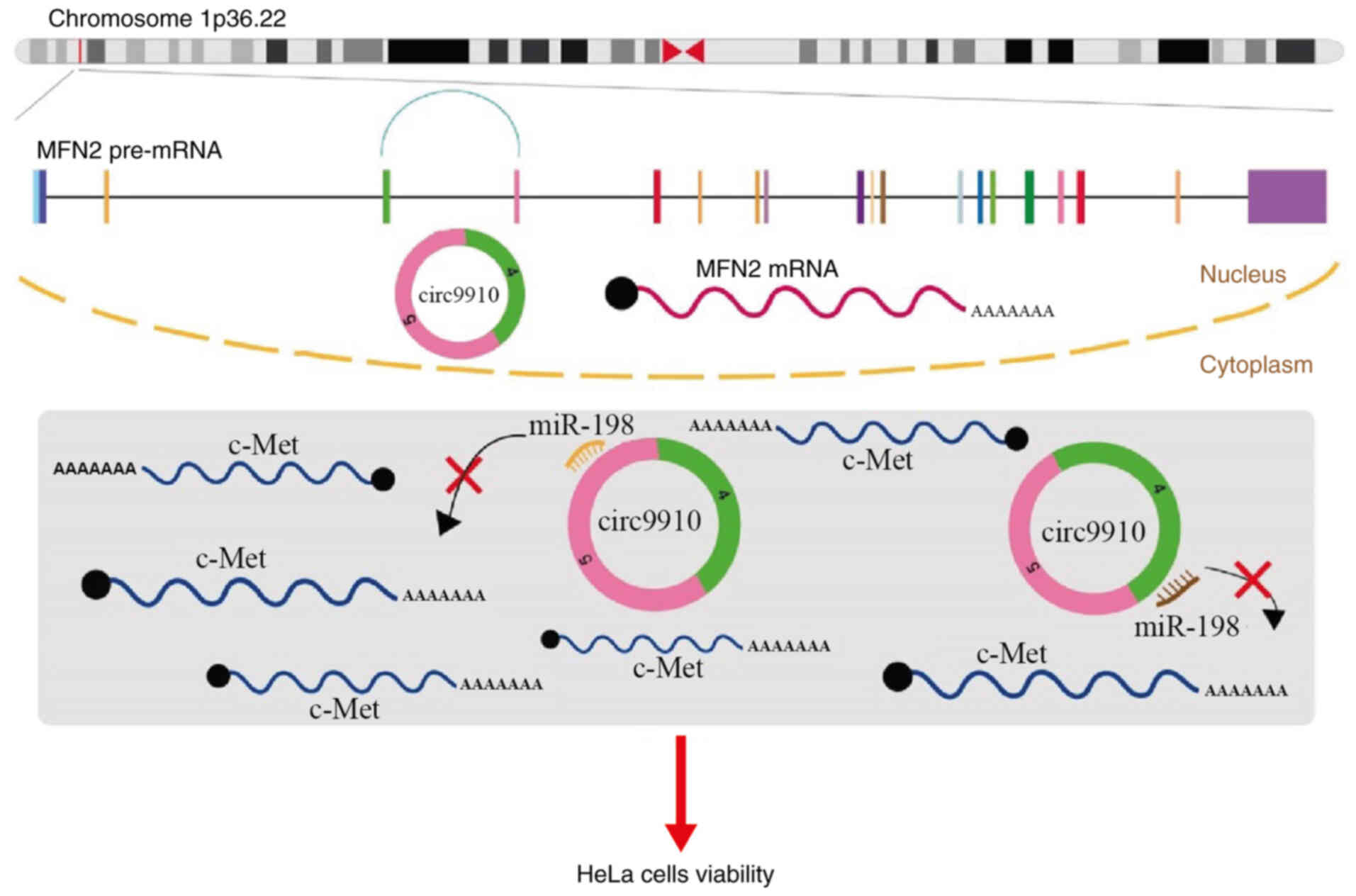
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bhatla N, Aoki D, Sharma DN and
Sankaranarayanan R: Cancer of the cervix uteri: 2021 update. Int J
Gynaecol Obstet. 155 (Suppl 1):S28–S44. 2021. View Article : Google Scholar
|
|
3
|
Volkova LV, Pashov AI and Omelchuk NN:
Cervical carcinoma: Oncobiology and biomarkers. Int J Mol Sci.
22:125712021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Alarcón-Romero LDC, Organista-Nava J,
Gómez-Gómez Y, Ortiz-Ortiz J, Hernández-Sotelo D, Del
Moral-Hernández O, Mendoza-Catalán MA, Antaño-Arias R,
Leyva-Vázquez MA, Sales-Linares N, et al: Prevalence and
distribution of human papillomavirus genotypes (1997–2019) and
their association with cervical cancer and precursor lesions in
women from Southern Mexico. Cancer Control.
