Introduction
Cervical cancer (CC) is the fourth most common
cancer in women worldwide and is considered a public health problem
in developing countries (1).
Although progress has been made in therapeutic methods, the overall
survival of patients with CC remains unsatisfactory due to
recurrence and metastasis (2).
Therefore, it is important to identify novel diagnostic and
prognostic biomarkers that allow the adequate management of
patients (3). Approximately 90% of
cases of CC are caused by long-term infection of high-risk human
papillomavirus (HPV), including the HPV-16 and HPV-18 genotypes
(4). In addition to viral
infection, epigenetic factors (DNA methylation, non-coding RNAs and
post-translational modifications of histones) are also related to
the development of CC (5).
In terms of epigenetic contribution, non-coding RNAs
(ncRNAs) serve key roles in CC (6).
Circular RNAs (circRNAs) are the most recent addition to the group
of endogenous ncRNAs (7). CircRNA
transcripts are generated through the back-splicing of the
precursor mRNA (8). Structurally,
they present a covalently closed circle shape as the 5′ and 3′ ends
are joined by a 3′,5′-phosphodiester bond (9). CircRNAs exhibit tissue-specific
expression (10) and are resistant
to digestion by Ribonuclease R, therefore, they are more stable
compared with their linear isoforms (11). CircRNAs are considered to be master
regulators of gene expression because of their ability to modulate
different mechanisms in cells, although these are currently not
fully understood (12). Using
high-throughput sequencing technologies and bioinformatics,
numerous novel differentially expressed circRNAs (DECs) in CC cell
lines and tissues have been identified (13). Previous research indicates that
these circular transcripts participate in the initiation and
progression of CC through modulation of biological processes such
as cell proliferation, migration, invasion, epithelial-mesenchymal
transition (EMT), metastasis and apoptosis (14,15).
In all types of cancer, circRNAs act as competing
endogenous RNAs (ceRNAs) by competitively binding microRNA
(miRNA/miR) response elements (MREs) that are present at the 5′ end
of miRNAs (16). Mature miRNAs can
regulate gene expression at the post-transcriptional level by
targeting specific mRNAs. miRNAs can bind to the 3′-untranslated
region of the mRNA, which consequently leads to mRNA downregulation
mediated by the RNA-induced silencing complex (17). CircRNAs mainly function as miRNA
sponges and inhibit the regulatory effects of the miRNA on their
target mRNA (18), thus forming a
circRNA/miRNA/mRNA regulatory network (19). This mechanism has been previously
described in CC (15). For example,
hsa_circ_0031288 exhibits high expression levels in cervical cancer
cells, and it has been reported that it acts as a sponge for
hsa-miR-139-3p and promotes increased expression levels of B cell
lymphoma 6 (Bcl-6) mRNA. Hsa_circ_0031288/hsa-miR-139-3p/Bcl-6
affects the proliferation, migration and invasion of HeLa cells
(20). Hsa_circ_0071474 expression
levels are increased in cervical cancer cells and hsa_circ_0071474
has been reported to bind to miR-137 to promote Kruppel-like factor
12 (KLF12) mRNA upregulation. The hsa_circ_0071474/miR-137/KLF12
network is important in tumor proliferation (21).
Despite the aforementioned studies, the expression
profiles and biological functions of numerous other circRNAs and
their roles in the initiation and progression of CC are still
unknown and need to be investigated to improve the current
understanding of their underlying mechanisms in CC. The present
study analyzed the effects of hsa_circ_0009910 knockdown on miR-198
and c-Met expressions levels, cell viability and apoptosis in HeLa
cells.
Materials and methods
Differential expression analysis
The expression microarray data (accession no.
GSE113696) were obtained from Gene Expression Omnibus (GEO;
http://www.ncbi.nlm.nih.gov/geo/). The
expression levels of circRNAs in five CC cell lines (HeLa, CaSki,
SiHa, C-33A and SW756) were compared with circRNA expression in
human cervical epithelial cells (HCerEpiC) to identify DECs. The
circRNA microarray had been performed using the Arraystar Human
circRNA Array (8×15K; Arraystar Inc.). The interactive online tool
GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/) was used to
perform differential expression analysis (22). The P-values were adjusted to reduce
the false positive rate using the Benjamini and Hochberg's false
discovery rate (FDR) method (23).
Finally, the DECs were selected based on the cut-off values of
logFC<-4.3 or >4.1 and FDR <0.1266.
Prediction of structure circRNAs
The genomic sequence and structural elements of
hsa_circ_0009910 were predicted using the Circular RNA Interactome
(https://circinteractome.nia.nih.gov/)
(24) and Cancer-Specific CircRNA
Database (https://gb.whu.edu.cn/CSCD/)
(25).
Gene Expression Profiling Interactive
Analysis (GEPIA)
The GEPIA database (http://gepia.cancer-pku.cn/) (26) was utilized to analyze the mRNA
expression level of mitofusin 2 (MFN2) and mesenchymal-epithelial
transition factor (c-Met or MET) in 306 biopsies from patients with
cervical squamous cell carcinoma and endocervical adenocarcinoma
(CESC) and 13 with healthy tissue. Expression data were normalized
and log2 transformed (TPM+1). P<0.05 was considered
statistically significant.
Construction of endogenous competing
RNA network
Due to potential circRNA-miRNA-mRNA-RNA-binding
proteins (RBP) regulation, interaction networks were constructed
for subsequent experimental analysis. The Circular RNA Interactome
(24) was used to predict
circRNA-miRNA and circRNA-RBP interactions; interactions with a
score of <90% were excluded.
Interactions between miRNAs-mRNAs were established
using the DIANA-TarBase v.8 software (version 8; http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=tarbasev8%2Findex)
(27) and miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/php/index.php)
(28). To increase the prediction
accuracy, target genes were selected using the following criteria:
i) The prediction score was >0.443; ii) ≥1 original article must
support the interaction; and iii) the article demonstrates the
miRNA-mRNA interaction with experimental evidence from reporter
assays, western blot or quantitative PCR. Finally, a
circRNA-miRNA-mRNA interaction network was constructed by combining
circRNA-miRNA, circRNA-RBP and miRNA-mRNA pairs.
Gene Ontology (GO) analysis
To predict the functional implications of mRNAs in
the network, GO analysis was performed using PANTHER (http://www.pantherdb.org/) (29). According to the program, pathways
with an FDR <0.05 were considered to indicate significantly
enriched pathways.
Cell culture
The HaCaT immortalized human keratinocyte cell line
and HPV-18 positive CC cell line (HeLa) were purchased from
American Type Culture Collection. The cells were cultured in
DMEM/F-12 (1:1) medium (Caisson Labs, Inc.) supplemented with 10%
fetal bovine serum (PAA Laboratories GmbH; GE Healthcare), 100 U/ml
penicillin and 100 µg/ml streptomycin (Caisson Labs, Inc.). The
cells were cultured at 37°C with 5% CO2.
Transfection
To perform the hsa_circ_0009910 knockdown, a small
interfering RNA (siRNA) targeting hsa_circ_0009910 (si-circ9910)
and siRNA negative control (si-NC) were synthesized by Integrated
DNA Technologies, Inc. (Table I).
HeLa cells were seeded in 6-well plates at 80% confluence and
subsequently transfected with si-circ9910 (50 nM) or siRNA negative
control using Lipofectamine® 3000 (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol,
and harvested 48 h later for further analysis.
 | Table I.Primer sequences. |
Table I.
Primer sequences.
| A, RT-qPCR |
|---|
|
|---|
| Gene | Sequence
(5′-3′) |
|---|
| hsa_circ_ | F:
AGGTTCTGGACGTCAAAGGTT |
| 0009910 | R:
TTGCATCGAGAGAAGAGCAGG |
| c-Met | F:
TATTTCCCAGATCATCCATTGCA |
|
| R:
AATGTAGGACTGGTCCGTCAAAA |
| GAPDH | F:
GACCCCTTCATTGACCTCAAC |
|
| R:
GTGGCAGTGATGGCATGGAC |
| miR-198 | F:
TCATTGGTCCAGAGGGGAGATAG |
|
| R:
GCAGGGTCCGAGGTATTC |
| RNU44 | F:
CCTGGATGATGATAAGCAAATG |
|
| R:
GTCAGTTAGAGCTAATTAAGACC |
|
| B,
siRNA |
|
| Gene | Sequence
(5′-3′) |
|
| si-circ9910 |
AGCAGGGACAUUGCGCGGCCA |
| si-NC |
CGUUAAUCGCGUAUAAUACGCGUA |
RNA extraction
Total RNA was extracted from HaCaT and HeLa cells
using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. The
concentration of RNA was determined by spectrophotometry using a
NanoDrop 2000c Spectrophotometer (NanoDrop Technologies; Thermo
Fisher Scientific, Inc.).
