Introduction
Lung cancer is the leading cause of cancer-related
deaths globally, with non-small cell lung cancer (NSCLC) making up
80–85% of cases (1). In both male
and female cases of malignant tumor-related deaths, ~21% are
attributed to lung cancer (1). The
risk factors for lung cancer include tobacco use, a family history
of the disease, exposure to radiation and the presence of chronic
lung conditions (2). Epidermal
growth factor receptor (EGFR) mutations are the most common
driver mutations in patients with NSCLC from the Asian population,
occurring in ~47% of cases (3). In
clinical practice, first-line therapy with EGFR tyrosine kinase
inhibitors (TKIs) is recommended, as it enhances the survival of
patients with advanced NSCLC with sensitive EGFR mutations
(4). There are three generations of
EGFR TKIs, each with distinct mechanisms of action.
First-generation EGFR TKIs, which include gefitinib, erlotinib and
icotinib, function as reversible inhibitors. Second-generation EGFR
TKIs, including afatinib and dacomitinib, are ErbB family blockers
(5). Third-generation EGFR TKIs,
such as osimertinib, almonertinib and furmonertinib, overcome the
resistance mechanisms posed by first- and second-generation
inhibitors by incorporating an acrylamide group, which alkylates
the Cys797 residue of the EGFR T790M mutation (6). However, drug resistance is inevitable,
even with the use of third-generation TKIs. The mechanisms reported
include changes in the EGFR signaling pathway, abnormal activation
of bypass and downstream signaling pathways and histological
transformation (7,8). Efforts are ongoing to clarify their
