoisomerase I inhibitor) have expanded the efficacy of targeted
treatments for patients with low HER2 expression. ADCs combine the
selectivity of targeted therapy with the cytotoxicity of
chemotherapy (19). Besides,
extensive ongoing and completed research on low HER2 expression has
confirmed that novel ADC treatments can benefit these patients,
necessitating accurate differentiation between HER2-0 and HER2 1+
and HER2 2+ cases (20–23).
Abnormal lipid metabolism is common in breast
cancer, which can impact various biological processes across
different cancer subtypes, including hormone receptor (HR)+, HER2 +
and triple-negative (24–26). Each subtype exhibits specific and
overlapping lipid dependencies. Numerous studies have demonstrated
that changes in lipid metabolism influence various aspects of
breast cancer, including cell growth, survival, adaptability,
resistance to treatment and the ability to spread (27–30).
Lipids, which are either non-polar or polar (amphipathic)
biomolecules, are synthesized within the cells or absorbed from the
surrounding environment (31).
Although lipidomic profiles have the potential for predicting and
diagnosing breast cancer, numerous challenges remain unresolved,
such as sample complexity and heterogeneity, as well as biomarker
validation.
The present study aimed to analyze the lipidomic
profiles of 30 patients with breast cancer, categorizing the
patients into four groups based on the HR and HER2 status.
Differences in lipid compositions among the groups were revealed
using liquid chromatography-mass spectrometry (LC-MS) and various
statistical methods, such as principal component analysis (PCA),
partial least squares-discriminant analysis (PLS-DA) and random
forest (RF) classification. These findings may enhance the
understanding of the link between breast cancer subtypes and lipid
metabolic changes as well as provide potential biomarkers and
therapeutic targets for breast cancer classification and treatment
in the future.
Materials and methods
Study population
The present study was conducted at the Department of
Breast and Thyroid Surgery, Shaoxing People's Hospital (Shaoxing,
China) between September, 2021 and December, 2023. In total, 30
patients with breast cancer were included in this study. The
patients were recruited prospectively and consecutively according
to the eligibility criteria as follows: i) Patients with breast
cancer confirmed via pathological examination; ii) patients who
could withstand study tests; iii) verbal informed consent obtained
from the patient, legal representative or responsible caregiver;
and iv) patients with a knowledgeable and reliable caregiver
accompanying them to all clinic visits during the study. The
exclusion criteria were as follows: i) Patients with other severe
disease; and ii) patients with comorbidities such as other types of
cancer, severe depression, severe renal or hepatic insufficiency
and severe cardiac or respiratory failure. The present study
followed the relevant principles of the Declaration of Helsinki.
The demographic and disease information of the patients was
obtained from the medical records, including age, sex, World Health
Organization (WHO) grade, Ki67 status, prognosis and lipid profiles
were collected from the patients' medical records.
Detection of markers via IHC or
FISH
HR status is defined as the presence or absence of
the estrogen receptor (ER) and progesterone receptor (PR) on the
surface of breast cancer cells. Tumors with these receptors are
known as HR+. In the IHC protocol, tissues are first fixed with
formaldehyde (10%, room temperature, 24 h), embedded in paraffin
and sectioned (5 µm) to create paraffin-embedded slides. These
slides are then deparaffinized and rehydrated. Antigen retrieval
was performed using a citrate buffer (100°C, 30 min) to enhance
antigen binding efficiency. Non-specific binding was minimized by
applying 5% goat serum (Beyotime Institute of Biotechnology; cat.
no. C0265). Endogenous peroxidase activity was blocked using a
hydrogen peroxide solution (2%, 10 min). The slides are
subsequently incubated at room temperature for 2 h with primary
antibodies targeting ER (Roche Diagnostics; clone: SP1; cat. no.
790-4325), PR (Roche Diagnostics; clone: 1E2; cat. no. 790-4296) or
HER2 (Roche Diagnostics; clone: 4B5; cat. no. 790-2991) at a 1:500
dilution, followed by incubation with a secondary antibody with
horseradish peroxidase (Thermo Fisher Scientific, Inc.; cat. no.
31430) at room temperature for 30 min at a 1:1,000 dilution.
Visualization was achieved using DAB (Roche Diagnostics; cat. no.
760-500) to detect the bound antibodies. An optical microscope
(Zeiss AG; OPMI PENTERO 900) was used to observe and collect
images, and the HALO platform (version 3.4; Indica Labs, Inc.) was
used to analyze the IHC staining. Patients with a HER2 IHC score of
2+ required further testing using FISH.
FISH utilized the same paraffin-embedded sections as
IHC, meaning the processes of tissue collection and fixation,
paraffin embedding and slide preparation were identical. The HER2
gene test kit (Anbiping Pharmaceutical Technology Co., Ltd.; cat.
no. 2408001) was used to further assess HER2 status. Denaturation,
hybridization, washing and restaining were performed according to
the manufacturer's instructions. The signals were then detected
using a fluorescence microscope. A ratio of HER2/chromosome 17
signal was classified as negative, while a ratio ≥2.0 was
classified as positive.
Lipidomic profiling
The lipidomic profiles of patients were established
through non-targeted LIPIDOMIC studies. The sample was added to
water and methyl-tert-butyl ether, followed by vortex mixing.
Methanol was then added and the mixture was vortexed again. The
mixture underwent ultrasonication for 20 min, incubated at room
temperature for 30 min and then centrifuged (14,000 × g, room
temperature, 15 min) to collect the supernatant. The organic phase
was dried under nitrogen, redissolved in a 90%
isopropanol/acetonitrile solution, vortexed and centrifuged (14,000
× g, room temperature, 15 min) again for analysis. The extracted
lipids were analyzed using high-throughput LC-MS with an Agilent
1290 liquid chromatography system (Agilent Technologies, Inc.)
connected to an Agilent 6550 iFunnel Q-TOF mass spectrometer
(Agilent Technologies, Inc.). LC-MS analysis was performed in both
positive and negative ion modes to capture comprehensive data to
ensure a detailed lipidomic profile. The key parameters of
electrospray ionization were as follows: Spray heater gas
temperature (nitrogen), 300°C; nebulizer pressure, 30 psi; gas flow
rate, 10 l/min.
Statistical analysis
Age differences among the four groups were analyzed
using analysis of variance (ANOVA). Post hoc comparisons were
conducted using Tukey's HSD test. Other variables such as
laterality, World Health Organization (WHO) grade (32) and Ki67 status were evaluated using
the Fisher test. WHO grading was used to assess the invasiveness
and growth rate of tumors and was classified into grades I to III
based on severity. P<0.05 was considered to indicate a
statistically significant difference. Lipidomics data were
processed and analyzed using the ‘MetaboAnalystR’ package (version
4.0) in R (version 4.3.0; R Foundation for Statistical Computing;
http://www.R-project.org/) (33). Differential lipids were identified
by integrating P-values from the unpaired t-test, variable
importance in projection (VIP) scores from the OPLS-DA model and
log fold change (logFC) criteria. The selection criteria were set
at P<0.05, VIP ≥1 and logFC ≥1 or logFC ≥-1, to ensure the
inclusion of only the most relevant lipids. Lipid pathway
enrichment analysis was conducted using the RaMP-DB database
(http://github.com/ncats/) (P<0.05),
linking differential lipids to potential biological functions.
Additionally, the relationships between lipids were assessed using
Spearman correlation (P<0.05), visualized in Cytoscape (version
3.9.1; http://cytoscape.org/) (34). Lipid centrality was determined using
the cytoHubba plugin, which was used to construct a lipid
interaction network diagram.
