Introduction
Thyroid carcinoma (THCA) is a cancerous tumor that
develops from the cells of the thyroid gland, specifically the
follicular or parafollicular epithelial cells. THCA is the most
prevalent form of cancer detected in the region encompassing the
head and neck (1). There has been a
notable increase in the prevalence of differentiated THCA worldwide
(2). The differentiated subtype of
THCA accounts for >90% of all cases and exhibits a favorable
prognosis, with a survival rate of >95% over a span of 10 years
(3). However, despite the generally
optimistic perspective commonly observed among the majority of
patients with differentiated THCA, there is a subgroup
characterized by tumor heterogeneity and harboring more aggressive
variants. Consequently, this has marked implications for subsequent
treatment strategies and overall rates of survival (4,5). The
limited exploration of potential mechanisms underlying the
invasiveness of THCA cases, particularly in terms of gene-specific
modulation of immune regulation, contributes to insufficient or
unsatisfactory management of the disease (6).
There has been notable progress in comprehending the
potential molecular mechanisms through which THCA may act as a
carcinogenic agent (7). Initiation
of THCA involves critical roles served by gene mutations, such as
PTEN, PIK3CA, RAS, TP53, BRAF and β-catenin mutations, in addition
to epigenetic alterations such as abnormal gene methylation
(8). The development of THCA is
characterized by the abnormal activation of different signaling
pathways at the molecular level (9). As the molecular pathogenesis of THCA
continues to be extensively studied, the integration of targeted
therapy based on biological factors is being slowly incorporated
into the realm of clinical practice (10). There is an immediate requirement for
novel biomarkers to assist in the identification, and in the
prediction of early detection and efficient clinical treatment
alternatives (11).
Protein phosphatase 1 regulatory subunit 3G
(PPP1R3G) is expressed at high levels in THCA. The PPP1R3G protein
is considered to possess glycogen binding activity and PP1 binding
activity (12); it is also
predicted to serve a role in the regulation of glycogen
biosynthetic process and act upstream of or within glucose
homeostasis (13). The PPP1R3G
protein, which is considered to be localized in the cytoplasm,
serves a positive regulatory role in glycogen synthase activity and
the biosynthesis of glycogen (14).
The diseases associated with PPP1R3G encompass myoclonic epilepsy
of Lafora and progressive myoclonus epilepsy (15). Gene Ontology (GO) annotations
pertaining to this gene involve ‘protein phosphatase binding and
glycogen binding’ (16). PPP1R3G
was also revealed to serve as an unfavorable prognostic biomarker
for lung adenocarcinoma (LUAD) and was associated with infiltration
of immune cells in the tumor microenvironment (17). The impact of this, however, remains
unclear in THCA. Therefore, the aim of the present study was to
assess the association between PPP1R3G expression and THCA
diagnosis, along with its molecular function and potential
connection with immune infiltration.
Materials and methods
Collection of data
The Cancer Genome Atlas (TCGA; portal.gdc.cancer.gov/) database was used to
obtain the gene expression data and clinical information for the
present study, encompassing 571 cases of THCA tissues and 59 cases
of adjacent non-cancerous tissues (GSE27155, GSE64912, GSE3467,
GSE163203, GSE33630, GSE153659, GSE53157, GSE165724). The TCGA data
were organized and processed using the Spliced Transcripts
Alignment to a Reference (STAR) method to extract Transcripts (Per
Kilobase of exon model) per Million mapped reads/ fragments
(TPM)-formatted information (18).
Differential mRNA expression analysis was performed using R (4.2.1;
The R Foundation), using criteria such as a minimum absolute
log2 fold-change of 1.5 and maximum adjusted P-value of
0.05. Co-expressed mRNAs with the target gene were also determined
using R language analysis. All procedures for processing data
followed the principles in the Declaration of Helsinki.
Analysis of Cox regression using both
univariate and multivariate approaches
The aim of the present study was to assess the
relationships between PPP1R3G, pathological stage,
Tumor-Node-Metastasis (TNM) stage (American Joint Committee on
Cancer/TNM Staging System, 8th Edition) (19), tumor grade and prognosis using Cox
regression analyses. Only the factors pertinent to the prognosis
were taken into consideration in the present analysis.
Additionally, the rms and survival receiver operating
characteristic (ROC) packages (pROC; version 1.18.0]) in Xiantao
Academic Online (https://www.xiantaozi.com/) were used to construct a
nomogram that accurately depicted the 2-, 5- and 10-year survival
rates of patients with THCA. Discrepancies in the overall survival
(OS) were evaluated using the Kaplan-Meier technique, accompanied
by a two-sided log-rank test. To assess the effectiveness of the
newly developed nomogram, calibration curve analysis was performed
in addition to calculating the consistency index.