Reverse transcription-qPCR
(RT-qPCR)
The expression levels of hsa_circ_0009910 and c-Met
were analyzed using RT-qPCR using the CYBRFast™ 1-Step RT-qPCR
Lo-ROX Kit (Tonbo™ Biosciences; Cytek® Biosciences)
according to the manufacturer's protocol. The following
thermocycling conditions were used for qPCR: Complementary DNA
(cDNA) synthesis at 50°C for 10 min, DNA polymerase activation at
95°C for 2 min, 40 cycles of denaturation at 95°C for 20 sec and
annealing and extension at 60°C for 30 sec. Reactions were
performed using the QuantStudio™ 3 Real-Time PCR System (Applied
Biosystems; Thermo Fisher Scientific, Inc.).
The expression level of miR-198 (accession no.
MI0000240) was assessed using the TaqMan® MicroRNA Assay
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The miR-198
cDNA was obtained using the TaqMan MicroRNA Reverse Transcription
Kit (Applied Biosystems; Thermo Fisher Scientific, Inc.) according
to the manufacturer's instructions and the reactions were performed
using a BioRad T100™ Thermal Cycler (Bio-Rad Laboratories, Inc.).
qPCR of miR-198 was performed using the TaqMan®
Universal PCR Master Mix (Applied Biosystem; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions and
were conducted using the QuantStudio 3 Real-Time PCR System
(Applied Biosystems; Thermo Fisher Scientific, Inc.). Data were
normalized using GAPDH and small nucleolar RNA, C/D box 44 (RNU44;
accession no. NR_002750) as internal controls and relative
expression were calculated using the 2−ΔΔCq method
(30). Primer sequences are shown
in Table I.
MTT assay
The MTT reduction assay was used to assess cell
viability (31). HeLa cells (10,000
cells/well) were seeded in 96-well plates. The next day, cells were
transfected with si-circ9910 for 48 h as aforementioned. After
transfection, 20 µl of MTT solution (5 mg MTT/ml PBS) was added to
each well and left to incubate for 3 h at 37°C. At termination of
the experiment, MTT was removed by aspiration and the cells were
treated with 100 µl of dimethyl sulfoxide (DMSO) to dissolve the
formazan crystals followed by gentle shaking of the microplate for
15 min. Absorbance was recorded at 570 nm using an Epoch
Spectrophotometer (BioTek; Agilent Technologies, Inc.). The
percentage of viability was calculated as follows: (Absorbance of
experimental group/absorbance of NT group) ×100%. The DMSO 5% and
A23187 10 µM groups served as positive controls for all viability
and apoptosis assays.
Neutral red uptake (NRU) assay
The NRU assay (cat. no. N4638; Sigma-Aldrich; Merck
KGaA) was used to determine the accumulation of the neutral red dye
in the lysosomes of viable cells (32). HeLa cells (10,000 cells/well) were
seeded in 96-well plates. The next day, cells were transfected with
si-circ9910 for 48 h as aforementioned. After transfection, cells
were incubated for 2 h at 37°C with 100 µl neutral red (40 µg
neutral red/ml DMEM). At termination of the experiment, the neutral
red solution was removed by aspiration and the cells were washed
twice with 150 µl PBS. Then, 150 µl of destain solution was added
to each well followed by gentle shaking of the microplate for 10
min. Absorbance was recorded at 540 nm using an Epoch
Spectrophotometer (BioTek; Agilent Technologies, Inc.). Results
were presented as a percentage of viability (calculated as
aforementioned in the MTT assay).
Viability and apoptosis assay
To assess cell viability and apoptosis, the
ApoLive-Glo™ Multiplex Assay (Promega Corporation) was used
according to the manufacturer's instructions. HeLa cells were
seeded in 96-well plates (10,000 cells/well). The next day, cells
were transfected with si-circ9910 for 48 h as aforementioned. After
transfection, 20 µl viability reagent was added to each well and
mixed by orbital shaking at 300–500 rpm for ~30 sec at room
temperature. Plates were incubated for 3 h at 37°C and fluorescence
measured at 400excitation/505emission nm to
determine viability. Subsequently, 100 µl Caspase-Glo®
3/7 reagent was added to the wells and mixed briefly by orbital
shaking at 300–500 rpm for ~30 sec at room temperature. Plates were
then left for 3 h at 37°C and luminescence was measured to
determine apoptosis levels. Fluorescence and luminescence were
measured using an Infinite M200 (Tecan Group, Ltd.) plate reader.
Results were presented as a percentage of viability and apoptosis
(calculated as aforementioned in the MTT assay).
Statistical analysis
All data were analyzed using the SigmaPlot (version
10.0; Systat Software Inc.) software. The results are presented as
the mean ± standard deviation. Unpaired Student's t-test was used
for comparative analysis between two groups and one-way analysis of
variance followed by Dunnett's post hoc test was used for
comparison among multiple groups. P<0.05 was considered to
indicate a statistically significant difference.
Results
DECs in CC
To identify novel circRNAs involved in the
initiation and progression of CC, the expression profiles of
circRNAs in HCerEpiC compared with HeLa cells in a previously
published circRNAs microarray dataset (accession no. GSE113696)
were analyzed. This analysis identified 25 DECs in HeLa cells with
potential roles in CC. A heat map was used to demonstrate the
expression patterns of the circRNAs identified in the HCerEpiC and
HeLa cell lines (Fig. 1A). Within
the top 25 DECs, 13 circRNAs were upregulated and 12 downregulated,
based on the cut-off values of logFC <-4.3 or >4.1 and FDR
<0.1266. The information of each circRNA was recorded (Table II) as follows: The parental gene
from which each circRNA was derived, circRNA type, expression level
and chromosomal location.
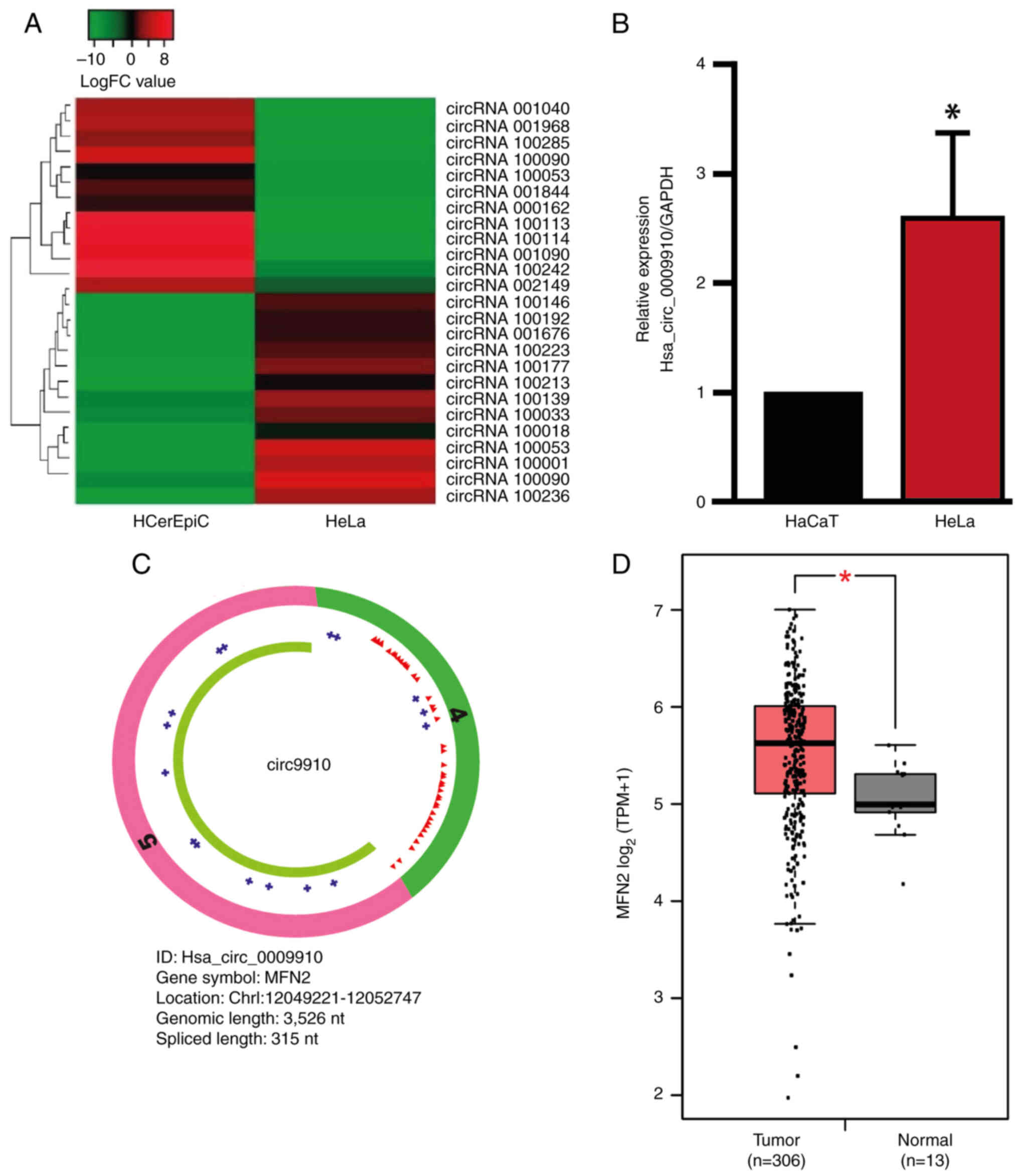 | Figure 1.Hsa_circ_0009910 is upregulated in
cervical cancer cells. (A) The heat map shows the expression of
downregulated and upregulated circRNAs in HeLa cells and HCerEpiC.