Results
Patient characteristics
The characteristics of the patients, including age,
sex, in situ carcinoma type, lymph node metastasis, HR
status and HER2 status are shown in Table I. The 30 patients with breast cancer
were divided into four groups based on the HR and HER2 status
according to the guidelines for breast cancer diagnosis and
treatment by the China Anti-Cancer Association (2024 edition)
(35): i) HR+ HER2-0: HR IHC
positive, HER2 IHC score of 0; ii) HR+ HER2-low: HR IHC positive,
HER2 IHC 1+ or 2+ and FISH negative; iii) HR+ HER2-positive (pos):
HR IHC positive, HER2 IHC 3+ or 2+ and FISH positive; and iv) HR-
HER2-pos: HR IHC negative, HER2 IHC 3+ or 2+ and FISH positive.
Fig. 1A displays representative HR
(ER and PR) and HER2 IHC images for the four groups of patients.
Fig. 1B displays representative
HER2 FISH images (negative and positive). ANOVA (with Tukey's HSD
post-hoc test) and Fisher analysis revealed no significant
differences in age, laterality or Ki67 status among the four
patient groups. However, a significant difference in the
distribution of WHO grades was observed among the four groups.
Nearly all patients in the HR- HER2-pos group were grade III,
whereas grades I–II were more prevalent in the HR+ HER2-low group
(Table I). The detailed patient
information is listed in Tables SI
and SII presents the follow-up
data for all patients, including recurrence status and the lipid
profiles at the time of the last examination.
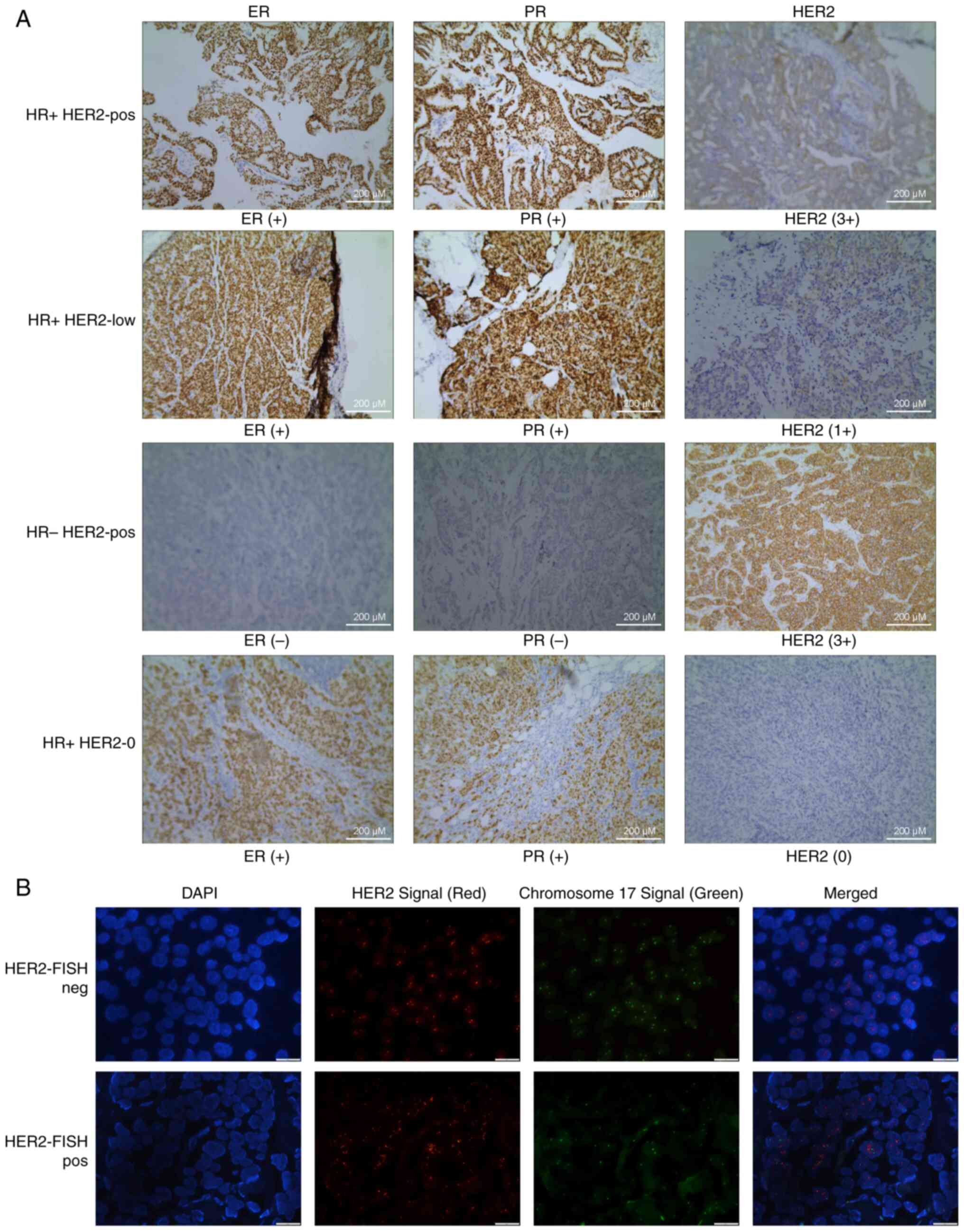 | Figure 1.Representative IHC and FISH images of
the HR and HER2 staining of samples from the four groups of
patients: (A) Representative ER and HER2 IHC images. HR+
HER2-pos (sample ID: 31), HR+ HER2-low (sample ID: 38),
HR− HER2-pos (sample ID: 43) and HR+ HER2-0
(sample ID: 51). Scale bar, 200 µm. (B) Representative HER2 FISH
images. HER2-FISH neg (sample ID: 42) and HER2-FISH pos (sample ID:
39). Scale bar, 20 µm. ER, estrogen receptor; FISH, fluorescence
in situ hybridization; HER2, human epidermal growth factor
receptor 2; HR, hormone receptor; pos, positive; IHC,
immunohistochemistry; PR, progesterone receptor. |
 | Table I.Baseline characteristic of the
included patients with breast cancer. |
Table I.
Baseline characteristic of the
included patients with breast cancer.
| Characteristic | HR+
HER2-pos, n=4 | HR+
HER2-low, n=15 | HR−
HER2-pos, n=7 | HR+
HER2-0, n=4 | P-value |
|---|
| Mean age (SD),
years | 54 (7.7) | 59.49 (10.7) | 59.5 (9.4) | 57.2 (8.3) | 0.1892 |
| Age, n (%) |
|
|
|
| 0.6283 |
| <40
years | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
|
| 40-59
years | 3 (75) | 8 (54) | 3 (43) | 3 (75) |
|
| ≥60
years | 1 (25) | 7 (46) | 4 (57) | 1 (25) |
|
| Laterality, n
(%) |
|
|
|
| 0.75 |
|
Left | 2 (50) | 7 (46) | 5 (71) | 2 (50) |
|
|
Right | 2 (50) | 8 (54) | 2 (29) | 2 (50) |
|
| WHO grade n
(%) |
|
|
|
|
<0.01a |
|
I–II | 3 (75) | 14 (94) | 0 (0) | 1 (25) |
|
|
III | 1 (25) | 1 (6) | 7 (100) | 2 (50) |
|
|
Unknown | 0 (0) | 0 (0) | 0 (0) | 1 (25) |
|
| Ki67, n (%) |
|
|
|
| 0.95 |
|
<20% | 1 (25) | 0 (0) | 0 (0) | 1 (25) |
|
|
≥20% | 3 (75) | 15 (100) | 7(100) | 3 (75) |
|
Lipidomic landscape
Changes in the lipid composition were detected using
high-throughput LC-MS analysis. Specifically,
lysophosphatidylcholine (LPC), triglycerides (TGs),
phosphatidylcholine (PC) and sphingomyelin (SM) lipids were
detected in patient plasma. Fig. 2A
provides a detailed display of the distribution of relative
abundance of lipid metabolites in all patients, with
TG(16:0-18:1-18:1)+NH4, TG(16:0-18:1-18:2)+NH4,
SM(d34:1)+H and TG(16:1-18:1-18:2)+NH4 showing higher relative
abundances. Furthermore, significant lipid feature differences
among the groups were evaluated using ANOVA. The top 25 lipid
features among the four groups are shown in Fig. 2B. For instance, significant
differences were observed in the levels of TG(4:0-11:2-12:3)+NH4
across the four groups. The levels in the HR- HER2-pos group were
notably lower compared with the other groups.