potential targetability; however, these strategies are still mostly
in the research phase. To the best of our knowledge, the present
report is the first to describe a case in which afatinib therapy
could overcome multiple EGFR mutations-mediated resistance
to a third-generation TKI (almonertinib).
Case report
Patient
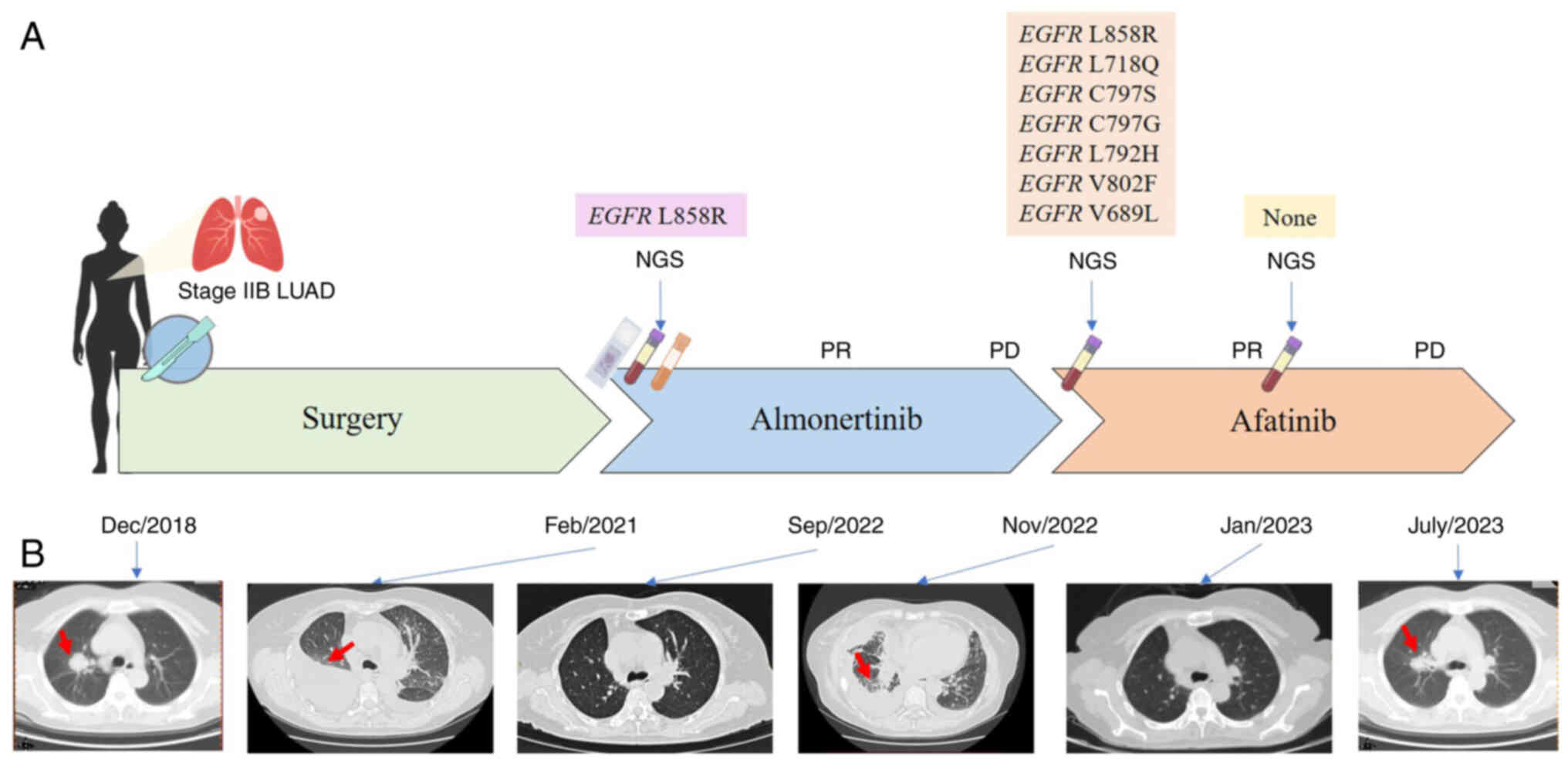
A 57-year-old Chinese female patient was referred to
Shenzhen People's Hospital (Shenzhen, China) due to the
identification of lung nodules in a routine physical examination in
December 2018. Positron emission tomography and CT revealed a
2.7×2.5×2.8-cm density mass in the upper lobe of the right lung
(Fig. 1A and B). The patient had no
underlying medical conditions. Subsequently, thoracoscopic radical
surgery was performed for right upper lung cancer, along with a
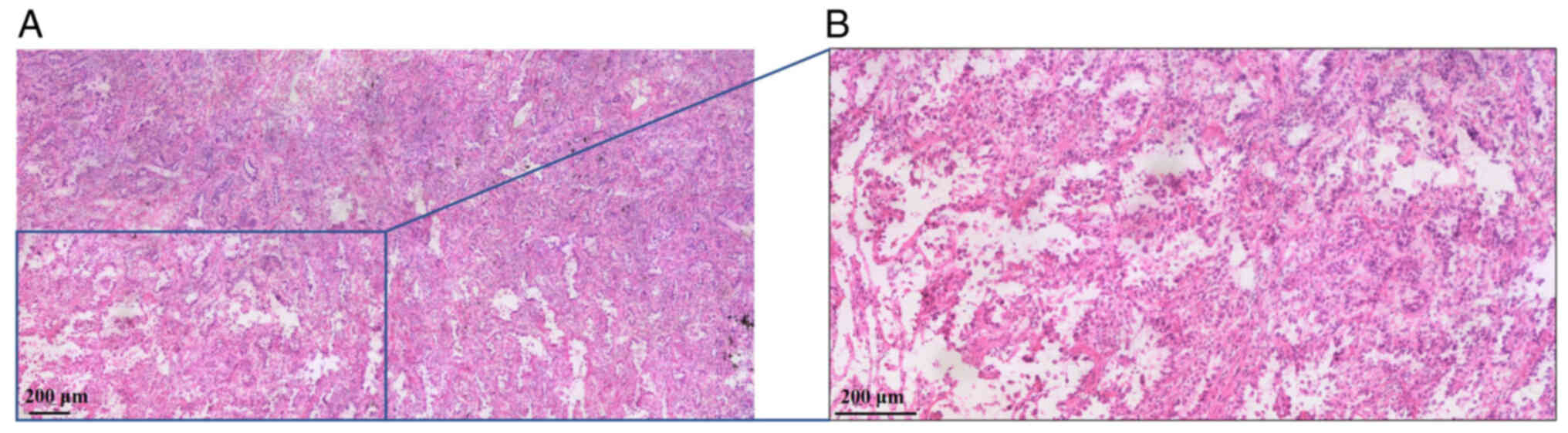
cauterization procedure for right pleural adhesion. Postoperative
pathology indicated invasive adenocarcinoma measuring 2.7×2.5×2.8
cm in diameter, which was mainly composed of papillary type
(Fig. 2). The patient was diagnosed
with stage IIB right upper lung infiltrating adenocarcinoma
(T1cN1M0) according to the 8th edition of TNM Staging System
(9). As the pathology of the
patient was clear and no residual lesions were found, and there
were no high-risk characteristics, no adjuvant treatment was
administered.
In February 2021, chest CT revealed multiple
metastases in both lungs with multiple lymph node metastases in the
mediastinum and both lung roots (Fig.
1B). Whole-body emission CT revealed multiple bone metastases
in the thoracic vertebrae, bilateral ribs, humerus and scapula.