(B) Hsa_circ_0009910 expression levels in HaCaT and HeLa cells. (C)
The hsa_circ_0009910 schematic shows exon 4 (green), exon 5 (pink),
miRNA response elements (red triangles), RBP-binding sites (blue
crosses) and an open reading frame (green line). (D) Comparison of
the MFN2 expression in CESC compared with normal tissue using data
retrieved from TCGA. The data are presented as the mean ± standard
deviation and from at least three independent experiments where
applicable. *P<0.05. circRNA, circular RNA; miRNA, microRNA;
RBP, RNA-binding protein; HCerEpiC, human cervical epithelial
cells; MFN2, mitofusin 2; nt, nucleotide; CESC, cervical squamous
cell carcinoma and endocervical adenocarcinoma; TMP, transcripts
per million. |
 | Table II.Basic characteristics of the 25
differentially expressed circRNAs in cervical cancer. |
Table II.
Basic characteristics of the 25
differentially expressed circRNAs in cervical cancer.
| circRNA | logFC | False discovery
rate | Expression | Type | Chromosome | Strand | Gene symbol |
|---|
|
Hsa_circ_0000379 | −10.45 | 0.044 | Down | Intronic | 12 | + | PLBD1 |
|
Hsa_circRNA_001968 | −10.45 | 0.044 | Down | / | / | / | / |
| Ha_circ_100285 | −10.81 | 0.044 | Down | / | / | / | / |
|
Hsa_circ_0008563 | 7.79 | 0.063 | Up | Exonic | 1 | - | ECE1 |
|
Hsa_circ_0009910 | 7.22 | 0.063 | Up | Exonic | 1 | + | MFN2 |
|
Hsa_circ_0000263 | −7.8 | 0.063 | Down | Exonic | 10 | + | TCONS_00017720 |
|
Hsa_circ_0012634 | 7.76 | 0.063 | Up | Exonic | 1 | - | TMEM59 |
|
Hsa_circ_0013222 | −7.23 | 0.063 | Down | Exonic | 1 | - | GCLM |
|
Hsa_circ_0000488 | −7.67 | 0.078 | Down | Intronic | 13 | - | DLEU2 |
|
Hsa_circ_100113 | −6.02 | 0.078 | Down | / | / | / | / |
|
Hsa_circ_0011279 | 5.86 | 0.078 | Up | Exonic | 1 | + | SERINC2 |
|
Hsa_circ_0011692 | 5.61 | 0.078 | Up | Exonic | 1 | - | STK40 |
|
Hsa_circ_0009581 | 5.45 | 0.078 | Up | Exonic | 1 | - | RERE |
|
Hsa_circ_0005866 | −5.6 | 0.078 | Down | Exonic | 1 | - | RPA2 |
|
Hsa_circRNA_001844 | −8.37 | 0.078 | Down | / | / | / | / |
|
Hsa_circ_0009189 | 6.65 | 0.078 | Up | Exonic | 1 | + | SAMD11 |
|
Hsa_circ_0012417 | 7.09 | 0.082 | Up | Exonic | 1 | - | EPS15 |
|
Hsa_circ_0011385 | 5.7 | 0.082 | Up | Exonic | 1 | + | EIF3I |
|
Hsa_circRNA_001090 | −10.53 | 0.088 | Down | / | / | / | / |
|
Hsa_circ_0000069 | 5.65 | 0.116 | Up | Exonic | 1 | - | STIL |
|
Hsa_circ_0012107 | 6.01 | 0.116 | Up | Exonic | 1 | + | ST3GAL3 |
|
Hsa_circ_0000423 | 6.04 | 0.116 | Up | Exonic | 12 | - | PPP1R12A |
|
Hsa_circRNA_100242 | −4.77 | 0.123 | Down | / | / | / | / |
|
Hsa_circ_0001627 | −4.21 | 0.126 | Down | Intronic | 6 | - | BACH2 |
|
Hsa_circ_0009361 | 4.17 | 0.126 | Up | Exonic | 1 | - | GNB1 |
Hsa_circ_0009910 is upregulated in CC
cells
In the circRNA microarray data, hsa_circ_0009910
(also known as circ9910) was identified as a DEC. To confirm this
finding, the expression levels of hsa_circ_0009910 in HaCaT and
HeLa cells were evaluated using RT-qPCR. The expression levels of
hsa_circ_0009910 were significantly upregulated in HeLa cells
compared with the HaCaT control cells (Fig. 1B), consistent with the data obtained
from the circRNAs microarray. Hsa_circ_0009910 has 315 nucleotides
and is generated from exon 4 and exon 5 of the MFN2 gene through
back-splicing (33). Structurally,
hsa_circ_0009910 contains multiple MREs and interacts with several
RBPs (Fig. 1C). Additionally, the
expression levels of the parental gene MFN2 in TCGA-retrieved data
were assessed. MFN2 is significantly upregulated in CESC samples
compared with normal cervical tissue (Fig. 1D). These results indicate that
hsa_circ_0009910 is upregulated in CC cells.
Construction of the ceRNA regulatory
network for hsa_circ_0009910
To identify potential functions of hsa_circ_0009910
in cancer, a circRNA-miRNA-mRNA interaction network mediated by
hsa_circ_0009910 was constructed. CircRNA-miRNA interactions were
predicted using the Circular RNA Interactome platform and
miRNA-mRNA interactions were identified using the DIANA-TarBase v.8
and miRTarBase software. The prediction software demonstrated that
hsa_circ_0009910 harbored 10 motifs that could interact with
miRNAs, which could in turn regulate 135 mRNAs. In addition,
hsa_circ_0009910 could interact with seven RBPs (Fig. S1).
As hsa_circ_0009910 could indirectly regulate 135
mRNAs, GO analysis was performed using the PANTHER classification
system to gain insight into the biological processes that may be
altered by hsa_circ_0009910 (Fig.
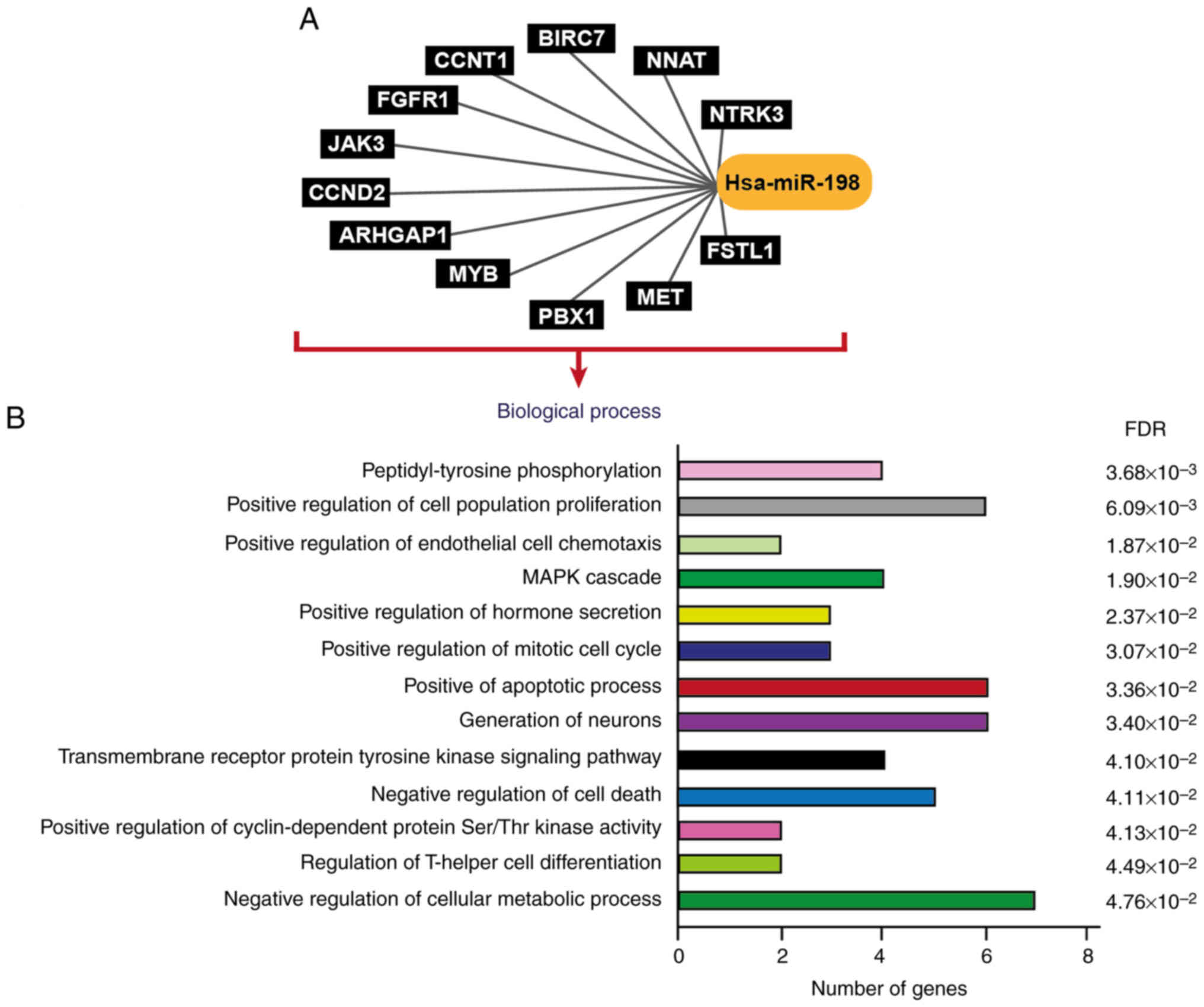
S2). Hsa_miR_198 (miR-198) is a notable miRNA regulated by
hsa_circ_0009910, due to the roles of miR-198 reported in prostate
cancer, oral squamous cell carcinoma, colorectal cancer and
glioblastoma (34–37). Numerous mRNAs were shown to be
potentially regulated by hsa-miR-198 (Fig. 2A) which participated in several
biological processes (Fig. 2B),
including the ‘positive regulation of cell population
proliferation’, the ‘positive regulation of mitotic cell cycle’ and
the ‘negative regulation of cell death’ as well as other processes
involved in the development of cancer. Thus, miR-198 was selected
as a biologically relevant target of hsa_circ_0009910 for further
analysis.