29:107327482211033312022. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fang J, Zhang H and Jin S: Epigenetics and
cervical cancer: From pathogenesis to therapy. Tumour Biol.
35:5083–5093. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Parashar D, Singh A, Gupta S, Sharma A,
Sharma MK, Roy KK, Chauhan SC and Kashyap VK: Emerging roles and
potential applications of non-coding RNAs in cervical cancer. Genes
(Basel). 13:12542022. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Saw PE, Xu X, Chen J and Song EW:
Non-coding RNAs: The new central dogma of cancer biology. Sci China
Life Sci. 64:22–50. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen LL: The biogenesis and emerging roles
of circular RNAs. Nat Rev Mol Cell Biol. 17:205–211. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Eger N, Schoppe L, Schuster S, Laufs U and
Boeckel JN: Circular RNA splicing. Adv Exp Med Biol. 1087:41–52.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lasda E and Parker R: Circular RNAs:
Diversity of form and function. RNA. 20:1829–1842. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Arnaiz E, Sole C, Manterola L,
Iparraguirre L, Otaegui D and Lawrie CH: CircRNAs and cancer:
Biomarkers and master regulators. Semin Cancer Biol. 58:90–99.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Huang J, Chen J and Huang Q: The profile
analysis of circular RNAs in cervical cancer. Medicine (Baltimore).
100:e274042021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tornesello ML, Faraonio R, Buonaguro L,
Annunziata C, Starita N, Cerasuolo A, Pezzuto F, Tornesello AL and
Buonaguro FM: The role of microRNAs, long non-coding RNAs, and
circular RNAs in cervical cancer. Front Oncol. 10:1502020.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bonelli P, Borrelli A, Tuccillo FM,
Buonaguro FM and Tornesello ML: The role of circRNAs in human
papillomavirus (HPV)-associated cancers. Cancers (Basel).
13:11732021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Panda AC: Circular RNAs act as miRNA
sponges. Adv Exp Med Biol. 1087:67–79. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Huntzinger E and Izaurralde E: Gene
silencing by microRNAs: Contributions of translational repression
and mRNA decay. Nat Rev Genet. 12:99–110. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kulcheski FR, Christoff AP and Margis R:
Circular RNAs are miRNA sponges and can be used as a new class of
biomarker. J Biotechnol. 238:42–51. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhu B, Gao J, Zhang Y, Liao B, Zhu S, Li
C, Liao J, Liu J, Jiang C and Zeng J: CircRNA/miRNA/mRNA axis
participates in the progression of partial bladder outlet
obstruction. BMC Urol. 22:1912022. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xu YJ, Yu H and Liu GX:
Hsa_circ_0031288/hsa-miR-139-3p/Bcl-6 regulatory feedback circuit
influences the invasion and migration of cervical cancer HeLa
cells. J Cell Biochem. 121:4251–4260. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen Y, Geng Y, Huang J, Xi D, Xu G, Gu W
and Shao Y: CircNEIL3 promotes cervical cancer cell proliferation
by adsorbing miR-137 and upregulating KLF12. Cancer Cell Int.
21:342021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets-update. Nucleic Acids Res. 41:D991–D995. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Green GH and Diggle PJ: On the operational
characteristics of the Benjamini and Hochberg false discovery rate
procedure. Stat Appl Genet Mol Biol. 6:Article272007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dudekula DB, Panda AC, Grammatikakis I, De
S, Abdelmohsen K and Gorospe M: CircInteractome: A web tool for
exploring circular RNAs and their interacting proteins and
microRNAs. RNA Biol. 13:34–42. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Feng J, Chen W, Dong X, Wang J, Mei X,
Deng J, Yang S, Zhuo C, Huang X, Shao L, et al: CSCD2: An
integrated interactional database of cancer-specific circular RNAs.