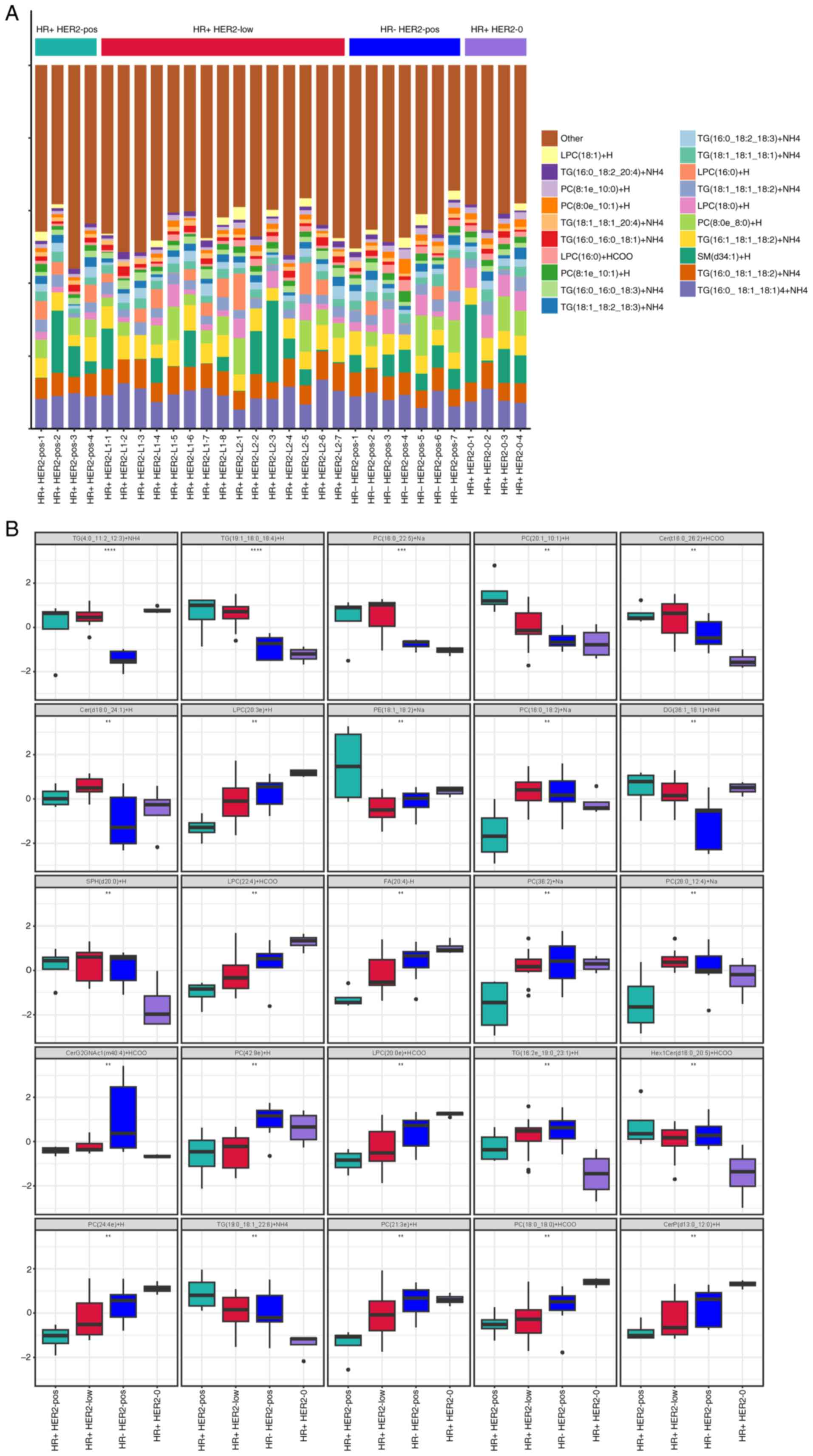 | Figure 2.Lipid profile variations among the
breast cancer subtypes. (A) Lipid composition profile showing the
prevalence of TG(16:0-18:1-18:1)+NH4 in the plasma of
patients with breast cancer, illustrating variations among the four
groups based on the HR and HER2 status. (B) A synthesized overview
of the lipid composition across the four patient groups,
highlighting the significant differences as identified by ANOVA.
**P<0.01, ***P<0.001, ****P<0.0001. Cer, ceramide;
CerG2GNAc1, N-acetylglucosaminyl(dihexosyl)ceramide; CerP,
ceramide-1-phosphate; DG, diacylglycerol; FA, fatty acid; HER2,
human epidermal growth factor receptor 2; Hex1Cer, hexosylceramide;
HR, hormone receptor; LPC, lysophosphatidylcholine; PC,
phosphatidylcholine; PE, phosphatidylethanolamine; pos, positive;
SM, sphingomyelin; SPH, sphingosine; TG, triglyceride. |
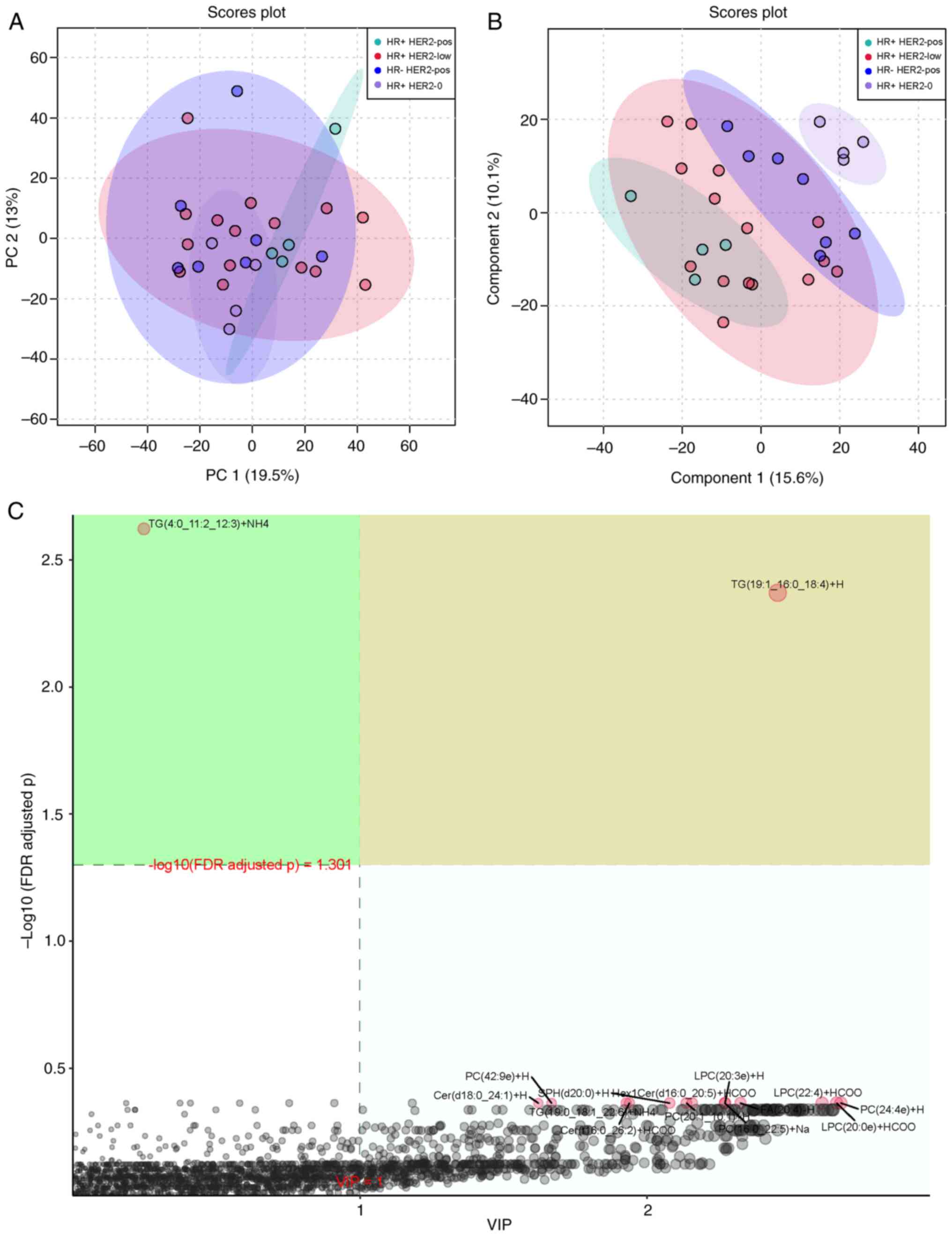
Multivariate statistical analysis
The differences in lipid profiles among the four
groups were also assessed using PCA (Fig. 3A) and PLS-DA (Fig. 3B). PCA revealed unique lipid
profiles in the HR+ HER2-pos and HR+ HER2-0 groups, while PLS-DA
distinguished the four groups. The VIP score plot for PLS-DA
indicated that TG(19:1-16:0-18:4)+H [VIP >1; -log10(FDR adjusted
P) >1.301] was the metabolite with the greatest impact in the
discriminant analysis (Fig. 3C).