Subsequent targeted next-generation sequencing (NGS) analysis of
425 cancer-related genes (Geneseeq122 Technology Inc.) identified
EGFR L858R, which had a mutation allelic frequency (MAF) of
17.3% in the plasma, 34.1% in the pleural fluid and 27.8% in the
tumor tissue of the upper lobe of the right lung (Table I). Subsequently, the patient was
treated with oral almonertinib (110 mg per day) in March 2021,
which is a third-generation EGFR TKI. The patient initially
achieved a partial response (PR) that was maintained for 20 months
(Fig. 1B).
 | Table I.Genetic alterations identified
through targeted next-generation sequencing in the primary tumor of
the upper lobe of the right lung, pleural fluid and serial plasma
circulating tumor DNA. |
Table I.
Genetic alterations identified
through targeted next-generation sequencing in the primary tumor of
the upper lobe of the right lung, pleural fluid and serial plasma
circulating tumor DNA.
|
|
| Baseline | Almonertinib
treatment (20 months later) | Afatinib treatment
(2 months later) |
|---|
|
|
|
|
|
|
|---|
| Gene | Alteration | FFPE, % | Pleural fluid,
% | Plasma, % | Plasma, % | Plasma, % |
|---|
| EGFR | L858R | 17.3 | 34.1 | 27.8 | 27.62 | - |
| EGFR | L718Q | - | - | - | 8.30 | - |
| EGFR | C797S | - | - | - | 6.55 | - |
| EGFR | C797G | - | - | - | 0.56 | - |
| EGFR | L792H | - | - | - | 0.36 | - |
| EGFR | V802F | - | - | - | 1.13 | - |
| EGFR | V689L | - | - | - | 26.41 | - |
In November 2022, chest CT of the patient revealed
that the size of the mass in the upper lobe of the right lung had
increased, which indicated progressive disease (PD; Fig. 1B). The patient has several small,
spread-out metastatic lesions in both lungs, making a puncture
biopsy unsuitable. To identify a more effective therapeutic
strategy, targeted NGS was performed using the plasma sample,
revealing the presence of EGFR L858R (MAF, 27.62%),
EGFR L718Q (MAF, 8.30%), EGFR C797S (MAF, 6.55%),
EGFR C797G (MAF, 0.56%), EGFR L792H (MAF, 0.36%),
EGFR V802F (MAF, 1.13%) and EGFR V689L (MAF, 26.41%;
Table I). The patient was then
switched to oral afatinib (40 mg per day), a second-generation EGFR
TKI, and achieved an initial PR, as indicated by chest CT 2 months
later, which revealed marked shrinkage of the lung lesions
(Fig. 1B).
In January 2023, follow-up genomic testing revealed
that all the genetic alterations of the tumor had disappeared.
However, in July 2023, the size of the mass in the upper lobe of
the right lung increased, which indicated PD (Fig. 1B). The patient reached a
progression-free survival (PFS) of 9 months. During this period,
the patient did not receive any other treatment. As the patient
refused chemotherapy, immunotherapy was planned for the patient.
However, due to economical difficulty, the follow-up treatment was
terminated.
Methods
Hematoxylin and eosin staining
Tissue samples were sliced and submerged in 10%
neutral buffered formalin. The fixation occurred at 25°C for 3–6 h.
After fixation, the tissue samples were dehydrated, embedded in
paraffin, and tissue sections were cut at 4 µm. The paraffin
sections were immersed in xylene for 10 min, xylene changed and
soaked for another 10 min to dissolve the wax. Samples were
rehydrated using a gradient of ethanol concentrations (anhydrous
ethanol, 95%, 85%; 70% ethanol), each immersion lasting 5 min. The
hydrated tissue sections were cleaned by immersion in PBS solution,
each immersion lasting 5 min, repeated three times. Subsequently,
tissues were stained in hematoxylin at room temperature for 10 min.
Afterwards, excess hematoxylin stain was rinsed with distilled
water. The samples were differentiated using 1% hydrochloric acid
in ethanol, and the sections were rinsed thoroughly with distilled
water. The bluing process was completed using 0.6% ammonia water,
rinsing with clean water, and then rinsing the sections thoroughly
with distilled water. The sections were immersed in eosin dye at
room temperature for 1 min. The sections were thoroughly rinsed
with distilled water, then dehydrated using a gradient of 80%
ethanol for 5 sec, 95% ethanol for 2 min and anhydrous ethanol for
2 min. The dehydrated tissue sections were immersed in in xylene
twice, each immersion lasting 4 min. Finally, tissue sections were
dried and sealed with neutral resin. Images were captured with the
Olympus BX43 light microscope (Olympus Corporation).