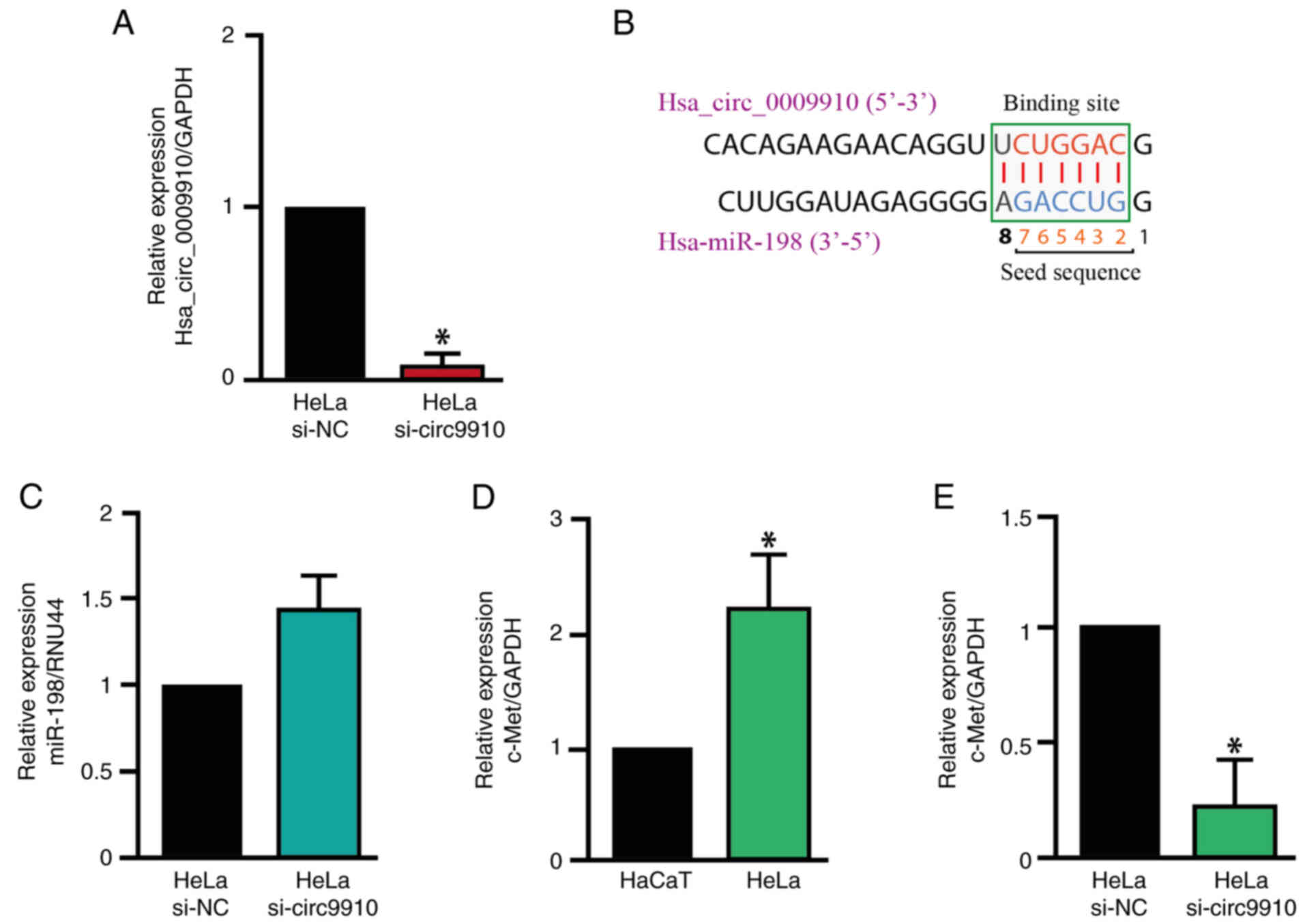
Hsa_circ_0009910 can act as a sponge
for miR-198
To investigate the role of hsa_circ_0009910 in
downstream gene expression and if it could serve as a miRNA sponge,
hsa_circ_0009910 knockdown in HeLa cells with si-circ9910 was
performed. The expression levels of hsa_circ_0009910 were reduced
by ~95% compared with the siRNA negative control in HeLa cells
(Fig. 3A). The region potential of
binding between hsa_circ_0009910 and miR-198 was identified
(Fig. 3B). Subsequently, the
expression levels of miR-198 in HeLa cells with hsa_circ_0009910
knockdown were evaluated. Knockdown of hsa_circ_0009910 led to a
notable increase in miR-198 expression levels in HeLa cells
compared with the siRNA negative control (Fig. 3C). Additionally, the expression
profiles of miR-198 in numerous normal tissues were evaluated using
data from the miRTarBase database, which demonstrated that cervical
tissue had the third highest expression levels of miR-198 of the 18
tissues assessed (Fig. S3).
c-Met is a potential target of
hsa_circ_0009910/miR-198
c-Met was chosen as a target gene from those
identified in the present study as previous studies have
demonstrated its molecular interactions with miR-198 (38–40)
and that it serves an important role in the development of CC
(41). Rho GTPase activating
protein 1 (ARHGAP1) and follistatin-like 1 (FSTL1) were not chosen
for further investigation primarily as, to the best of our
knowledge, there is no experimental evidence of its ARHGAP1
interaction with miR-198 and a relationship of mutual
transcriptional regulation between FSTL1 and miR-198 has been
reported (42,43). Therefore, only c-Met was further
investigated in the present study. c-Met was shown to be
upregulated in CC using the TCGA database (Fig. S4) and the expression levels of
c-Met were significantly increased in HeLa cells compared with that
in HaCaT cells (Fig. 3D).
Therefore, the influence of the hsa_circ_0009910/miR-198 axis on
c-Met expression levels was evaluated. In this regard,
hsa_circ_0009910 knockdown significantly decreased the expression
levels of c-Met in HeLa cells (Fig.
3E).