Nucleic Acids Res. 50:D1179–D1183. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tang Z, Li C, Kang B, Gao G and Zhang Z:
GEPIA: A web server for cancer and normal gene expression profiling
and interactive analyses. Nucleic Acids Res. 45:W98–W102. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Karagkouni D, Paraskevopoulou MD,
Chatzopoulos S, Vlachos IS, Tastsoglou S, Kanellos I, Papadimitriou
D, Kavakiotis I, Maniou S, Skoufos G, et al: DIANA-TarBase v8: A
decade-long collection of experimentally supported miRNA-gene
interactions. Nucleic Acids Res. 46:D239–D245. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Huang HY, Lin YC, Cui S, Huang Y, Tang Y,
Xu J, Bao J, Li Y, Wen J, Zuo H, et al: miRTarBase update 2022: An
informative resource for experimentally validated miRNA-target
interactions. Nucleic Acids Res. 50:D222–D230. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mi H, Muruganujan A, Casagrande JT and
Thomas PD: Large-scale gene function analysis with the PANTHER
classification system. Nat Protoc. 8:1551–1566. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kumar P, Nagarajan A and Uchil PD:
Analysis of cell viability by the MTT assay. Cold Spring Harb
Protoc. 2018:62018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Repetto G, del Peso A and Zurita JL:
Neutral red uptake assay for the estimation of cell
viability/cytotoxicity. Nat Protoc. 3:1125–1131. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kadkhoda S, Taslimi R, Noorbakhsh F,
Darbeheshti F, Bazzaz JT, Ghafouri-Fard S and Shakoori A:
Importance of Circ0009910 in colorectal cancer pathogenesis as a
possible regulator of miR-145 and PEAK1. World J Surg Oncol.
19:2652021. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ray J, Hoey C, Huang X, Jeon J, Taeb S,
Downes MR, Boutros PC and Liu SK: MicroRNA-198 suppresses prostate
tumorigenesis by targeting MIB1. Oncol Rep. 42:1047–1056.
2019.PubMed/NCBI
|
|
35
|
Kang Y, Zhang Y and Sun Y: MicroRNA-198
suppresses tumour growth and metastasis in oral squamous cell
carcinoma by targeting CDK4. Int J Oncol. 59:392021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li LX, Lam IH, Liang FF, Yi SP, Ye LF,
Wang JT, Guo WW and Xu M: MiR-198 affects the proliferation and
apoptosis of colorectal cancer through regulation of
ADAM28/JAK-STAT signaling pathway. Eur Rev Med Pharmacol Sci.
23:1487–1493. 2019.PubMed/NCBI
|
|
37
|
Nie E, Jin X, Wu W, Yu T, Zhou X, Shi Z,
Zhang J, Liu N and You Y: MiR-198 enhances temozolomide sensitivity
in glioblastoma by targeting MGMT. J Neurooncol. 133:59–68. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Shi Y, Fang N, Li Y, Guo Z, Jiang W, He Y,
Ma Z and Chen Y: Circular RNA LPAR3 sponges microRNA-198 to
facilitate esophageal cancer migration, invasion, and metastasis.
Cancer Sci. 111:2824–2836. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tan S, Li R, Ding K, Lobie PE and Zhu T:
miR-198 inhibits migration and invasion of hepatocellular carcinoma
cells by targeting the HGF/c-MET pathway. FEBS Lett. 585:2229–2234.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Xu F, Ni M, Li J, Cheng J, Zhao H, Zhao J,
Huang S and Wu X: Circ0004390 promotes cell proliferation through
sponging miR-198 in ovarian cancer. Biochem Biophys Res Commun.
526:14–20. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Miekus K, Pawlowska M, Sekuła M, Drabik G,
Madeja Z, Adamek D and Majka M: MET receptor is a potential
therapeutic target in high grade cervical cancer. Oncotarget.
6:10086–10101. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Sundaram GM, Common JE, Gopal FE, Srikanta
S, Lakshman K, Lunny DP, Lim TC, Tanavde V, Lane EB and Sampath P:
‘See-saw’ expression of microRNA-198 and FSTL1 from a single
transcript in wound healing. Nature. 495:103–106. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Sundaram GM, Quah S, Guang LG and Sampath
P: HuR enhances FSTL1 transcript stability to promote invasion and
metastasis of squamous cell carcinoma. Am J Cancer Res.
11:4981–4993. 2021.PubMed/NCBI
|
|
44
|
Qian L, Yu S, Chen Z, Meng Z, Huang S and
Wang P: The emerging role of circRNAs and their clinical
significance in human cancers. Biochim Biophys Acta Rev Cancer.
1870:247–260. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Chen L and Shan G: CircRNA in cancer:
Fundamental mechanism and clinical potential. Cancer Lett.
505:49–57. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wu F and Zhou J: CircAGFG1 promotes
cervical cancer progression via miR-370-3p/RAF1 signaling. BMC
Cancer. 19:10672019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Rong X, Gao W, Yang X and Guo J:
Downregulation of hsa_circ_0007534 restricts the proliferation and
invasion of cervical cancer through regulating miR-498/BMI-1
signaling. Life Sci. 235:1167852019. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhang X, Zhang Q, Zhang K, Wang F, Qiao X
and Cui J: Circ SMARCA5 inhibited tumor metastasis by interacting
with SND1 and downregulating the YWHAB gene in cervical cancer.