Figs. S1 and S2 display the PCA and PLS-DA for the six
pairwise combinations among the four groups.
Importance and variation analysis
The RF approach highlighted
phosphatidylinositol-3,4,5-trisphosphate (PIP3)
(21:2)+NH4 as a crucial lipid feature for sample
grouping accuracy (Fig. 4A).
Fig. S3 displays the importance of
different metabolites in distinguishing the six pairwise
combinations among the four groups. The results showed that
TG(50:13)+NH4, TG(18:3e-18:3-21:1)+Na, PC(36:5)+H,
TG(4:0-11:2-12:3)+NH4, TG(18:0-20:4-22:6)+NH4
and DG(16:0-20:4)+H were important for distinguishing HR+ HER2-pos
and HR- HER2-low, HR+ HER2-pos and HR- HER2-pos, HR+ HER2-pos and
HR+ HER2-0, HR+ HER2-low and HR- HER2-pos, HR+ HER2-low and HR+
HER2-0 as well as HR+ HER2-pos and HR+ HER2-0, respectively.
Additionally, the abundance of the top-ranked lipid features is
shown in a heatmap, providing a visual representation of lipid
feature distribution across groups (Fig. 4B). These analyses were integrated
into a single figure to display the importance and abundance
variations of the lipid features.
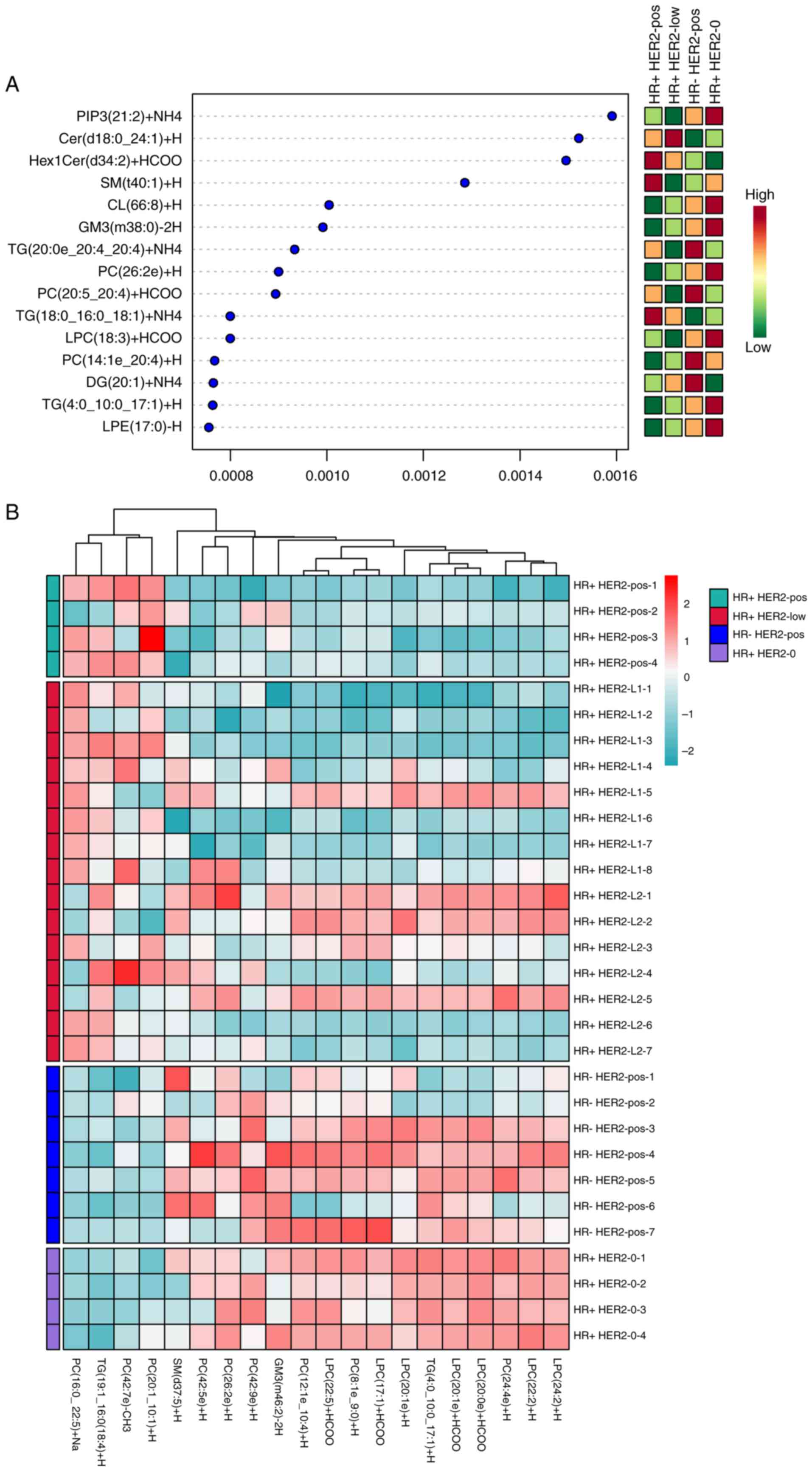 | Figure 4.Key lipid features in breast cancer
subtyping via random forest analysis. (A) RF analysis highlighting
PIP3(21:2)+NH4 as a crucial lipid feature for accurately grouping
patients, showcasing its potential as a biomarker for breast cancer
subtypes. (B) Heatmap displaying the abundance of top-ranked lipid
features across the four groups, providing insights into the
distribution variations and potential metabolic distinctions. Cer,
ceramide; CL, cardiolipin; DG, diacylglycerol; GM3, monosialo
ganglioside GM3; HER2, human epidermal growth factor receptor 2;
Hex1Cer, hexosylceramide; HR, hormone receptor; LPC,
lysophosphatidylcholine; LPE, lysophosphatidylethanolamine; PC,
phosphatidylcholine; PIP3,
phosphatidylinositol-3,4,5-trisphosphate; pos, positive; SM,
sphingomyelin; TG, triglyceride. |
Advanced analysis of the differential
lipids
Fatty acids with different carbon chain lengths have
different properties and functions. Lipid carbon chain length and
the number of double bonds are closely related to lipid oxidation
and function (36). The potential
oxidation of lipids is lower when the carbon chain is longer or the
number of double bonds is lower (37). Herein, only the number of carbon
atoms and double bonds of the differential lipids were evaluated to
identify significant correlations between the four groups (Fig. 5A). A heatmap based on the classified
lipids further explored these relationships, emphasizing the
physical and chemical properties of lipids that contribute to lipid
group classification (Fig. 5B). The
results indicated that TGs, including TG(20:4e-12:3-12:3),
TG(10:0-18:2-18:2) and TG(19:1-16:0-16:0), as well as DGs such as
DG(34:5e), DG(18:3-18:2) and DG(16:0-18:3), were the most distinct
lipids promoting group classifications.