Analysis of functional enrichment
The differentially expressed genes that co-expressed
with PPP1R3G were identified and were represented as a heatmap
using the DESeq2 package [1.36.0] in Xiantao Academic Online. The
analysis of functional enrichment was performed using GO and Kyoto
Encyclopedia of Genes and Genomes (KEGG) enrichment analysis, as
well as Gene Set Enrichment Analysis (GSEA). The association
between the expression levels of PPP1R3G and pathological stage, as
well as clinical parameters in patients with THCA, was assessed by
analyzing the RNA sequencing (RNA-seq) data from TCGA databases
using R. GSEA was used for gene expression profiling followed by
functional cluster analysis. Differential genes were detected using
clusterProfiler (version 4.4.4] package via Xiantao Academic
Online, with a significance threshold of normalized enrichment
score (NES) >1.5 and P<0.05 for identifying significantly
enriched signaling pathways. The results of the enrichment analysis
were visualized using the ggplot2 package [3.3.6] in Xiantao
Academic Online.
Immune cell infiltration of
single-sample (ss)GSEA
The gene expression profiles of clinical samples
from patients with THCA were analyzed to assess the levels of
specific immune cell types. To evaluate immune infiltration, ssGSEA
was employed using the ‘GSVA’ R package (version 1.46.0] in Xiantao
Academic Online. Additionally, Spearman's correlation coefficient
was used to assess the potential correlation between the expression
of PPP1R3G and infiltration of immune cells. Furthermore, the
Wilcoxon rank-sum test was used to assess possible associations
between PPP1R3G expression and infiltration levels of different
immune cell types.
Immunohistochemistry
An immunohistochemical assessment was performed on
paraffin-embedded samples collected from patients with newly
diagnosed THCA in Zibo Central Hospital (Zibo, China) between July
2019 and December 2022 to evaluate the expression levels of PPP1R3G
in these treatment-naive patients (21 males and 47 females, with an
average age of 46.2 years). Individuals who had previously
undergone chemotherapy or radiotherapy were excluded. The
immunohistochemical assessment was performed using 10%
formalin-fixed (overnight at room temperature), paraffin-embedded
sections of human thyroid tissues. The tissue sections (5 µm) were
dewaxed by heating at 55°C for a duration of 30 min and underwent
two washes with xylene, each lasting for 15 min. Subsequently, the
sections were rehydrated through a series of ethanol washes, each
taking 5 min. To facilitate antigen unmasking, the sections were
placed in an enamel cylinder containing sodium citrate (pH 6.0)
with a concentration of 10 mmol/l and heated using a gas cooker at
95°C for a period of 5 min. Following this step, endogenous
peroxidase activity was deactivated by treating the sections with
hydrogen peroxide at a concentration of 3% for half an hour. The
sections were then incubated with 4% fetal bovine serum (cat. no.
26170035; Thermo Fisher Scientific, Inc.) at 37°C for half an hour
before being exposed to specific rabbit polyclonal antibodies
targeting PPP1R3G antibody (diluted at a ratio of 1:1,000; cat. no.
NBP2-3417, Novus Biologicals, USA). This incubation process took
place overnight at a temperature of 4°C. The control tumor slides
were treated with PBS) serving as a control group The sections were
rinsed with PBS and subjected to a 30-min incubation at 37°C using
biotinylated goat anti-rabbit secondary antibody (1:1,000; cat. no.
31460; Thermo Fisher). To visualize the positive expression, the
substrate 3′3-diaminobenzidine (DAB) tetrachloride, dissolved in
heated water, was introduced. The presence of PPP1R3G was
determined based on its positive localization in cytoplasm regions.
A combination of the percentage of tumor cells exhibiting positive
staining and the intensity of staining was used, following
previously described methods (20).
A fluorescence microscope was employed for image acquisition, and
the quantitative analysis of protein expression levels was
conducted using Image-Pro Plus software 6.0 (Media Cybernetics,
Inc.). The PPP1R3G tissues were quantified based on the percentage
of positive cells that were serially counted in one microscopic
field. The cell counting was repeated in five randomly selected
microscopic fields at a magnification of ×100.
Reverse transcription-quantitative PCR
(RT-qPCR)
The total cellular RNA was extracted from 20 cases
of THCA tissues and paired para-cancer normal tissues collected
from patients with newly diagnosed THCA from Zibo Central Hospital
in July 2024 using TRIzol (Sangon Biotech Co., Ltd.). These
patients were recruited specifically for the purpose of RT-qPCR.