DNA extraction and targeted
enrichment
FFPE genomic DNA was purified using the QIAamp DNA
FFPE Tissue Kit (Qiagen). cfDNA was extracted using the NucleoSpin
Plasma XS kit (Macherey Nagel) with optimized manufacturer's
protocols. Fresh tissue DNA and whole blood DNA were extracted
using the DNeasy Blood & Tissue kit (Qiagen GmbH) according to
the manufacturer's protocols. The DNA was quantified using the
dsDNA HS Assay Kit on a Qubit Fluorometer (Thermo Fisher
Scientific, Inc.). Sequencing libraries were prepared using the
KAPA Hyper Prep Kit (KAPA Biosystems; Roche Diagnostics), as
described previously (10). Indexed
DNA libraries were pooled together for probe-based hybridization
(11) capture of the targeted gene
regions covering 437 cancer-related genes. The finial libraries
were quantified by qPCR using the KAPA Library Quantification Kit
(KAPA Biosystems; Roche Diagnostics) for sequencing.
Sequencing data processing
Paired-end sequencing of the 300 bp amplicon was
performed using the Illumina HiSeq4000 platform (Illumina, Inc.),
followed by data analysis as previously described (12). The mean coverage depth was >100×
for the whole blood control samples, and >300× for tumor tissues
after removing PCR duplicates. For cfDNA samples, the original
targeted sequencing depth was >3,000×. The final concentration
of the library was determined based on the sample throughput and
sample quality. In brief, sequencing data were analyzed by
Trimmomatic (13) to remove
low-quality (quality <15) or N bases, and were then mapped to
the human reference genome, hg19, using the Burrows-Wheeler Aligner
(BWA-mem, v0.7.12; http://github.com/lh3/bwa/tree/master/bwakit). PCR
duplicates were removed by Picard (available at http://broadinstitute.github.io/picard/). The Genome
Analysis Toolkit (GATK 3.4.0; http://software.broadinstitute.org/gatk/) was used to
perform local realignments around indels and base quality
reassurance. Gene fusions were identified by FACTERA (14). Somatic SNPs and indels were analyzed
by VarScan2 (15) and Mutect2, with
the mutant allele frequency cutoff at 2% for tissue samples and a
minimum of three unique mutant reads. Common SNPs were excluded
using dbSNP (v137) if they were present in >1% population
frequency in the 1000 Genomes Project or the Exome Aggregation
Consortium (ExAC) 65,000 exomes database. The resulting mutation
list was further filtered by an in-house list of recurrent
artifacts based on a normal pool of whole blood samples.
Discussion
Almonertinib is a third-generation EGFR TKI with
demonstrated activity against EGFR-sensitizing and T790M
mutations. Its design is a modified version of osimertinib, in
which the methyl group on the indole ring is replaced with a
cyclopropyl group. This alteration enhances its ability to bind
with EGFR T790M and improves its transport through the
blood-brain barrier (8,16). Despite its efficacy, acquired
resistance to almonertinib inevitably develops. A previous study
reported that the resistance patterns to almonertinib are similar
to those of osimertinib (17). The
resistance mechanisms include the following: Loss of the T790M
mutation, maintenance of the T790M mutation, EGFR mutations
(C797S, G724S and L718Q), activation of alternative pathways and
histological transformation (6,17,18).
In the present case, most acquired resistance mutations to
almonertinib were also reported, including L718Q, C797S/G, L792H
and V802F. Mutation at EGFR L718 has been identified as a
factor contributing to resistance against osimertinib, both in
vitro and in vivo. The L718 mutation may mediate drug
resistance by causing a substitution at the L718 residue in the
ATP-binding site of the EGFR kinase domain. This alteration can
lead to steric hindrance that can obstruct osimertinib-EGFR binding
(19). The EGFR C797S mutation
involves a change from cysteine to serine at codon 797 within the
ATP-binding site. This alteration leads to the loss of the covalent
bond between osimertinib and the mutant EGFR (20). Mutations in L792 can create steric
interference with a methoxy group on the phenyl ring of
osimertinib, disrupting its ability to bind to the kinase domain
(21). The V802F mutation can
displace the first helix adjacent to the hinge region in comparison
with the wild-type EGFR, leading to minimal effects on
osimertinib binding (22).
Furthermore, EGFR V689L in exon 18 was also observed in the
present report, which has been reported to likely be associated
with EGFR TKI sensitivity (23).
However, its role in mediating third-generation TKI resistance has
not been established yet.
Studies have reported cases of patients who received
afatinib after progression on third-generation TKIs (osimertinib or
almonertinib; Table II). Among 28
patients, most of them had EGFR 19del or L858R mutations,
several accompanied by T790M mutation, prior to receiving
third-generation TKI treatment (24–35).