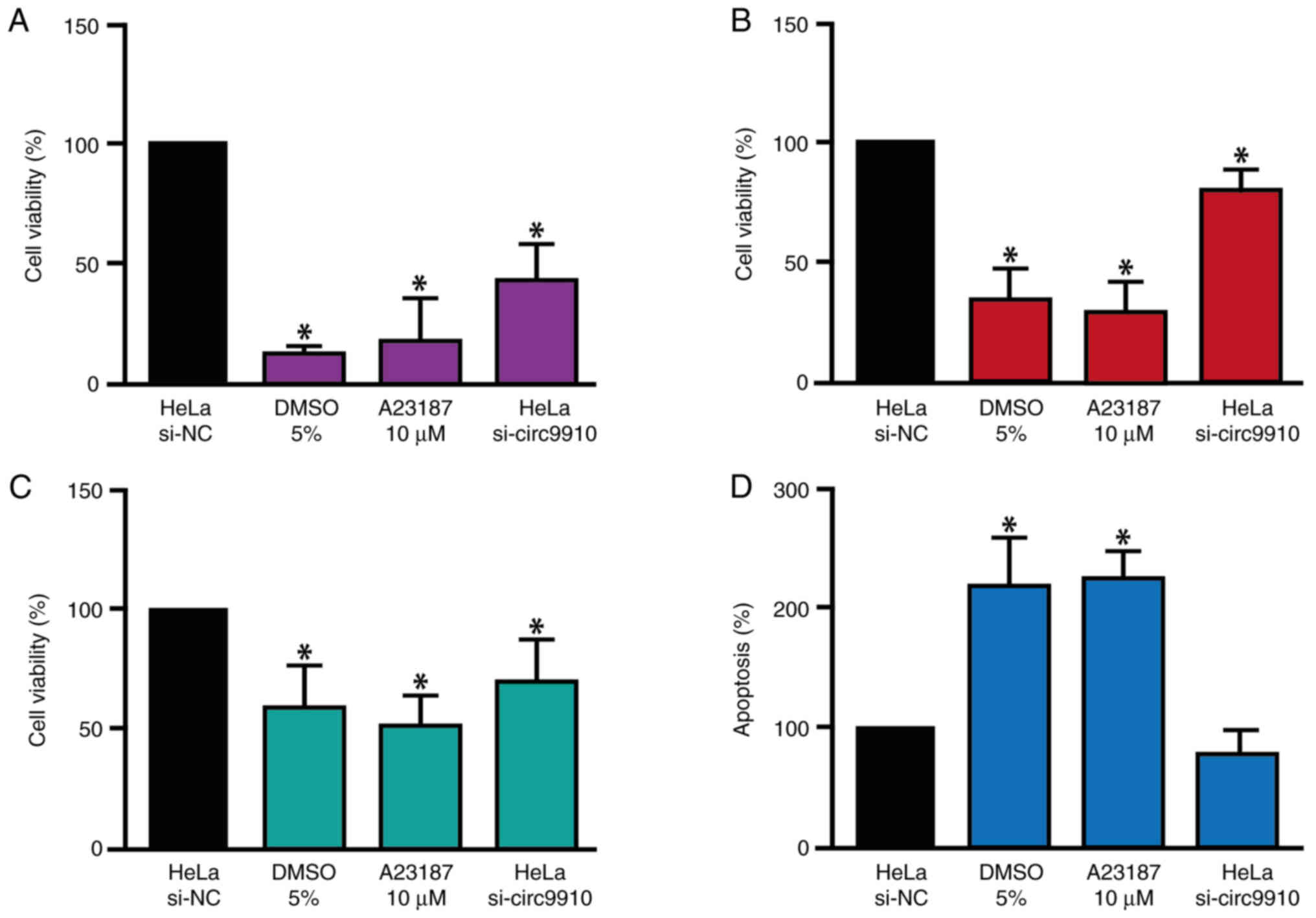
Knockdown of hsa_circ_0009910
decreases viability and does not affect apoptosis in HeLa
cells
To investigate the functional role of
hsa_circ_0009910/miR-198/c-Met axis, hsa_circ_0009910 knockdown was
performed in HeLa cells and the cell viability and apoptosis was
analyzed. The MTT, NRU and ApoLive-Glo Multiplex assays indicated
that the knockdown of hsa_circ_0009910 significantly decreased the
viability of HeLa cells (Fig.
4A-C), meanwhile, apoptosis levels were not affected (Fig. 4D).
Knockdown of hsa_circ_0009910 does not
affect the mRNA level of E6 and E7
Additionally, as HeLa cells were infected with
HPV-18, and its E6 and E7 oncoproteins maintain the cancerous
phenotype and prevent apoptosis, the effect of hsa_circ_0009910
knockdown on E6 and E7 expression was analyzed. Knockdown of
hsa_circ_0009910 did not affect the mRNA level of E6 and E7 in HeLa
cells (Fig. S5).
Discussion
CircRNAs are important molecules in the initiation
and progression of different types of human cancer (44). CircRNAs regulate gene expression
through different mechanisms, as they function as sponges for
miRNAs, interact with RBPs, act as scaffolds for protein complexes,
regulate gene transcription and certain circRNAs are translated
into small proteins (45). In CC, a
set of abnormally expressed circRNAs have been identified that may
function as oncogenes (46,47) or tumor suppressors (48,49).
However, research on the circRNAs in CC is currently in the early
stages and the understanding of mechanisms used by circRNAs to
promote cancer is limited.
In the present study, a potential regulatory network
in HeLa constituting hsa_circ_0009910, miR-198 and c-Met cells was
identified. Previous studies have reported several similar
regulatory networks in certain types of cancer (50,51)
including CC (52,53). A regulatory network in CC controlled
by hsa_circ_0001400 has been previously described, which
participates in the pathogenesis of CC by sponging hsa-miR-326,
resulting in Akt mRNA upregulation, a key molecule in the PI3K-Akt
signaling pathway. The hsa_circ_0001400/miR-326/Akt network exerts
its oncogenic effect by maintaining the cell cycle active,
promoting migration and inhibiting cancer cell apoptosis (54). By contrast, hsa_circ_0132980, also
known as circSLC26A4, is an oncogenic circRNA that acts as a
molecular sponge of miR-1287-5p to promote an increase in homeobox
genes A7 (HOXA7) mRNA levels. The
hsa_circ_0132980/miR-1287-5p/HOXA7 network facilitates CC
progression by modulating the proliferation, invasion and tumor
growth of cancer cells (55).
Another example of a regulatory network includes hsa_circ_0058514,
also known as circAGFG1, which functions as a miR-370-3p sponge and
promotes an increase in raf-1 proto-oncogene, serine/threonine
kinase (RAF1) mRNA expression levels. RAF1 serves a key role in the
phosphorylation and activation of MEK1/2 and ERK1/2 proteins. The
hsa_circ_0058514/miR-370-3p/RAF1 network promotes the proliferation
and migration of CC cells (46).
Furthermore, as hsa_circ_0009910 has binding sites for numerous
miRNAs, it participates in other regulatory networks. In
osteosarcoma, hsa_circ_0009910 functions as a sponge of miR-449a
and consequently promotes the upregulation of IL-6 receptor mRNA,
which is involved in the regulation of JAK1/STAT3 signaling pathway
(56). In acute myeloid leukemia,
hsa_circ_0009910 can bind to miR-5195-3p to promote growth factor
receptor-bound protein 10 mRNA upregulation, and consequently
influences the proliferation and apoptosis of cancer cells
(57). Over the last decade, the
importance of regulatory networks between circRNAs, miRNAs and
their target mRNAs has been evidenced by an increasing role in
numerous human diseases and cellular processes.
The present study demonstrated that hsa_circ_0009910
knockdown increased the expression levels of miR-198 and decreased
the expression levels of c-Met mRNA in HeLa cells. Hsa_circ_0009910
may function as a sponge of miR-198, which results in the decreased
of c-Met mRNA expression levels. To the best of our knowledge,
there has been no evidence previously that miR-198 is sponged by
hsa_circ_0009910, although it has been reported that
hsa_circ_0009910 functions as a sponge for miR-145 (33,58),
miR-335-5p (59), miR-34a-5p
(60) and miR-20a-5p (61). In addition, this exonic circRNA is
primarily localized in the cytoplasm of cancer cells, which
supports its function as a sponge for miRNAs (60). By contrast, it has been reported
that c-Met mRNA is a target of miR-198 in esophageal cancer
(38), hepatocellular carcinoma
(39), ovarian cancer (40) and osteosarcoma (62). c-Met, also known as
mesenchymal-epithelial transition factor or hepatocyte growth
factor receptor, is a receptor tyrosine kinase located on the cell
membrane of epithelial and endothelial cells (63). c-Met activation promotes the
activation of signaling pathways that are related to biological
processes such as cell proliferation, survival, apoptosis,
migration and invasion (64).
In addition to the potential regulation of miR-198
by hsa_circ_0009910, the previous research suggests that the
regulation of this miRNA is more complex. Studies have reported
that lncSChLAP1 (65), circ0004390
(40), circRNA LPAR3 (38), circ_0002060 (66), circ_ERBB2 (67), circRNA AKT3 (68) and circ_0005198 (69) can regulate miR-198 expression levels
in cancer.
Similarly, post-transcriptional regulation of c-Met
mRNA is complex. In addition to the miR-198/c-Met interaction, it
has been reported that c-Met can be regulated by other miRNAs in
CC, including to miR-1 (70),
miR-23b-3p (71), miR-454-3p
(72) and miR-876-5p (73). This may explain why c-Met expression
levels in hsa_circ_0009910 knockdown cells were notably decreased.