The PrimeScript™ RT Master Mix Kit (cat. no. RR036A; Takara Bio,
Inc.) was used to reverse-transcribe total RNA into cDNA according
to the manufacturer's instructions. The qPCR protocol included
denaturation at 94°C for 2 min, followed by cycles of 94°C for 30
sec, 54°C for 30 sec and 72°C for 35 sec (total of 30 cycles). Gene
expression analysis was performed using FastStart Universal SYBR
Master mix (Roche Diagnostics GmbH). The relative expression levels
were determined using the 2−ΔΔCq method (21) and normalized to β-actin. The primer
sequences used were as follows: β-actin forward,
5′-CTCCATCCTGGCCTCGCTGT-3′ and reverse 5′-GCTGTCACCTTCACCGTTCC-3′;
and PPP1R3G forward, 5′-GCGCTACACCTTTACCGAGT-3′ and reverse,
5′-TGGCTCTTTCTTGGCATCCC-3′.
Statistical analysis
Statistical analysis was performed using R (version
3.5.1; The R Foundation). The unpaired t-test, Welch's t-test and
the Wilcoxon rank-sum test were used to analyze differences among
the different groups. To evaluate the diagnostic value of PPP1R3G
expression, ROC curve analysis was performed. Univariate and
multivariate Cox regression analyses were used to determine the
significance of PPP1R3G as a prognostic factor in patients with
THCA. The Kaplan-Meier method and log-rank test were applied to
assess OS in both high and low expression groups of PPP1R3G,
followed by multivariate analysis using a Cox proportional hazard
regression model. Furthermore, the association between PPP1R3G
expression and clinical indicators in patients with THCA was
assessed using χ2 test or Fisher's exact test. P<0.05
was considered to indicate a statistically significant
difference.
Results
Prognosis-associated and
differentially expressed genes identified in THCA
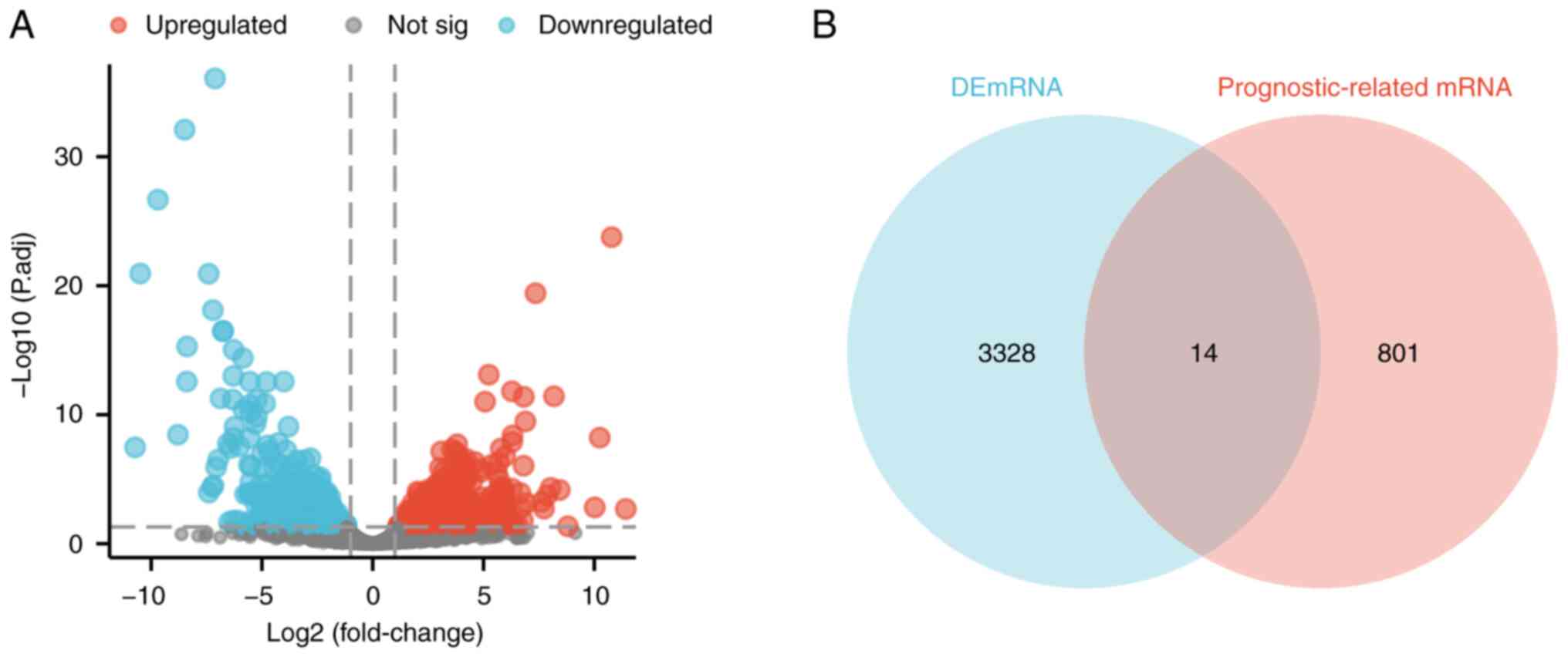
In the present study, a comprehensive analysis of
TCGA database was performed to identify genes that exhibited
differential expression in THCA (n=571) and adjacent non-cancerous
tissues (n=59) (Fig. 1A).