Osimertinib was most often given as second-line therapy. The median
PFS was 8 months. After progression, 13 patients exhibited
resistance mechanisms dependent on the ErbB family, including
EGFR L718Q and L718V, EGFR R776H, and EGFR
C797S mutations, as well as amplification of Erb-B2 receptor
tyrosine kinase 2 (ERBB2). Certain patients had other mutations,
such as EGFR G724S, EGFR P794L, ERBB2
amplification, MAP2K1 K57T and AKT2 amplification. Afatinib
was most commonly given as monotherapy in the fourth-line treatment
setting for 13 patients. It was used in combination with cetuximab
for 11 patients, with bevacizumab for 2 patients and with apatinib
for 2 patients. Excluding patients whose PFS was not completely
recorded, the remaining patients had a median PFS of 3.8 months
(24–35). The patient in the present report had
multiple EGFR mutations and benefited from afatinib after
almonertinib failed. Afatinib is designed to be a multitarget
inhibitor that can irreversibly bind to the ATP-binding site of the
EGFR tyrosine kinase domain, specifically at Cys797 of EGFR, Cys805
of HER2 and Cys803 of HER4. This binding effectively blocks the
downstream transduction signaling pathways (36). A preclinical study reported that
19del, L858R and L718Q mutations were highly sensitive to
second-generation TKIs, such as afatinib (31). This has been further validated in
other clinical cases, with patients carrying EGFR L858R/L718Q
mutations experiencing a PFS of 4–6 months under these treatments
(31). The patient in the present
report had multiple acquired EGFR mutations, including
L718Q, C797S/G, L792H, V802F and V689L, and showed a sustained
response to afatinib monotherapy. This is in line with previous
findings that have indicated that afatinib can be effective in
patients with uncommon EGFR mutations (37). Therefore, afatinib could be a
promising option following third-generation EGFR TKI treatment in
these patients.
 | Table II.Literature review of afatinib
treatment after progression on third-generation EGFR TKIs in
patients with EGFR-mutated non-small cell lung cancer. |
Table II.
Literature review of afatinib
treatment after progression on third-generation EGFR TKIs in
patients with EGFR-mutated non-small cell lung cancer.
| First author/s,
year | Patient no. | Age, years | Sex | Smoker | EGFR mutation | Third-generation
TKI treatment | Line of
treatment | PFS, months | Resistance
mechanism | Afatinib
treatment | Line of
treatment | PFS, months | (Refs.) |
|---|
| Van Kempen et
al, 2018 | 1 | 36 | F | No | Exon 19
deletion | Osimertinib | 4 | 19 | EGFR P794L | Afatinib | 5 | >3.8 | (24) |
| Fang et al,
2020 | 2 | 55 | M | Yes | Exon 19
deletion | Osimertinib | 3 | 3 | EGFR G724S | Afatinib | 4 | >3.8 | (25) |
| Liu et al,
2019 | 3 | 65 | F | NA | L858R, T790M | Osimertinib | 2 | 9 | EGFR L718Q | Afatinib | 3 | 4 | (26) |
| Fang et al,
2019 | 4 | 45 | M | Yes | L858R, T790M | Osimertinib | 2 | 8 | EGFR L718V | Afatinib | 3 | >6 | (27) |
| Yang et al,
2020 | 5 | 69 | F | No | L858R, T790M | Osimertinib | 3 | 14 | EGFR L718Q | Afatinib | 4 | 4 | (28) |
| Minari et
al, 2021 | 6 | 51 | M | No | Exon 19 deletion,
T790M | Osimertinib | 2 | 8 | EGFR G724S | Afatinib | 5 | >2 | (29) |
| Zhao et al,
2021 | 7 | 69 | M | No | Exon 19 deletion,
T790M | Osimertinib | 5 | 16 | EGFR C797S | Afatinib +
apatinib | 7 | 10 | (30) |
| Zhang et al,
2022 | 8 | 72 | M | Yes | L858R | Almonertinib +
bisphosphonates | 1 | 12 | EGFR L718Q | Afatinib +
cetuximab | 3 | 7 | (31) |
| Song et al,
2022 | 9 | 55 | F | No | L858R, T790M | Osimertinib | 2 | 10 | EGFR L718V | Afatinib +
apatinib | 4 | >18 | (32) |
| Nozaki et
al, 2022 | 10 | 68 | M | Yes | L858R | Osimertinib | 1 | 2 | High TMB | Afatinib | 2 | 5 | (33) |
| Aredo et al,
2022 | 11 | NA | NA | No | L858R | Osimertinib | 1 | 6.6 | EGFR L718Q, EGFR
L718V | Afatinib | 2 | 2.5 | (34) |
|
| 12 | NA | NA | No | L861Q | Osimertinib | 1 | 8.3 | Not tested | Afatinib | 2 | 19.6 |
|
|
| 13 | NA | NA | No | Exon 19
deletion | Osimertinib +
bevacizumab | 5 | 8 | ERBB2 amp | Afatinib | 7 | 1.8 |
|
|
| 14 | NA | NA | No | L858R | Osimertinib | 1 | 1.7 | None detected | Afatinib +
cetuximab | 3 | 1.3 |
|
|
| 15 | NA | NA | No | L858R | Osimertinib | 2 | 1.4 | Not tested | Afatinib +
cetuximab | 4 | 2 |
|
|