Therefore, the abnormal expression and function of c-Met results
from dysregulation of a set of miRNAs, and these miRNAs in turn may
be regulated by long ncRNAs or circRNAs. This highlights the
importance of further research into regulatory networks in CC.
The knockdown of hsa_circ_0009910 decreases cell
viability, potentially through the modulation of the miR-198/c-Met
axis. This result could be explained based by two hypotheses.
First, hsa_circ_0009910 knockdown increases expression levels of
miR-198, which may allow this miR-198 to function as a negative
regulator of its target mRNAs, including the c-Met oncogene.
Second, the decrease of c-Met expression levels mRNA may result in
the disruption of certain signaling pathways that control cellular
proliferation and cell viability such as the PI3K/Akt, Ras/MAPK,
JAK/STAT, Wnt/β-catenin, FAK/Src and NF-κB pathways (63).
By contrast, hsa_circ_0009910 knockdown did not
increase in HeLa cell apoptosis. However, it has been reported in
acute myeloid leukemia and osteosarcoma that the knockdown of
hsa_circ_0009910 decreases Bcl-2 expression levels and increases
Bax expression levels, which are anti- and pro-apoptosis proteins,
respectively (56,57,61).
Furthermore, in chronic myeloid leukemia, the knockdown of
hsa_circ_0009910 promotes the activation of caspase-3 (60). By contrast, the circRNA_0000285
(74), circRNA_0001400 (54) and circ-ATAD1 (75) positively regulate apoptosis in CC
cells. These data suggest that hsa_circ_0009910-mediated regulation
of apoptosis may be cell-type specific and that in HeLa cells,
apoptosis is modulated by mechanisms that do not include
hsa_circ_0009910, or at least not under the experimental conditions
used in the present study. Additionally, HeLa cells are infected
with HPV-18, a highly oncogenic HPV (76). The E6 and E7 oncoproteins maintain
the cancerous phenotype in HeLa cells and experimental evidence
suggests that the elimination of E6 and E7 are necessary events to
induce apoptosis in HeLa cells (77). Certain apoptosis-inducing agents,
such as N-benzylcinnamide (PT-3), in HeLa indirectly cause a
decrease in E6 or E7 oncoprotein expression levels (78). This may explain why apoptosis did
not increase in the present model as, although the expression
levels of hsa_circ_0009910 were decreased, E6 and E7 of HPV-18
expression and activity may have persisted.
Due to their function and regulatory activity in
different types of cancer, including CC, hsa_circ_0009910 (58), miR-198 (79) and c-Met (41,71)
may be considered biomarkers or therapeutic targets. A challenge in
CC continues to be the identification of biomarkers for early
diagnosis in precancerous lesions (80). In this sense, hsa_circ_0009910,
miR-198 and c-Met may be markers for the early diagnosis of CC in
the future. However, further investigations using precancerous
cervical lesion samples and animal models are needed to assess
their potential use as biomarkers. A limitation of the present
study is that the interaction between hsa_circ_0009910 and miR-198
was not demonstrated, and this would be necessary to confirm that
miR-198 is sponged by hsa_circ_0009910.
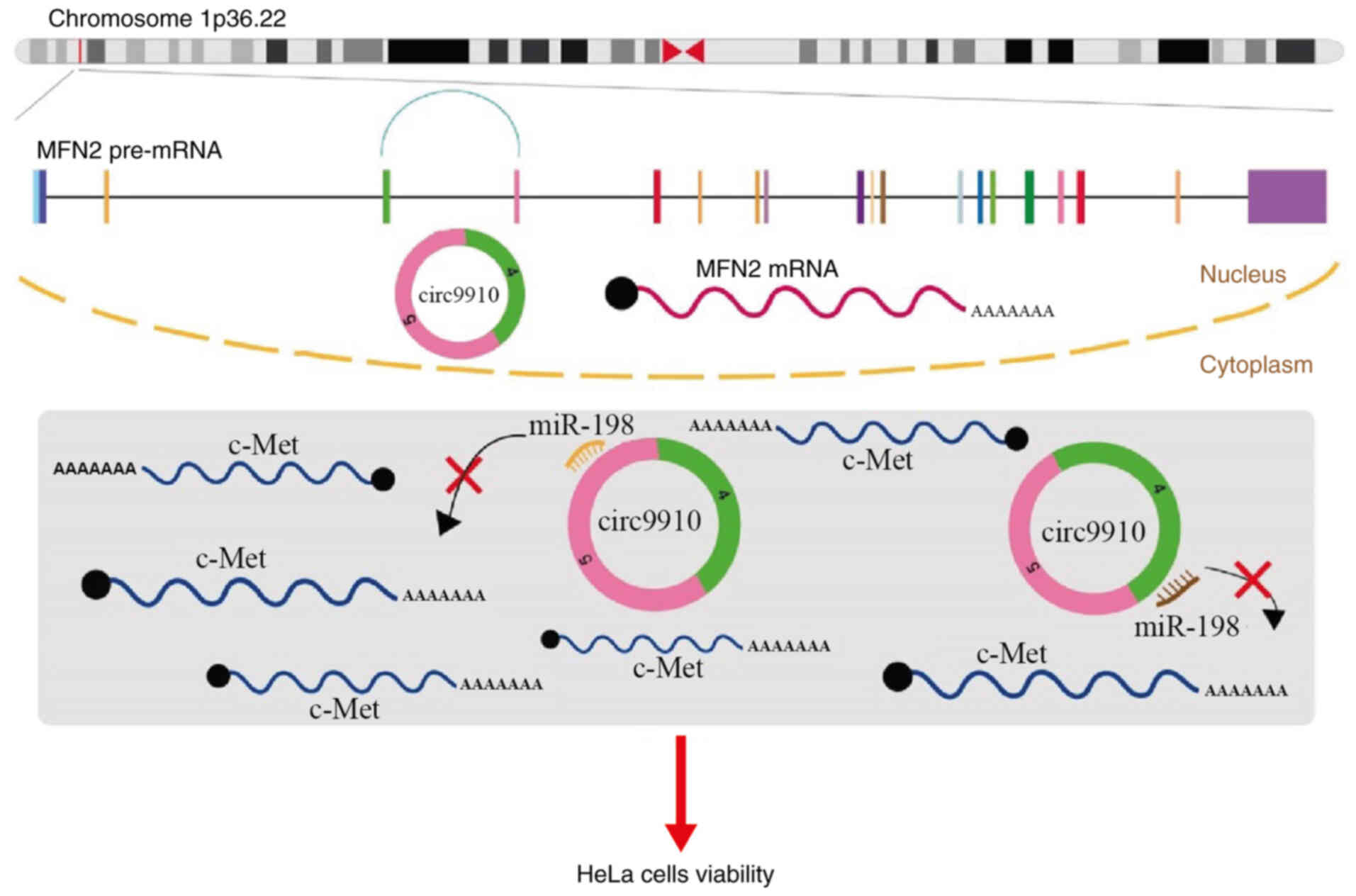
To summarize the present findings, hsa_circ_0009910
functioned as a sponge for miR-198 and promoted an increase in
c-Met expression levels, and as this regulatory network decreased
the cell viability of HeLa cells (Fig.
5). Taken together, these results indicate that
hsa_circ_0009910 could be a molecular sponge of miR-198 and
contribute to the upregulation of c-Met expression levels. The
hsa_circ_0009910/miR-198/c-Met interaction network affects cell
viability but not apoptosis in HeLa cells. Based on this mechanism,
hsa_circ_0009910 may be a promising biomarker for CC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Council of
Humanities, Sciences, and Technologies (grant no. 242812).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
BXTM and JLL designed and performed the experiments.
AOS, GFT, YCC and DHS conceived and designed the study. BXTM and
DHS confirm the authenticity of all the raw data. BXTM, YCC, JLL
and DHS wrote the manuscript. All authors read and approved the
final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bhatla N, Aoki D, Sharma DN and
Sankaranarayanan R: Cancer of the cervix uteri: 2021 update. Int J
Gynaecol Obstet. 155 (Suppl 1):S28–S44. 2021. View Article : Google Scholar
|
|
3
|
Volkova LV, Pashov AI and Omelchuk NN:
Cervical carcinoma: Oncobiology and biomarkers. Int J Mol Sci.
22:125712021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Alarcón-Romero LDC, Organista-Nava J,
Gómez-Gómez Y, Ortiz-Ortiz J, Hernández-Sotelo D, Del
Moral-Hernández O, Mendoza-Catalán MA, Antaño-Arias R,
Leyva-Vázquez MA, Sales-Linares N, et al: Prevalence and
distribution of human papillomavirus genotypes (1997–2019) and
their association with cervical cancer and precursor lesions in
women from Southern Mexico. Cancer Control.
29:107327482211033312022. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fang J, Zhang H and Jin S: Epigenetics and
cervical cancer: From pathogenesis to therapy. Tumour Biol.