Additionally, by integrating prognostic data from TCGA, a set of
genes was successfully identified that was associated with the
prognosis of patients with THCA. Furthermore, the intersection
between prognosis-related genes and differentially expressed genes
in THCA was determined. Notably, overexpression of 14 specific
genes, namely PPP1R3G, SYT5, PCDHA5, OTX1, DMRT1, SMTNL2, FCRLB,
FAM155A, SPRED3, RPL13AP12, CRNDE, THBS4-AS1, AL445649.1 and
AC008875.1, was notably associated with an unfavorable prognosis in
patients diagnosed with THCA (Fig.
1B).
Level of PPP1R3G expression
demonstrates a significantly strong predictive value for patients
with THCA
To assess the potential of PPP1R3G in influencing
THCA expression patterns and identifying new therapeutic strategies
for cancer treatment, RNA-seq data were collected from 33 tumor
projects available in TCGA database. These data were then organized
and processed using the STAR method to extract TPM-formatted
information. The expression levels of PPP1R3G mRNA were evaluated
in several malignant tumor types and the findings indicated that,
compared with that in normal tissues, PPP1R3G displayed
significantly elevated expression levels in a range of cancer
types, such as THCA, lung squamous cell carcinoma, kidney clear
cell carcinoma, LUAD, glioblastoma, kidney renal papillary cell
carcinoma, pancreatic cancer, and head and neck squamous cell
carcinoma. By contrast, lower expression levels of PPP1R3G were
observed in gastric adenocarcinoma, breast infiltrating carcinoma,
bile duct carcinoma, prostate adenocarcinoma, endometrial cancer,
pheochromocytoma and paraganglioma, and renal chromophobe cell
carcinoma compared with those in both normal tissues and the
corresponding adjacent normal tissue samples (Fig. 2A and B). These findings indicate
that PPP1R3G exhibits varying expression patterns across different
types of cancer. Furthermore, analysis of TCGA database revealed a
significant increase in the expression level of PPP1R3G in THCA
tissues (n=512) compared with that in normal tissues (n=59)
(Fig. 2C). In addition, paired
samples of normal tissue adjacent to the cancerous tissue were
obtained from TCGA database for further assessment. A comparative
analysis of the expression of PPP1R3G was performed in tumor tissue
(n=59) and its corresponding normal thyroid tissues (n=59) among
patients with THCA. The results revealed a significant increase in
PPP1R3G levels in THCA tissues, compared with that in normal
tissues (Fig. 2D). Subsequently, a
prognostic evaluation for THCA was performed by analyzing the value
of PPP1R3G. The categorization of the high and low expression group
was determined based on the median level of expression. The
assessment of TCGA database revealed that patients with high levels
of PPP1R3G expression (n=256) were associated with a significantly
diminished OS rate in comparison with those with low PPP1R3G
expression (n=256) (Fig. 2E). This
was demonstrated by the results obtained from the Kaplan-Meier
curve analysis, further reinforcing its prognostic predictive
capacity for patients with THCA.