Cell Transplant. 30:9636897209837862021. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Xu J, Zhang Y, Huang Y, Dong X, Xiang Z,
Zou J, Wu L and Lu W: circEYA1 functions as a sponge of miR-582-3p
to suppress cervical adenocarcinoma tumorigenesis via upregulating
CXCL14. Mol Ther Nucleic Acids. 22:1176–1190. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhang X, Wang S, Wang H, Cao J, Huang X,
Chen Z, Xu P, Sun G, Xu J, Lv J and Xu Z: Circular RNA circNRIP1
acts as a microRNA-149-5p sponge to promote gastric cancer
progression via the AKT1/mTOR pathway. Mol Cancer. 18:202019.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Huang G, Liang M, Liu H, Huang J, Li P,
Wang C, Zhang Y, Lin Y and Jiang X: CircRNA hsa_circRNA_104348
promotes hepatocellular carcinoma progression through modulating
miR-187-3p/RTKN2 axis and activating Wnt/β-catenin pathway. Cell
Death Dis. 11:10652020. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Liu J, Wang D, Long Z, Liu J and Li W:
CircRNA8924 promotes cervical cancer cell proliferation, migration
and invasion by competitively binding to MiR-518d-5p /519-5p family
and modulating the expression of CBX8. Cell Physiol Biochem.
48:173–184. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Qu X, Zhu L, Song L and Liu S:
circ_0084927 promotes cervical carcinogenesis by sponging miR-1179
that suppresses CDK2, a cell cycle-related gene. Cancer Cell Int.
20:3332020. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Cai Y, Li C, Peng F, Yin S, Liang H, Su J,
Li L, Yang A, Liu H, Yang C, et al: Downregulation of
hsa_circRNA_0001400 helps to promote cell apoptosis through
disruption of the circRNA_0001400-miR-326 sponge in cervical cancer
cells. Front Genet. 12:7791952021. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Ji F, Du R, Chen T, Zhang M, Zhu Y, Luo X
and Ding Y: Circular RNA circSLC26A4 accelerates cervical cancer
progression via miR-1287-5p/HOXA7 axis. Mol Ther Nucleic Acids.
19:413–420. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Deng N, Li L, Gao J, Zhou J, Wang Y, Wang
C and Liu Y: Hsa_circ_0009910 promotes carcinogenesis by promoting
the expression of miR-449a target IL6R in osteosarcoma. Biochem
Biophys Res Commun. 495:189–196. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Wang D, Ming X, Xu J and Xiao Y:
Circ_0009910 shuttled by exosomes regulates proliferation, cell
cycle and apoptosis of acute myeloid leukemia cells by regulating
miR-5195-3p/GRB10 axis. Hematol Oncol. 39:390–400. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Li Y, Lin S and An N: Hsa_circ_0009910:
Oncogenic circular RNA targets microRNA-145 in ovarian cancer
cells. Cell Cycle. 19:1857–1868. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Li HW and Liu J: Circ_0009910 promotes
proliferation and metastasis of hepatocellular carcinoma cells
through miR-335-5p/ROCK1 axis. Eur Rev Med Pharmacol Sci.
24:1725–1735. 2020.PubMed/NCBI
|
|
60
|
Cao HX, Miao CF, Sang LN, Huang YM, Zhang
R, Sun L and Jiang ZX: Circ_0009910 promotes imatinib resistance
through ULK1-induced autophagy by sponging miR-34a-5p in chronic
myeloid leukemia. Life Sci. 243:1172552020. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Ping L, Jian-Jun C, Chu-Shu L, Guang-Hua L
and Ming Z: Silencing of circ_0009910 inhibits acute myeloid
leukemia cell growth through increasing miR-20a-5p. Blood Cells Mol
Dis. 75:41–47. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Georges S, Calleja LR, Jacques C, Lavaud
M, Moukengue B, Lecanda F, Quillard T, Gabriel MT, Cartron PF,
Baud'huin M, et al: Loss of miR-198 and −206 during primary tumor
progression enables metastatic dissemination in human osteosarcoma.