| 16 | NA | NA | No | L858R | Osimertinib | 2 | 36.4 | EGFR R776H | Afatinib +
cetuximab | 4 | 2.5 |
|
|
| 17 | NA | NA | No | Dupl. exons 18 | Osimertinib | 4 | 2.9 | Not tested | Afatinib +
cetuximab | 6 | 1.3 |
|
|
| 18 | NA | NA | No | Exon 19
deletion | Osimertinib +
bevacizumab | 2 | 4.7 | MAP2K1 K57T | Afatinib +
cetuximab | 4 | 1.2 |
|
|
| 19 | NA | NA | No | Exon 19
deletion | Osimertinib | 2 | 2 | Not tested | Afatinib +
cetuximab | 4 | 1.4 |
|
|
| 20 | NA | NA | No | Exon 19
deletion | Osimertinib +
bevacizumab | 2 | 1.7 | Not tested | Afatinib +
cetuximab | 5 | 1.9 |
|
|
| 21 | NA | NA | No | L858R | Osimertinib | 5 | 7.9 | EGFR C797S | Afatinib +
cetuximab | 7 | 3.8 |
|
|
| 22 | NA | NA | No | L858R | Osimertinib | 2 | 7 | AKT2 amp | Afatinib +
cetuximab | 5 | 4.3 |
|
|
| 23 | NA | NA | No | L858R | Osimertinib | 2 | 19.4 | Not tested | Afatinib +
cetuximab | 4 | 5.6 |
|
|
| 24 | NA | NA | No | L858R | Osimertinib | 2 | 2.6 | Not tested | Afatinib +
bevacizumab | 4 | 2.9 |
|
|
| 25 | NA | NA | No | L858R | Osimertinib | 2 | 8.8 | Not tested | Afatinib +
bevacizumab | 5 | 5.5 |
|
| Sanchis-Borja et
al, 2024 | 26 | 61 | M | No | L858R, T790M | Osimertinib | 2 | 38.6 | EGFR L718Q | Afatinib | 3 | 7.2 | (35) |
|
| 27 | 58 | F | No | L858R, T790M,
G719X | Osimertinib | 3 | 41.5 | EGFR L718Q | Afatinib | 4 | 6.1 |
|
|
| 28 | 65 | M | Yes | Exon 19 deletion,
T790M | Osimertinib | 2 | 9 | EGFR L718Q | Afatinib | 4 | 1.9 |
|
It is important to acknowledge the limitations of
the single-case presentation of the present report. The
effectiveness and side effects of almonertinib and afatinib need to
be further assessed in larger cohorts. Moreover, the histological
test results during the afatinib treatment are missing as only
imaging and genetic tests were performed. In the present case, the
EGFR V689L mutation may have served as a potential
resistance mechanism to almonertinib; however, further preclinical
studies and clinical evidence are required to support this.
In conclusion, to the best of our knowledge, the
present report describes the first case of successful treatment of
NSCLC with multiple acquired EGFR mutations using afatinib
after the patient developed resistance to almonertinib. The patient
received afatinib treatment for ~9 months and achieved a sustained
PR without any significant side effects. The present case suggests
that afatinib may overcome almonertinib resistance and could serve
as a promising treatment option for similar patients. However,
further investigation is required to determine any additional
resistance mechanisms related to EGFR TKIs.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The sequencing results and raw data generated in the
present study may be found in the BioProject database under
accession numbers PRJNA1174043 or at the following URLs: https://www.ncbi.nlm.nih.gov/sra/PRJNA1174043.
Authors' contributions
FY designed this study and collected the data for
this case report. JL conceived the present study, analyzed and
interpreted of data. MX acquired data. BP made substantial
contributions to conception and design. FY and BP confirm the
authenticity of all the raw data. All authors have read and
approved the final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Written informed consent to publish the clinical
details and images were obtained from the patient.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD, Fuchs HE and Jemal
A: Cancer statistics, 2022. CA Cancer J Clin. 72:7–33. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen P, Liu Y, Wen Y and Zhou C: Non-small
cell lung cancer in China. Cancer Commun (Lond). 42:937–970. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Midha A, Dearden S and McCormack R: EGFR
mutation incidence in non-small-cell lung cancer of adenocarcinoma
histology: A systematic review and global map by ethnicity
(mutMapII). Am J Cancer Res. 5:2892–2911. 2015.PubMed/NCBI
|
|
4
|
Xue J, Li B, Wang Y, Huang Z, Liu X, Guo
C, Zheng Z, Liang N, Le X and Li S: Efficacy and safety of
epidermal growth factor receptor (EGFR)-tyrosine kinase inhibitor
combination therapy as first-line treatment for patients with
advanced EGFR-mutated, non-small cell lung cancer: A systematic
review and bayesian network meta-analysis. Cancers (Basel).