35:5083–5093. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Parashar D, Singh A, Gupta S, Sharma A,
Sharma MK, Roy KK, Chauhan SC and Kashyap VK: Emerging roles and
potential applications of non-coding RNAs in cervical cancer. Genes
(Basel). 13:12542022. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Saw PE, Xu X, Chen J and Song EW:
Non-coding RNAs: The new central dogma of cancer biology. Sci China
Life Sci. 64:22–50. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen LL: The biogenesis and emerging roles
of circular RNAs. Nat Rev Mol Cell Biol. 17:205–211. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Eger N, Schoppe L, Schuster S, Laufs U and
Boeckel JN: Circular RNA splicing. Adv Exp Med Biol. 1087:41–52.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lasda E and Parker R: Circular RNAs:
Diversity of form and function. RNA. 20:1829–1842. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Arnaiz E, Sole C, Manterola L,
Iparraguirre L, Otaegui D and Lawrie CH: CircRNAs and cancer:
Biomarkers and master regulators. Semin Cancer Biol. 58:90–99.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Huang J, Chen J and Huang Q: The profile
analysis of circular RNAs in cervical cancer. Medicine (Baltimore).
100:e274042021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tornesello ML, Faraonio R, Buonaguro L,
Annunziata C, Starita N, Cerasuolo A, Pezzuto F, Tornesello AL and
Buonaguro FM: The role of microRNAs, long non-coding RNAs, and
circular RNAs in cervical cancer. Front Oncol. 10:1502020.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bonelli P, Borrelli A, Tuccillo FM,
Buonaguro FM and Tornesello ML: The role of circRNAs in human
papillomavirus (HPV)-associated cancers. Cancers (Basel).
13:11732021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Panda AC: Circular RNAs act as miRNA
sponges. Adv Exp Med Biol. 1087:67–79. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Huntzinger E and Izaurralde E: Gene
silencing by microRNAs: Contributions of translational repression
and mRNA decay. Nat Rev Genet. 12:99–110. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kulcheski FR, Christoff AP and Margis R:
Circular RNAs are miRNA sponges and can be used as a new class of
biomarker. J Biotechnol. 238:42–51. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhu B, Gao J, Zhang Y, Liao B, Zhu S, Li
C, Liao J, Liu J, Jiang C and Zeng J: CircRNA/miRNA/mRNA axis
participates in the progression of partial bladder outlet
obstruction. BMC Urol. 22:1912022. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xu YJ, Yu H and Liu GX:
Hsa_circ_0031288/hsa-miR-139-3p/Bcl-6 regulatory feedback circuit
influences the invasion and migration of cervical cancer HeLa
cells. J Cell Biochem. 121:4251–4260. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen Y, Geng Y, Huang J, Xi D, Xu G, Gu W
and Shao Y: CircNEIL3 promotes cervical cancer cell proliferation
by adsorbing miR-137 and upregulating KLF12. Cancer Cell Int.
21:342021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets-update. Nucleic Acids Res. 41:D991–D995. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Green GH and Diggle PJ: On the operational
characteristics of the Benjamini and Hochberg false discovery rate
procedure. Stat Appl Genet Mol Biol. 6:Article272007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dudekula DB, Panda AC, Grammatikakis I, De
S, Abdelmohsen K and Gorospe M: CircInteractome: A web tool for
exploring circular RNAs and their interacting proteins and
microRNAs. RNA Biol. 13:34–42. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Feng J, Chen W, Dong X, Wang J, Mei X,
Deng J, Yang S, Zhuo C, Huang X, Shao L, et al: CSCD2: An
integrated interactional database of cancer-specific circular RNAs.
Nucleic Acids Res. 50:D1179–D1183. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tang Z, Li C, Kang B, Gao G and Zhang Z:
GEPIA: A web server for cancer and normal gene expression profiling
and interactive analyses. Nucleic Acids Res. 45:W98–W102. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Karagkouni D, Paraskevopoulou MD,
Chatzopoulos S, Vlachos IS, Tastsoglou S, Kanellos I, Papadimitriou
D, Kavakiotis I, Maniou S, Skoufos G, et al: DIANA-TarBase v8: A
decade-long collection of experimentally supported miRNA-gene
interactions. Nucleic Acids Res. 46:D239–D245. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Huang HY, Lin YC, Cui S, Huang Y, Tang Y,
Xu J, Bao J, Li Y, Wen J, Zuo H, et al: miRTarBase update 2022: An
informative resource for experimentally validated miRNA-target
interactions. Nucleic Acids Res. 50:D222–D230. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mi H, Muruganujan A, Casagrande JT and
Thomas PD: Large-scale gene function analysis with the PANTHER
classification system. Nat Protoc. 8:1551–1566. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kumar P, Nagarajan A and Uchil PD:
Analysis of cell viability by the MTT assay. Cold Spring Harb
Protoc. 2018:62018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Repetto G, del Peso A and Zurita JL:
Neutral red uptake assay for the estimation of cell
viability/cytotoxicity. Nat Protoc. 3:1125–1131. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kadkhoda S, Taslimi R, Noorbakhsh F,
Darbeheshti F, Bazzaz JT, Ghafouri-Fard S and Shakoori A:
Importance of Circ0009910 in colorectal cancer pathogenesis as a
possible regulator of miR-145 and PEAK1. World J Surg Oncol.
19:2652021. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ray J, Hoey C, Huang X, Jeon J, Taeb S,
Downes MR, Boutros PC and Liu SK: MicroRNA-198 suppresses prostate
tumorigenesis by targeting MIB1. Oncol Rep. 42:1047–1056.
2019.PubMed/NCBI
|
|
35
|
Kang Y, Zhang Y and Sun Y: MicroRNA-198
suppresses tumour growth and metastasis in oral squamous cell
carcinoma by targeting CDK4. Int J Oncol. 59:392021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li LX, Lam IH, Liang FF, Yi SP, Ye LF,
Wang JT, Guo WW and Xu M: MiR-198 affects the proliferation and
apoptosis of colorectal cancer through regulation of
ADAM28/JAK-STAT signaling pathway. Eur Rev Med Pharmacol Sci.
23:1487–1493. 2019.PubMed/NCBI
|
|
37
|
Nie E, Jin X, Wu W, Yu T, Zhou X, Shi Z,
Zhang J, Liu N and You Y: MiR-198 enhances temozolomide sensitivity
in glioblastoma by targeting MGMT. J Neurooncol. 133:59–68. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Shi Y, Fang N, Li Y, Guo Z, Jiang W, He Y,
Ma Z and Chen Y: Circular RNA LPAR3 sponges microRNA-198 to
facilitate esophageal cancer migration, invasion, and metastasis.
Cancer Sci. 111:2824–2836. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tan S, Li R, Ding K, Lobie PE and Zhu T:
miR-198 inhibits migration and invasion of hepatocellular carcinoma
cells by targeting the HGF/c-MET pathway. FEBS Lett. 585:2229–2234.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Xu F, Ni M, Li J, Cheng J, Zhao H, Zhao J,
Huang S and Wu X: Circ0004390 promotes cell proliferation through
sponging miR-198 in ovarian cancer. Biochem Biophys Res Commun.
526:14–20. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Miekus K, Pawlowska M, Sekuła M, Drabik G,
Madeja Z, Adamek D and Majka M: MET receptor is a potential
therapeutic target in high grade cervical cancer. Oncotarget.
6:10086–10101. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Sundaram GM, Common JE, Gopal FE, Srikanta
S, Lakshman K, Lunny DP, Lim TC, Tanavde V, Lane EB and Sampath P:
‘See-saw’ expression of microRNA-198 and FSTL1 from a single
transcript in wound healing. Nature. 495:103–106. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Sundaram GM, Quah S, Guang LG and Sampath
P: HuR enhances FSTL1 transcript stability to promote invasion and
metastasis of squamous cell carcinoma. Am J Cancer Res.
11:4981–4993. 2021.PubMed/NCBI
|
|
44
|
Qian L, Yu S, Chen Z, Meng Z, Huang S and
Wang P: The emerging role of circRNAs and their clinical
significance in human cancers. Biochim Biophys Acta Rev Cancer.
1870:247–260. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Chen L and Shan G: CircRNA in cancer:
Fundamental mechanism and clinical potential. Cancer Lett.
505:49–57. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wu F and Zhou J: CircAGFG1 promotes
cervical cancer progression via miR-370-3p/RAF1 signaling. BMC
Cancer. 19:10672019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Rong X, Gao W, Yang X and Guo J:
Downregulation of hsa_circ_0007534 restricts the proliferation and
invasion of cervical cancer through regulating miR-498/BMI-1
signaling. Life Sci. 235:1167852019. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhang X, Zhang Q, Zhang K, Wang F, Qiao X
and Cui J: Circ SMARCA5 inhibited tumor metastasis by interacting
with SND1 and downregulating the YWHAB gene in cervical cancer.