Oncotarget. 9:35726–35741. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Zhang Y, Xia M, Jin K, Wang S, Wei H, Fan
C, Wu Y, Li X, Li X, Li G, et al: Function of the c-Met receptor
tyrosine kinase in carcinogenesis and associated therapeutic
opportunities. Mol Cancer. 17:452018. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Raj S, Kesari KK, Kumar A, Rathi B, Sharma
A, Gupta PK, Jha SK, Jha NK, Slama P, Roychoudhury S and Kumar D:
Molecular mechanism(s) of regulation(s) of c-MET/HGF signaling in
head and neck cancer. Mol Cancer. 21:312022. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Li Y, Luo H, Xiao N, Duan J, Wang Z and
Wang S: Long noncoding RNA SChLAP1 accelerates the proliferation
and metastasis of prostate cancer via targeting miR-198 and
promoting the MAPK1 pathway. Oncol Res. 26:131–143. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Ji Y, Liu J, Zhu W and Ji J: circ_0002060
enhances doxorubicin resistance in osteosarcoma by regulating the
miR-198/ABCB1 axis. Cancer Biother Radiopharm. 38:585–595.
2023.PubMed/NCBI
|
|
67
|
Zhong JX, Kong YY, Luo RG, Xia GJ, He WX,
Chen XZ, Tan WW, Chen QJ, Huang YY and Guan YX: Circular RNA
circ-ERBB2 promotes HER2-positive breast cancer progression and
metastasis via sponging miR-136-5p and miR-198. J Transl Med.
19:4552021. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Huang X, Li Z, Zhang Q, Wang W, Li B, Wang
L, Xu Z, Zeng A, Zhang X, Zhang X, et al: Circular RNA AKT3
upregulates PIK3R1 to enhance cisplatin resistance in gastric
cancer via miR-198 suppression. Mol Cancer. 18:712019. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Deng Y, Zhu H, Xiao L, Liu C and Meng X:
Circ_0005198 enhances temozolomide resistance of glioma cells
through miR-198/TRIM14 axis. Aging (Albany NY). 13:2198–2211. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Cheng Y, Yang M and Peng J: Correlation
the between the regulation of miRNA-1 in c-Met-induced EMT and
cervical cancer progression. Oncol Lett. 17:3341–3349.
2019.PubMed/NCBI
|
|
71
|
Campos-Viguri GE, Peralta-Zaragoza O,
Jiménez-Wences H, Longinos-González AE, Castañón-Sánchez CA,
Ramírez-Carrillo M, Camarillo CL, Castañeda-Saucedo E,
Jiménez-López MA, Martínez-Carrillo DN and Fernández-Tilapa G:
MiR-23b-3p reduces the proliferation, migration and invasion of
cervical cancer cell lines via the reduction of c-Met expression.
Sci Rep. 10:32562020. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Guo Y, Tao M and Jiang M: MicroRNA-454-3p
inhibits cervical cancer cell invasion and migration by targeting
c-Met. Exp Ther Med. 15:2301–2306. 2018.PubMed/NCBI
|
|
73
|
Guo Q, Li L, Bo Q, Chen L, Sun L and Shi
H: Long noncoding RNA PITPNA-AS1 promotes cervical cancer
progression through regulating the cell cycle and apoptosis by
targeting the miR-876-5p/c-MET axis. Biomed Pharmacother.
128:1100722020. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Zhang S, Xu Y and Zheng Q: circRNA_0000285
knockdown suppresses viability and promotes apoptosis of cervical
cancer cells by sponging microRNA-654-3p. Bioengineered.
13:5251–5261. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Fei Z, Qin L, Luo F and Yu Y: CircRNA
circ-ATAD1 is upregulated in cervical squamous cell carcinoma and
regulates cell proliferation and apoptosis by suppressing the
maturation of miR-218. Reprod Sci. 28:2982–2988. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Xiao CY, Fu BB, Li ZY, Mushtaq G, Kamal
MA, Li JH, Tang GC and Xiao SS: Observations on the expression of
human papillomavirus major capsid protein in HeLa cells. Cancer
Cell Int. 15:532015. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Qi Z, Xu X, Zhang B, Li Y, Liu J, Chen S,
Chen G and Huo X: Effect of simultaneous silencing of HPV-18 E6 and
E7 on inducing apoptosis in HeLa cells. Biochem Cell Biol.
88:697–704. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Xiong Y, Chen L and Luo P:
N-Benzylcinnamide induces apoptosis in HPV16 and HPV18 cervical
cancer cells via suppression of E6 and E7 protein expression. IUBMB
Life. 67:374–379. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Wang X, Zhu Y and Xie Q: The promising
role and prognostic value of miR-198 in human diseases. Am J Transl
Res. 14:2749–2766. 2022.PubMed/NCBI
|
|
80
|
Kahraman A and Dirilenoğlu F: Assessing
the diagnostic value of CAIX and ProEx-C in cervical squamous
intraepithelial lesions. Pathol Res Pract. 253:1550292024.
View Article : Google Scholar : PubMed/NCBI
|