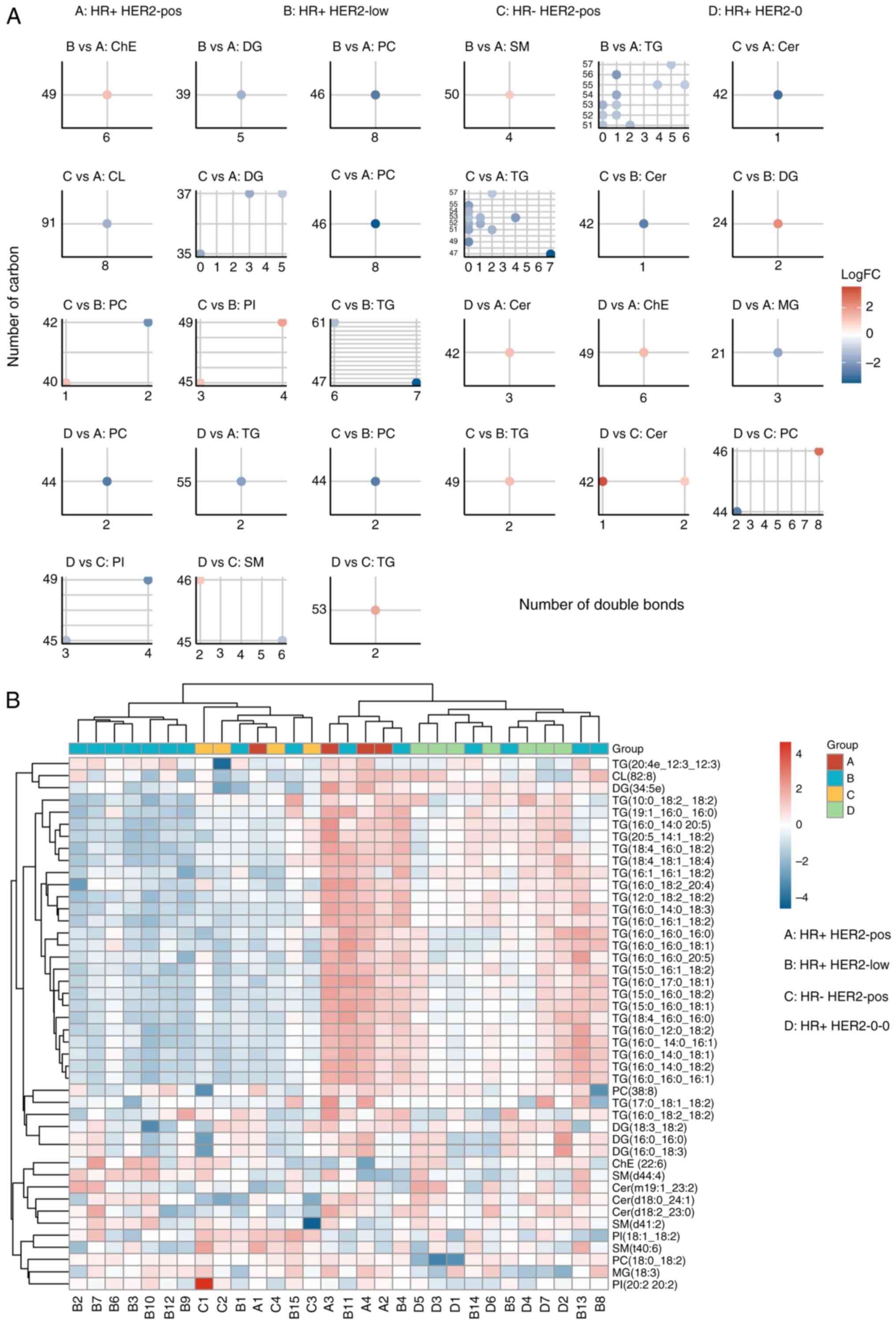 | Figure 5.Correlation between the lipid
structural properties and the breast cancer subtypes. (A)
Statistical analysis of the number of carbon atoms and double bonds
in the differential lipids, showing significant correlations with
group classification, suggesting that the structural properties of
the lipids are potentially influential in cancer phenotypes. (B)
Heatmap based on classified lipids, emphasizing the physical and
chemical properties of lipids that contribute to group
classifications, and highlighting the structural diversity and its
implications. Cer, ceramide; CL, cardiolipin; ChE, cholesterol
ester; DG, diacylglycerol; FC, fold change; HER2, human epidermal
growth factor receptor 2; HR, hormone receptor; MG,
monoacylglycerol; PC, phosphatidylcholine; PI,
phosphatidylinositol; pos, positive; SM, sphingomyelin; TG,
triglyceride. |
Correlation and pathway enrichment
analysis of the differential lipids
A correlation heatmap of the differential lipid
classes revealed that specific lipid types were positively
correlated with one another (Fig.
6A). Blue and red indicate positive and negative correlations,
with deeper shades indicating stronger associations. SMs were
negatively correlated with TGs, whereas ceramides were positively
correlated with TGs. Network analysis indicated that TGs were
extensively correlated with other lipids (Fig. 6B). Pathway enrichment analysis
showed that differential lipids were associated with various
pathways, particularly the ‘Assembly of active LPL and LIPC lipase
complexes’, which had a high fold enrichment and statistical
significance (Fig. 6C).