14:48942022. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li D, Ambrogio L, Shimamura T, Kubo S,
Takahashi M, Chirieac LR, Padera RF, Shapiro GI, Baum A,
Himmelsbach F, et al: BIBW2992, an irreversible EGFR/HER2 inhibitor
highly effective in preclinical lung cancer models. Oncogene.
27:4702–4711. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chhouri H, Alexandre D and Grumolato L:
Mechanisms of acquired resistance and tolerance to EGFR targeted
therapy in non-small cell lung cancer. Cancers (Basel). 15:5042023.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mu Y, Hao X, Xing P, Hu X, Wang Y, Li T,
Zhang J, Xu Z and Li J: Acquired resistance to osimertinib in
patients with non-small-cell lung cancer: mechanisms and clinical
outcomes. J Cancer Res Clin Oncol. 146:2427–2433. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang JCH, Camidge DR, Yang CT, Zhou J, Guo
R, Chiu CH, Chang GC, Shiah HS, Chen Y, Wang CC, et al: Safety,
efficacy, and pharmacokinetics of almonertinib (HS-10296) in
pretreated patients with EGFR-mutated advanced NSCLC: A
multicenter, open-label, phase 1 trial. J Thorac Oncol.
15:1907–1918. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hwang JK, Page BJ, Flynn D, Passmore L,
McCaul E, Brady J, Yang IA, Marshall H, Windsor M, Bowman RV, et
al: Validation of the eighth edition TNM lung cancer staging
system. J Thorac Oncol. 15:649–654. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shu Y, Wu X, Tong X, Wang X, Chang Z, Mao
Y, Chen X, Sun J, Wang Z, Hong Z, et al: Circulating tumor DNA
mutation profiling by targeted next generation sequencing provides
guidance for personalized treatments in multiple cancer types. Sci
Rep. 7:5832017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hockenhull K, Ortega-Franco A and Califano
R: Pembrolizumab plus platinum-based chemotherapy for squamous
non-small cell lung cancer: The new kid on the block. Transl Lung
Cancer Res. 10:3850–3854. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yang Z, Yang N, Ou Q, Xiang Y, Jiang T, Wu
X, Bao H, Tong X, Wang X, Shao YW, et al: Investigating novel
resistance mechanisms to third-generation EGFR tyrosine kinase
inhibitor osimertinib in non-small cell lung cancer patients. Clin
Cancer Res. 24:3097–3107. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bolger AM, Lohse M and Usadel B:
Trimmomatic: A flexible trimmer for Illumina sequence data.
Bioinformatics. 30:2114–2120. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Newman AM, Bratman SV, Stehr H, Lee LJ,
Liu CL, Diehn M and Alizadeh AA: FACTERA: A practical method for
the discovery of genomic rearrangements at breakpoint resolution.
Bioinformatics. 30:3390–3393. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Koboldt DC, Zhang Q, Larson DE, Shen D,
McLellan MD, Lin L, Miller CA, Mardis ER, Ding L and Wilson RK:
VarScan 2: Somatic mutation and copy number alteration discovery in
cancer by exome sequencing. Genome Res. 22:568–576. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nagasaka M, Zhu VW, Lim SM, Greco M, Wu F
and Ou SI: Beyond osimertinib: The development of third-generation
EGFR tyrosine kinase inhibitors for advanced EGFR+ NSCLC. J Thorac
Oncol. 16:740–763. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tian M, Lu Z, Chen S, Lu G, Bu F, Deng W
and Ding R: 1014P Resistance landscape to almonertinib in
EGFR-mutated NSCLC. Ann Oncol. 33 (Suppl 7):S10172022. View Article : Google Scholar
|
|
18
|
Kwon Y, Kim M, Jung HS, Kim Y and Jeoung
D: Targeting autophagy for overcoming resistance to anti-EGFR
treatments. Cancers (Basel). 11:13742019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bersanelli M, Minari R, Bordi P, Gnetti L,
Bozzetti C, Squadrilli A, Lagrasta CA, Bottarelli L, Osipova G,
Capelletto E, et al: L718Q Mutation as new mechanism of acquired
resistance to AZD9291 in EGFR-mutated NSCLC. J Thorac Oncol.