Cell Transplant. 30:9636897209837862021. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Xu J, Zhang Y, Huang Y, Dong X, Xiang Z,
Zou J, Wu L and Lu W: circEYA1 functions as a sponge of miR-582-3p
to suppress cervical adenocarcinoma tumorigenesis via upregulating
CXCL14. Mol Ther Nucleic Acids. 22:1176–1190. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhang X, Wang S, Wang H, Cao J, Huang X,
Chen Z, Xu P, Sun G, Xu J, Lv J and Xu Z: Circular RNA circNRIP1
acts as a microRNA-149-5p sponge to promote gastric cancer
progression via the AKT1/mTOR pathway. Mol Cancer. 18:202019.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Huang G, Liang M, Liu H, Huang J, Li P,
Wang C, Zhang Y, Lin Y and Jiang X: CircRNA hsa_circRNA_104348
promotes hepatocellular carcinoma progression through modulating
miR-187-3p/RTKN2 axis and activating Wnt/β-catenin pathway. Cell
Death Dis. 11:10652020. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Liu J, Wang D, Long Z, Liu J and Li W:
CircRNA8924 promotes cervical cancer cell proliferation, migration
and invasion by competitively binding to MiR-518d-5p /519-5p family
and modulating the expression of CBX8. Cell Physiol Biochem.
48:173–184. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Qu X, Zhu L, Song L and Liu S:
circ_0084927 promotes cervical carcinogenesis by sponging miR-1179
that suppresses CDK2, a cell cycle-related gene. Cancer Cell Int.
20:3332020. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Cai Y, Li C, Peng F, Yin S, Liang H, Su J,
Li L, Yang A, Liu H, Yang C, et al: Downregulation of
hsa_circRNA_0001400 helps to promote cell apoptosis through
disruption of the circRNA_0001400-miR-326 sponge in cervical cancer
cells. Front Genet. 12:7791952021. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Ji F, Du R, Chen T, Zhang M, Zhu Y, Luo X
and Ding Y: Circular RNA circSLC26A4 accelerates cervical cancer
progression via miR-1287-5p/HOXA7 axis. Mol Ther Nucleic Acids.
19:413–420. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Deng N, Li L, Gao J, Zhou J, Wang Y, Wang
C and Liu Y: Hsa_circ_0009910 promotes carcinogenesis by promoting
the expression of miR-449a target IL6R in osteosarcoma. Biochem
Biophys Res Commun. 495:189–196. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Wang D, Ming X, Xu J and Xiao Y:
Circ_0009910 shuttled by exosomes regulates proliferation, cell
cycle and apoptosis of acute myeloid leukemia cells by regulating
miR-5195-3p/GRB10 axis. Hematol Oncol. 39:390–400. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Li Y, Lin S and An N: Hsa_circ_0009910:
Oncogenic circular RNA targets microRNA-145 in ovarian cancer
cells. Cell Cycle. 19:1857–1868. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Li HW and Liu J: Circ_0009910 promotes
proliferation and metastasis of hepatocellular carcinoma cells
through miR-335-5p/ROCK1 axis. Eur Rev Med Pharmacol Sci.
24:1725–1735. 2020.PubMed/NCBI
|
|
60
|
Cao HX, Miao CF, Sang LN, Huang YM, Zhang
R, Sun L and Jiang ZX: Circ_0009910 promotes imatinib resistance
through ULK1-induced autophagy by sponging miR-34a-5p in chronic
myeloid leukemia. Life Sci. 243:1172552020. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Ping L, Jian-Jun C, Chu-Shu L, Guang-Hua L
and Ming Z: Silencing of circ_0009910 inhibits acute myeloid
leukemia cell growth through increasing miR-20a-5p. Blood Cells Mol
Dis. 75:41–47. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Georges S, Calleja LR, Jacques C, Lavaud
M, Moukengue B, Lecanda F, Quillard T, Gabriel MT, Cartron PF,
Baud'huin M, et al: Loss of miR-198 and −206 during primary tumor
progression enables metastatic dissemination in human osteosarcoma.
Oncotarget. 9:35726–35741. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Zhang Y, Xia M, Jin K, Wang S, Wei H, Fan
C, Wu Y, Li X, Li X, Li G, et al: Function of the c-Met receptor
tyrosine kinase in carcinogenesis and associated therapeutic
opportunities. Mol Cancer. 17:452018. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Raj S, Kesari KK, Kumar A, Rathi B, Sharma
A, Gupta PK, Jha SK, Jha NK, Slama P, Roychoudhury S and Kumar D:
Molecular mechanism(s) of regulation(s) of c-MET/HGF signaling in
head and neck cancer. Mol Cancer. 21:312022. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Li Y, Luo H, Xiao N, Duan J, Wang Z and
Wang S: Long noncoding RNA SChLAP1 accelerates the proliferation
and metastasis of prostate cancer via targeting miR-198 and
promoting the MAPK1 pathway. Oncol Res. 26:131–143. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Ji Y, Liu J, Zhu W and Ji J: circ_0002060
enhances doxorubicin resistance in osteosarcoma by regulating the
miR-198/ABCB1 axis. Cancer Biother Radiopharm. 38:585–595.
2023.PubMed/NCBI
|
|
67
|
Zhong JX, Kong YY, Luo RG, Xia GJ, He WX,
Chen XZ, Tan WW, Chen QJ, Huang YY and Guan YX: Circular RNA
circ-ERBB2 promotes HER2-positive breast cancer progression and
metastasis via sponging miR-136-5p and miR-198. J Transl Med.
19:4552021. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Huang X, Li Z, Zhang Q, Wang W, Li B, Wang
L, Xu Z, Zeng A, Zhang X, Zhang X, et al: Circular RNA AKT3
upregulates PIK3R1 to enhance cisplatin resistance in gastric
cancer via miR-198 suppression. Mol Cancer. 18:712019. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Deng Y, Zhu H, Xiao L, Liu C and Meng X:
Circ_0005198 enhances temozolomide resistance of glioma cells
through miR-198/TRIM14 axis. Aging (Albany NY). 13:2198–2211. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Cheng Y, Yang M and Peng J: Correlation
the between the regulation of miRNA-1 in c-Met-induced EMT and
cervical cancer progression. Oncol Lett. 17:3341–3349.
2019.PubMed/NCBI
|
|
71
|
Campos-Viguri GE, Peralta-Zaragoza O,
Jiménez-Wences H, Longinos-González AE, Castañón-Sánchez CA,
Ramírez-Carrillo M, Camarillo CL, Castañeda-Saucedo E,
Jiménez-López MA, Martínez-Carrillo DN and Fernández-Tilapa G:
MiR-23b-3p reduces the proliferation, migration and invasion of
cervical cancer cell lines via the reduction of c-Met expression.
Sci Rep. 10:32562020. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Guo Y, Tao M and Jiang M: MicroRNA-454-3p
inhibits cervical cancer cell invasion and migration by targeting
c-Met. Exp Ther Med. 15:2301–2306. 2018.PubMed/NCBI
|
|
73
|
Guo Q, Li L, Bo Q, Chen L, Sun L and Shi
H: Long noncoding RNA PITPNA-AS1 promotes cervical cancer
progression through regulating the cell cycle and apoptosis by
targeting the miR-876-5p/c-MET axis. Biomed Pharmacother.
128:1100722020. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Zhang S, Xu Y and Zheng Q: circRNA_0000285
knockdown suppresses viability and promotes apoptosis of cervical
cancer cells by sponging microRNA-654-3p. Bioengineered.
13:5251–5261. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Fei Z, Qin L, Luo F and Yu Y: CircRNA
circ-ATAD1 is upregulated in cervical squamous cell carcinoma and
regulates cell proliferation and apoptosis by suppressing the
maturation of miR-218. Reprod Sci. 28:2982–2988. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Xiao CY, Fu BB, Li ZY, Mushtaq G, Kamal
MA, Li JH, Tang GC and Xiao SS: Observations on the expression of
human papillomavirus major capsid protein in HeLa cells. Cancer
Cell Int. 15:532015. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Qi Z, Xu X, Zhang B, Li Y, Liu J, Chen S,
Chen G and Huo X: Effect of simultaneous silencing of HPV-18 E6 and
E7 on inducing apoptosis in HeLa cells. Biochem Cell Biol.
88:697–704. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Xiong Y, Chen L and Luo P:
N-Benzylcinnamide induces apoptosis in HPV16 and HPV18 cervical
cancer cells via suppression of E6 and E7 protein expression. IUBMB
Life. 67:374–379. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Wang X, Zhu Y and Xie Q: The promising
role and prognostic value of miR-198 in human diseases. Am J Transl
Res. 14:2749–2766. 2022.PubMed/NCBI
|
|
80
|
Kahraman A and Dirilenoğlu F: Assessing
the diagnostic value of CAIX and ProEx-C in cervical squamous
intraepithelial lesions. Pathol Res Pract. 253:1550292024.
View Article : Google Scholar : PubMed/NCBI
|