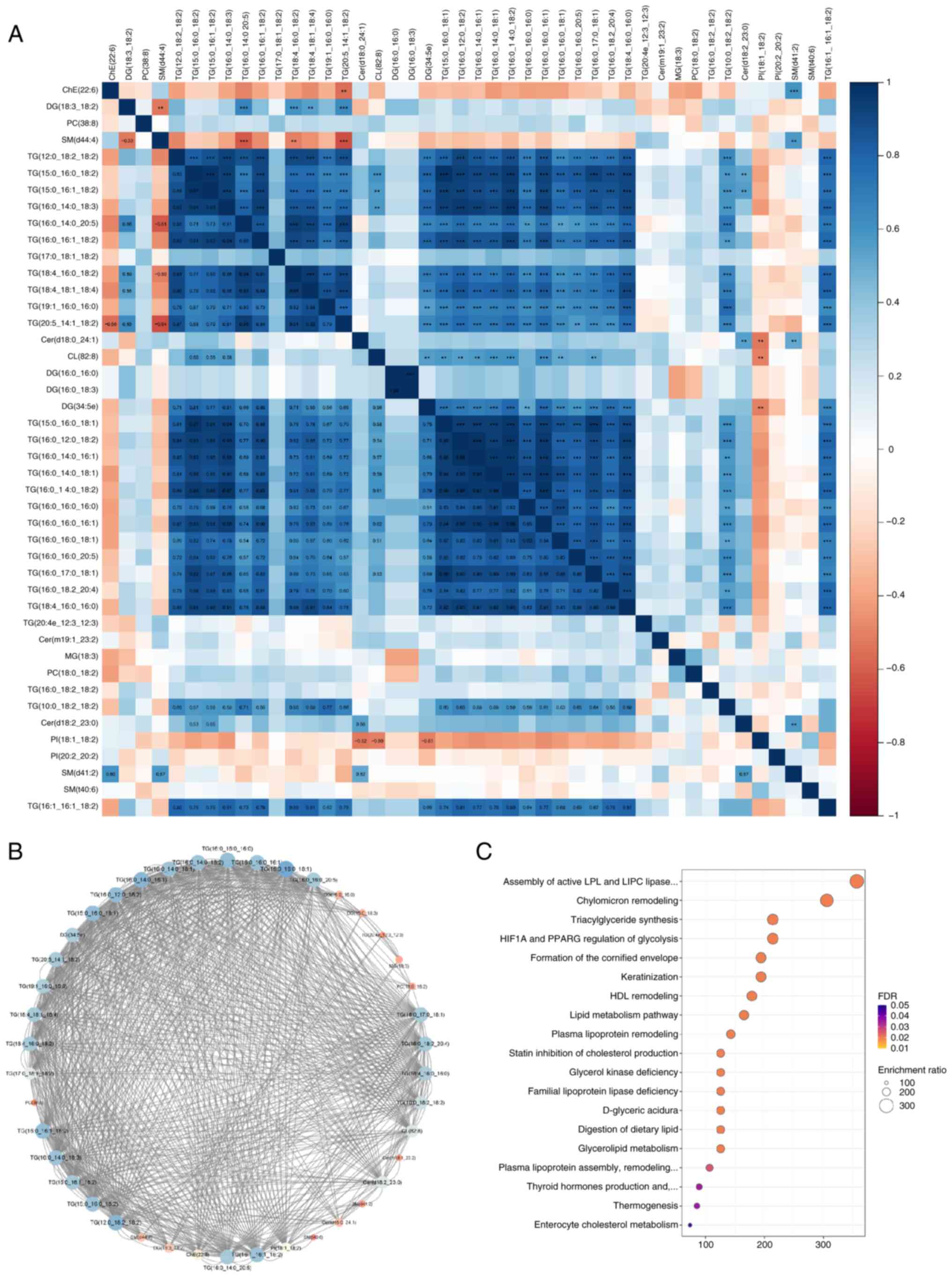 | Figure 6.Network and pathway analyses of
lipidomic data in breast cancer. (A) Clustering heatmap of
differential lipid classes, revealing positive correlations among
specific lipid types, illustrating interconnected lipid metabolic
processes and their relevance to breast cancer. (B) Network
analysis showing extensive correlations between TG and other lipids
within the lipidome, underscoring the complex interactions in lipid
metabolism in breast cancer. (C) Pathway enrichment analysis
identifying significant pathways such as the ‘Assembly of active
LPL and LIPC lipase complexes,’ highlighted by their statistical
significance and high fold enrichment, indicating key metabolic
pathways involved in breast cancer. **P<0.01, ***P<0.001.
Cer, ceramide; CL, cardiolipin; ChE, cholesterol ester; DG,
diacylglycerol; FDR, false discovery rate; MG, monoacylglycerol;
PC, phosphatidylcholine; PI, phosphatidylinositol; SM,
sphingomyelin; TG, triglyceride. |
Discussion
Fatty acid metabolism dysregulation plays a crucial
role in the malignant transformation of various cancer types
(38–42), including breast cancer (43,44).
Key metabolic enzymes involved in fatty acid synthesis and
oxidation have notable roles in the proliferation, migration and
invasion of breast cancer cells (43). Fatty acid metabolism involves
multiple pathways, including fatty acid transport, storage in lipid
droplets in the form of TGs and cholesterol esters, mobilization
from phospholipids and TGs as well as fatty acid oxidation. Most
human cells meet their fatty acid requirements by utilizing dietary
fatty acids. De novo fatty acid synthesis pathways are only
crucial in the liver, breast tissue and adipose tissue (45). The de novo fatty acid
synthesis pathway has different roles in normal and cancerous
tissues, making it an attractive therapeutic target (38).
A previous review showed that the lipid profile of
tumors may be used to distinguish HER2 +, luminal and BRCA-mutated
tumors (46). In the present study,
distinct lipidomic profiles associated with different breast cancer
subtypes, defined by HR and HER2 status, were delineated. The
grouping into four groups (HR+ HER2-0, HR+ HER2-low, HR+ HER2-pos
and HR- HER2-pos) allowed for a nuanced analysis of how lipidomic
landscapes vary with these biomarkers. Notably, the lipid profile
diversity, particularly the prevalence of TGs, such as
TG(16:0-18:1-18:1)+NH4, underscored the potential
biological variance among these groups. Lipid metabolism is
involved in cancer biology, influencing various processes, such as
cell membrane composition, energy storage and signaling
pathways.
A recent study (47)
showed that lipidomics-based phenotype heterogeneity can be used to
classify cancer types where genetic analysis alone is insufficient.
In the present study, the composition of PCs and TGs and their
relationships with the HR/HER2 phenotypes in breast cancer were
discovered. The results also underscored the significance of TGs,
while indicating that DGs are less important. The correlation and
pathway enrichment analyses indicated the broader biological
implications of the study findings. The clustering heatmap and
network analysis of differential lipids highlighted interconnected
lipid metabolism pathways, particularly the assembly of active
lipase complexes. Such pathways are crucial for lipid processing
and can be integral in understanding the metabolic reprogramming in
cancer cells. A recent study (48)
indicated that lipoprotein lipase is associated with poor prognosis
in breast cancer, indicating that the LPC pathway is crucial in
breast cancer.
The present study still has certain limitations that
cannot be ignored, which should be addressed in future larger
cohorts and longitudinal designs. First, it must be acknowledged
that the small sample size is a significant limitation, rendering
the present study a pilot study. Although some key lipids have been
identified in the present study, the relatively small number of
samples limits the statistical power and generalizability,
preventing a direct assessment of how these specific lipids
influence breast cancer cells. Larger cohorts are necessary to
validate the potential of lipidomic analysis in the clinical
context of breast cancer. To further understand the direct effects
of these lipids, we plan to examine their roles in regulating cell
proliferation, migration and invasion using breast cancer cell
lines with different HR/HER2 phenotypes, thereby potentially
identifying novel therapeutic targets. Furthermore, exploring the
correlation between lipid profiles and clinical outcomes is
essential. The lipid profiles and clinical outcomes of patients are
shown in Table SII; however, the
small sample size limits the validity of further analysis of the
relationship between these two characteristics. The preliminary
results of the present study provide valuable insights into the
metabolic variations across different subtypes of breast cancer.
Therefore, our future research will focus on examining the
relationships and differences between lipid profiles and survival
outcomes across various breast cancer subtypes.