11:e121–e123. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Leonetti A, Sharma S, Minari R, Perego P,
Giovannetti E and Tiseo M: Resistance mechanisms to osimertinib in
EGFR-mutated non-small cell lung cancer. Br J Cancer. 121:725–737.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ou SI, Cui J, Schrock AB, Goldberg ME, Zhu
VW, Albacker L, Stephens PJ, Miller VA and Ali SM: Emergence of
novel and dominant acquired EGFR solvent-front mutations at Gly796
(G796S/R) together with C797S/R and L792F/H mutations in one EGFR
(L858R/T790M) NSCLC patient who progressed on osimertinib. Lung
Cancer. 108:228–231. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lin L, Lu Q, Cao R, Ou Q, Ma Y, Bao H, Wu
X, Shao Y, Wang Z and Shen B: Acquired rare recurrent EGFR
mutations as mechanisms of resistance to Osimertinib in lung cancer
and in silico structural modelling. Am J Cancer Res. 10:4005–4015.
2020.PubMed/NCBI
|
|
23
|
Ricciuti B, Baglivo S, De Giglio A and
Chiari R: Afatinib in the first-line treatment of patients with
non-small cell lung cancer: Clinical evidence and experience. Ther
Adv Respir Dis. 12:17534666188086592018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
van Kempen LC, Wang H, Aguirre ML, Spatz
A, Kasymjanova G, Vilacha JF, Groves MR, Agulnik J and Small D:
Afatinib in osimertinib-resistant EGFR ex19del/T790M/P794L mutated
NSCLC. J Thorac Oncol. 13:e161–e163. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fang W, Huang Y, Gan J, Zheng Q and Zhang
L: Emergence of EGFR G724S after progression on osimertinib
responded to afatinib monotherapy. J Thorac Oncol. 15:e36–e37.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu J, Jin B, Su H, Qu X and Liu Y:
Afatinib helped overcome subsequent resistance to osimertinib in a
patient with NSCLC having leptomeningeal metastasis baring acquired
EGFR L718Q mutation: A case report. BMC Cancer. 19:7022019.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fang W, Gan J, Huang Y, Zhou H and Zhang
L: Acquired EGFR L718V mutation and loss of T790M-mediated
resistance to osimertinib in a patient with NSCLC who responded to
afatinib. J Thorac Oncol. 14:e274–e275. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yang X, Huang C, Chen R and Zhao J:
Resolving resistance to osimertinib therapy with afatinib in an
NSCLC patient with EGFR L718Q mutation. Clin Lung Cancer.
21:e258–e260. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Minari R, Leonetti A, Gnetti L, Zielli T,
Ventura L, Bottarelli L, Lagrasta C, La Monica S, Petronini PG,
Alfieri R and Tiseo M: Afatinib therapy in case of EGFR G724S
emergence as resistance mechanism to osimertinib. Anticancer Drugs.
32:758–762. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhao Y, Chen Y, Huang H, Li X, Shao L and
Ding H: Significant benefits of afatinib and apatinib in a
refractory advanced NSCLC patient resistant to osimertinib: A case
report. Onco Targets Ther. 14:3063–3067. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang G, Yan B, Guo Y, Yang H, Li X and Li
J: Case report: A patient with the rare third-generation
TKI-resistant mutation EGFR L718Q who responded to afatinib plus
cetuximab combination therapy. Front Oncol. 12:9956242022.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Song Z, Ren G, Wang X, Du H, Sun Y and Hu
L: Durable clinical benefit from afatinib in a lung adenocarcinoma
patient with acquired EGFR L718V mutation-mediated resistance
towards osimertinib: A case report and literature review. Ann
Palliat Med. 11:1126–1134. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Nozaki K, Watanabe S, Nishio K, Sakai K
and Kikuchi T: Effectiveness of afatinib in an NSCLC patient with
EGFR mutation and early progression to osimertinib: a case report.
Transl Cancer Res. 11:295–298. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Aredo JV, Wakelee HA, Neal JW and Padda
SK: Afatinib after progression on osimertinib in EGFR-mutated
non-small cell lung cancer. Cancer Treat Res Commun. 30:1004972022.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sanchis-Borja M, Guisier F, Swalduz A,
Curcio H, Basse V, Maritaz C, Chouaid C and Auliac JB:
Characterization of patients with EGFR mutation-positive NSCLC
following emergence of the osimertinib resistance mutations, L718Q
or G724S: A multicenter retrospective observational study in
France. Onco Targets Ther. 17:439–448. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Karachaliou N, Fernandez-Bruno M, Bracht
JWP and Rosell R: EGFR first- and second-generation TKIs-there is
still place for them in EGFR-mutant NSCLC patients. Transl Cancer
Res. 8 (Suppl 1):S23–S47. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yang JCH, Schuler M, Popat S, Miura S,
Heeke S, Park K, Märten A and Kim ES: Afatinib for the treatment of
NSCLC harboring uncommon EGFR mutations: A database of 693 cases. J
Thorac Oncol. 15:803–815. 2020. View Article : Google Scholar : PubMed/NCBI
|