In summary, several lipid molecules were
significantly different and could be used to distinguish between
breast cancer groups based on the HR and HER2 status. These key
lipid features include: i) TG: TG(16:0-18:1-18:1)+NH4,
prominent lipid composition; TG(16:0-18:1-18:2)+NH4,
another major lipid composition; TG(16:1-18:1-18:2)+NH4,
featured prominently in lipidomic profiles;
TG(50:13)+NH4, important for distinguishing between the
HR+ HER2-pos and HR+ HER2-low groups; TG(18:3e-18:3-21:1)+Na,
crucial for distinguishing between the HR+ HER2-pos and HR-
HER2-pos groups; TG(4:0-11:2-12:3)+NH4, significant for
distinguishing between the HR+ HER2-low and HR- HER2-pos groups;
TG(18:0-20:4-22:6)+NH4, important for distinguishing
between the HR+ HER2-low and HR+ HER2-0 groups;
TG(20:4e-12:3-12:3), TG(10:0-18:2-18:2) and TG(19:1-16:0-16:0),
distinct lipids for group classifications based on carbon chain
length and the number of double bonds. ii) PC: PC(36:5)+H,
important for distinguishing between HR+ HER2-pos and HR+ HER2-0
groups. iii) SM: SM (d34:1)+H, prominent lipid feature in HR+
HER2-0 group. iv) DG: DG(16:0-20:4)+H, significant for
distinguishing between the HR+ HER2-pos and HR+ HER2-0 groups;
DG(34:5e), DG(18:3-18:2) and DG(16:0-18:3), most distinct DGs for
group classifications. v) PIP: PIP3(21:2)+NH4, crucial
lipid feature for sample grouping accuracy.
In conclusion, the comprehensive analysis of
lipidomic variations across different breast cancer subtypes offers
valuable insights into the metabolic alterations associated with
cancer progression and phenotype. Therefore, the findings of the
present study may improve the development of tailored therapeutic
strategies that target specific metabolic pathways in breast
cancer.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was supported by the Shaoxing Public Welfare
Application Research Program Project (grant no. 2020A13009).
Availability of data and materials
The lipidomics data generated in the present study
may be found in the MetaboLights repository under the accession
number MTBLS10858 or at the following URL: https://www.ebi.ac.uk/metabolights/MTBLS10858.
All other data generated in the present study may be requested from
the corresponding author.
Authors' contributions
XQ was responsible for data curation and formal
analysis, ensuring accurate data representation and preliminary
analysis. JH and XJ managed the methods and project implementation,
selecting and implementing appropriate analytical techniques to
ensure smooth progress of the project. JZ handled the validation
process, critically assessing the accuracy and reliability of the
data and analyses. SH (the corresponding author), conceptualized
the study, developed the methodology and led the manuscript
writing, framing the study's findings within the broader context of
lipid metabolism and cancer progression. All authors read and
approved the final version of the manuscript. XQ and SH confirm the
authenticity of all the raw data.
Ethics approval and consent to
participate
This study was approved by the Ethics Committee of
Shaoxing People's Hospital (Shaoxing, China; approval no.
2021-K-Y-53-01), and the study followed relevant guidelines and
regulations. Verbal informed consent was obtained from all
individual participants involved in the study, and the requirement
for written informed consent was waived by the Ethics
Committee.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
ADC
|
antibody-drug conjugates
|
|
ANOVA
|
analysis of variance
|
|
DG
|
diacylglycerol
|
|
FISH
|
fluorescence in situ
hybridization
|
|
HER2
|
human epidermal growth factor receptor
2
|
|
HR
|
hormone receptor
|
|
IHC
|
immunohistochemistry
|
|
LC-MS
|
liquid chromatography-mass
spectrometry
|
|
LPC
|
lysophosphatidylcholine
|
|
PC
|
phosphatidylcholine
|
|
PCA
|
principal component analysis
|
|
PIP3
|
phosphatidylinositol-3,4,5-trisphosphate
|
|
PLS-DA
|
partial least squares discriminant
analysis
|
|
SM
|
sphingomyelin
|
|
TG
|
triglycerides
|
|
VIP
|
variable importance in projection
|
References
|
1
|
Ferlay J, Ervik M, Lam F, Laversanne M,
Colombet M, Mery L, Piñeros M, Znaor A, Soerjomataram I and Bray F:
Global cancer observatory: Cancer today. International Agency for
Research on Cancer; Lyon: 2020
|
|
2
|
Wolf-Yadlin A, Kumar N, Zhang Y,
Hautaniemi S, Zaman M, Kim HD, Grantcharova V, Lauffenburger DA and
White FM: Effects of HER2 overexpression on cell signaling networks
governing proliferation and migration. Mol Syst Biol. 2:542006.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Eladdadi A and Isaacson D: A mathematical
model for the effects of HER2 overexpression on cell proliferation
in breast cancer. Bull Math Biol. 70:1707–1729. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tagliabue E, Agresti R, Carcangiu ML,
Ghirelli C, Morelli D, Campiglio M, Martel M, Giovanazzi R, Greco
M, Balsari A and Ménard S: Role of HER2 in wound-induced breast
carcinoma proliferation. Lancet. 362:527–533. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Freudenberg JA, Wang Q, Katsumata M,
Drebin J, Nagatomo I and Greene MI: The role of HER2 in early
breast cancer metastasis and the origins of resistance to
HER2-targeted therapies. Exp Mol Pathol. 87:1–11. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li YM, Pan Y, Wei Y, Cheng X, Zhou BP, Tan
M, Zhou X, Xia W, Hortobagyi GN, Yu D and Hung MC: Upregulation of
CXCR4 is essential for HER2-mediated tumor metastasis. Cancer Cell.
6:459–469. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Harris C, Ward R, Dobbins T, Drew A and
Pearson S: The efficacy of HER2-targeted agents in metastatic
breast cancer: A meta-analysis. Ann Oncol. 22:1308–1317. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lin NU and Winer EP: Brain metastases: The
HER2 paradigm. Clin Cancer Res. 13:1648–1655. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Merkhofer EC, Cogswell P and Baldwin AS:
Her2 activates NF-kappaB and induces invasion through the canonical
pathway involving IKKalpha. Oncogene. 29:1238–1248. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Luo X, He Y, Tang H, Cao Y, Gao M, Liu B
and Hu Z: Effects of HER2 on the invasion and migration of gastric
cancer. Am J Transl Res. 11:7604–7613. 2019.PubMed/NCBI
|
|
11
|
Al-Juboori SI, Vadakekolathu J, Idri S,
Wagner S, Zafeiris D, Pearson J, Almshayakhchi R, Caraglia M,
Desiderio V, Miles AK, et al: PYK2 promotes HER2-positive breast
cancer invasion. J Exp Clin Cancer Res. 38:1–14. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chi F, Wu R, Jin X, Jiang M and Zhu X:
HER2 induces cell proliferation and invasion of non-small-cell lung
cancer by upregulating COX-2 expression via MEK/ERK signaling
pathway. Onco Targets Ther. 5:2709–2716. 2016.PubMed/NCBI
|
|
13
|
Alameddine RS, Otrock ZK, Awada A and
Shamseddine A: Crosstalk between HER2 signaling and angiogenesis in
breast cancer: Molecular basis, clinical applications and
challenges. Curr Opin Oncol. 25:313–324. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Vogl G, Bartel H, Dietze O and
Hauser-Kronberger C: HER2 is unlikely to be involved in directly
regulating angiogenesis in human breast cancer. Appl
Immunohistochem Mol Morphol. 14:138–145. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tarantino P, Hamilton E, Tolaney SM,
Cortes J, Morganti S, Ferraro E, Marra A, Viale G, Trapani D,
Cardoso F, et al: HER2-low breast cancer: Pathological and clinical
landscape. J Clin Oncol. 38:1951–1962. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yan M, Schwaederle M, Arguello D, Millis
SZ, Gatalica Z and Kurzrock R: HER2 expression status in diverse
cancers: Review of results from 37,992 patients. Cancer Metastasis
Rev. 34:157–164. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Maximiano S, Magalhaes P, Guerreiro MP and
Morgado M: Trastuzumab in the treatment of breast cancer. BioDrugs.
30:75–86. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Vu T and Claret FX: Trastuzumab: Updated
mechanisms of action and resistance in breast cancer. Front Oncol.
2:622012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Modi S, Jacot W, Yamashita T, Sohn J,
Vidal M, Tokunaga E, Tsurutani J, Ueno NT, Prat A, Chae YS, et al:
Trastuzumab deruxtecan in previously treated HER2-low advanced
breast cancer. N Engl J Med. 387:9–20. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Skidmore L, Sakamuri S, Knudsen NA, Hewet
AG, Milutinovic S, Barkho W, Biroc SL, Kirtley J, Marsden R, Storey
K, et al: ARX788, a site-specific anti-HER2 antibody-drug
conjugate, demonstrates potent and selective activity in HER2-low
and T-DM1-resistant breast and gastric cancers. Mol Cancer Ther.
19:1833–1843. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ogitani Y, Aida T, Hagihara K, Yamaguchi
J, Ishii C, Harada N, Soma M, Okamoto H, Oitate M, Arakawa S, et
al: DS-8201a, a novel HER2-targeting ADC with a novel DNA
Topoisomerase I inhibitor, demonstrates a promising antitumor
efficacy with differentiation from T-DM1. Clin Cancer Res.
22:5097–5108. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Dai LJ, Ma D, Xu YZ, Li M, Li YW, Xiao Y,
Jin X, Wu SY, Zhao YX, Wang H, et al: Molecular features and
clinical implications of the heterogeneity in Chinese patients with
HER2-low breast cancer. Nat Commun. 14:51122023. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
de Nonneville A, Houvenaeghel G, Cohen M,
Sabiani L, Bannier M, Viret F, Gonçalves A and Bertucci F:
Pathological complete response rate and disease-free survival after
neoadjuvant chemotherapy in patients with HER2-low and HER2-0
breast cancers. Eur J Cancer. 176:181–188. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu L, Kawashima M, Sugimoto M, Sonomura
K, Pu F, Li W, Takeda M, Goto T, Kawaguchi K, Sato TA and Toi M:
Discovery of lipid profiles in plasma-derived extracellular
vesicles as biomarkers for breast cancer diagnosis. Cancer Sci.
114:4020–4031. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Vitaliti A, Roccatani I, Iorio E, Perta N,
Gismondi A, Chirico M, Pisanu ME, Di Marino D, Canini A, De Luca A
and Rossi L: AKT-driven epithelial-mesenchymal transition is
affected by copper bioavailability in HER2 negative breast cancer
cells via a LOXL2-independent mechanism. Cell Oncol (Dordr).
46:93–115. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xiao Y, Ma D, Yang YS, Yang F, Ding JH,
Gong Y, Jiang L, Ge LP, Wu SY, Yu Q, et al: Comprehensive
metabolomics expands precision medicine for triple-negative breast
cancer. Cell Res. 32:477–490. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li S, Zeng H, Fan J, Wang F, Xu C, Li Y,
Tu J, Nephew KP and Long X: Glutamine metabolism in breast cancer
and possible therapeutic targets. Biochem Pharmacol.
210:1154642023. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shen L, Huang H, Li J, Chen W, Yao Y, Hu
J, Zhou J, Huang F and Ni C: Exploration of prognosis and
immunometabolism landscapes in ER+ breast cancer based on a novel
lipid metabolism-related signature. Front Immunol. 14:11994652023.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Azam A and Sounni NE: Lipid metabolism
heterogeneity and crosstalk with mitochondria functions drive
breast cancer progression and drug resistance. Cancers (Basel).
14:62672022. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zipinotti Dos Santos D, de Souza JC,
Pimenta TM, da Silva Martins B, Junior RSR, Butzene SMS, Tessarolo
NG, Cilas PML Jr, Silva IV and Rangel LBA: The impact of lipid
metabolism on breast cancer: A review about its role in
tumorigenesis and immune escape. Cell Commun Signal. 21:1612023.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ward AV, Anderson SM and Sartorius CA:
Advances in analyzing the breast cancer lipidome and its relevance
to disease progression and treatment. J Mammary Gland Biol
Neoplasia. 26:399–417. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
World Health Organization (WHO), . WHO
classification of tumours editorial board: Breast tumours. WHO;
Geneva: 2019
|
|
33
|
Chong J, Soufan O, Li C, Caraus I, Li S,
Bourque G, Wishart DS and Xia J: MetaboAnalyst 4.0: Towards more
transparent and integrative metabolomics analysis. Nucleic Acids
Res. 46:W486–W494. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Duan Y, Du Y, Gu Z, Zheng X and Wang C:
prognostic value, immune signature, and molecular mechanisms of the
PHLDA family in pancreatic adenocarcinoma. Int J Mol Sci.
23:103162022. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Breast Cancer Professional Committee of
the Chinese Anti-Cancer Association and Breast Tumor Group of the
Oncology Branch of the Chinese Medical Association, . Guidelines
for breast cancer diagnosis and treatment by China Anti-cancer
Association (2024 edition) (Chinese version). China Oncology.
33:1092–1187. 2023.
|
|
36
|
Zhang R, Yang M, Hou X, Hou R, Wang L, Shi
L, Zhao F, Liu X, Meng Q, Wang L and Zhang L: Characterization and
difference of lipids and metabolites from Jianhe White Xiang and
Large White pork by high-performance liquid chromatography-tandem
mass spectrometry. Food Res Int. 162:1119462022. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang Z, Liao Q, Sun Y, Pan T, Liu S, Miao
W, Li Y, Zhou L and Xu G: Lipidomic and transcriptomic analysis of
the longissimus muscle of luchuan and duroc pigs. Front Nutr.
8:6676222021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Koundouros N and Poulogiannis G:
Reprogramming of fatty acid metabolism in cancer. Br J Cancer.
122:4–22. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yousuf U, Sofi S, Makhdoomi A and Mir MA:
Identification and analysis of dysregulated fatty acid metabolism
genes in breast cancer subtypes. Med Oncol. 39:2562022. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhang F and Du G: Dysregulated lipid
metabolism in cancer. World J Biol Chem. 3:167–174. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang M, Han J, Xing H, Zhang H, Li Z,
Liang L, Li C, Dai S, Wu M, Shen F and Yang T: Dysregulated fatty
acid metabolism in hepatocellular carcinoma. Hepat Oncol.
3:241–251. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhou X, Huang F, Ma G, Wei W, Wu N and Liu
Z: Dysregulated ceramides metabolism by fatty acid 2-hydroxylase
exposes a metabolic vulnerability to target cancer metastasis.
Signal Transduct Target Ther. 7:3702022. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Monaco ME: Fatty acid metabolism in breast
cancer subtypes. Oncotarget. 8:29487–29500. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Qin L, An N, Yuan B, Zhu Q and Feng Y: The
metabolomic characteristics and dysregulation of fatty acid esters
of hydroxy fatty acids in breast cancer. Metabolites. 13:11082023.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Weiss L, Hoffmann GE, Schreiber R, Andres
H, Fuchs E, Körber E and Kolb HJ: Fatty-acid biosynthesis in man, a
pathway of minor importance. Purification, optimal assay
conditions, and organ distribution of fatty-acid synthase. Biol
Chem Hoppe Seyler. 367:905–912. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ribas HT, Sogayar MC, Dolga AM,
Winnischofer SM and Trombetta-Lima M: Lipid profile in breast
cancer: From signaling pathways to treatment strategies. Biochimie.
219:118–129. 2023. View Article : Google Scholar
|
|
47
|
Aramaki S, Tsuge S, Islam A, Eto F,
Sakamoto T, Oyama S, Li W, Zhang C, Yamaguchi S, Takatsuka D, et
al: Lipidomics-based tissue heterogeneity in specimens of luminal
breast cancer revealed by clustering analysis of mass spectrometry
imaging: A preliminary study. PLoS One. 18:e02831552023. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Bavis MM, Nicholas AM, Tobin AJ, Christian
SL and Brown RJ: The breast cancer microenvironment and lipoprotein
lipase: Another negative notch for a beneficial enzyme? FEBS Open
Bio. 13:586–596. 2023. View Article : Google Scholar : PubMed/NCBI
|