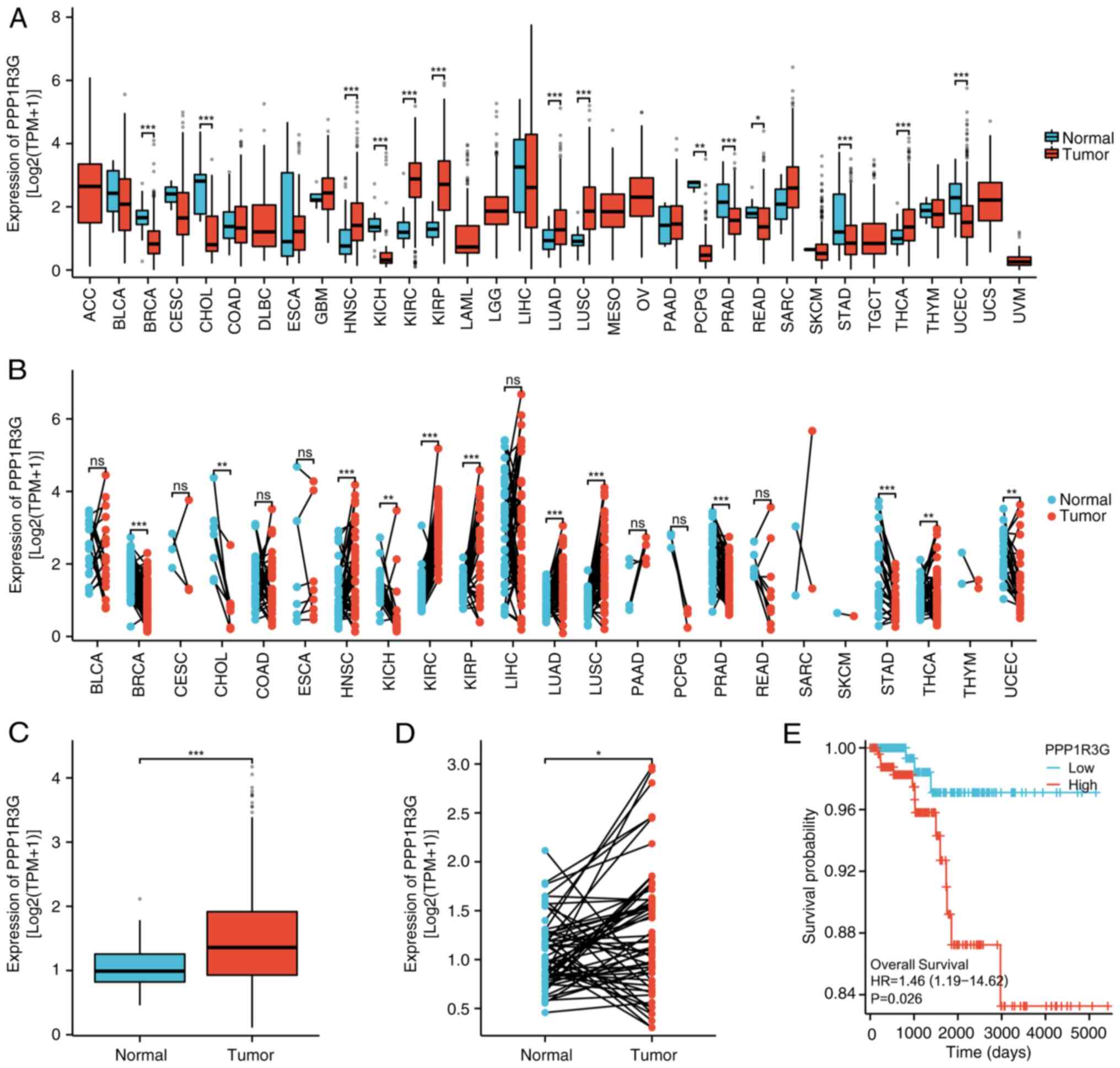 | Figure 2.Diagnostic predictive value of PPP1R3G
expression in patients with THCA. (A) Comparative analysis was
performed to evaluate the mRNA expression levels of PPP1R3G in
different types of malignancy, comparing tumor tissues with
adjacent normal tissues. (B) mRNA levels of PPP1R3G were quantified
using RNA-seq from TCGA database data obtained from tumor samples
and their corresponding normal tissues. (C) Dissimilar mRNA
expression of PPP1R3G was observed when comparing normal thyroid
tissues (n=59) with those impacted by THCA (n=512), based on the
analysis of the RNA-seq data. (D) Further assessment of PPP1R3G
mRNA expression was performed using RNA-seq data, using matched
samples of THCA (n=59) and non-cancerous thyroid tissues (n=59).
(E) Kaplan-Meier curve showing the comparison of overall survival
among different subgroups of patients with THCA based on their mRNA
expression levels of PPP1R3G. *P<0.05; **P<0.01;
***P<0.001. THCA, thyroid carcinoma; PPP1R3G, protein
phosphatase 1 regulatory subunit 3G; RNA-seq, RNA sequencing; TCGA,
The Cancer Genome Atlas; TPM, transcripts per million; HR, hazard
ratio; ns, not significant. |
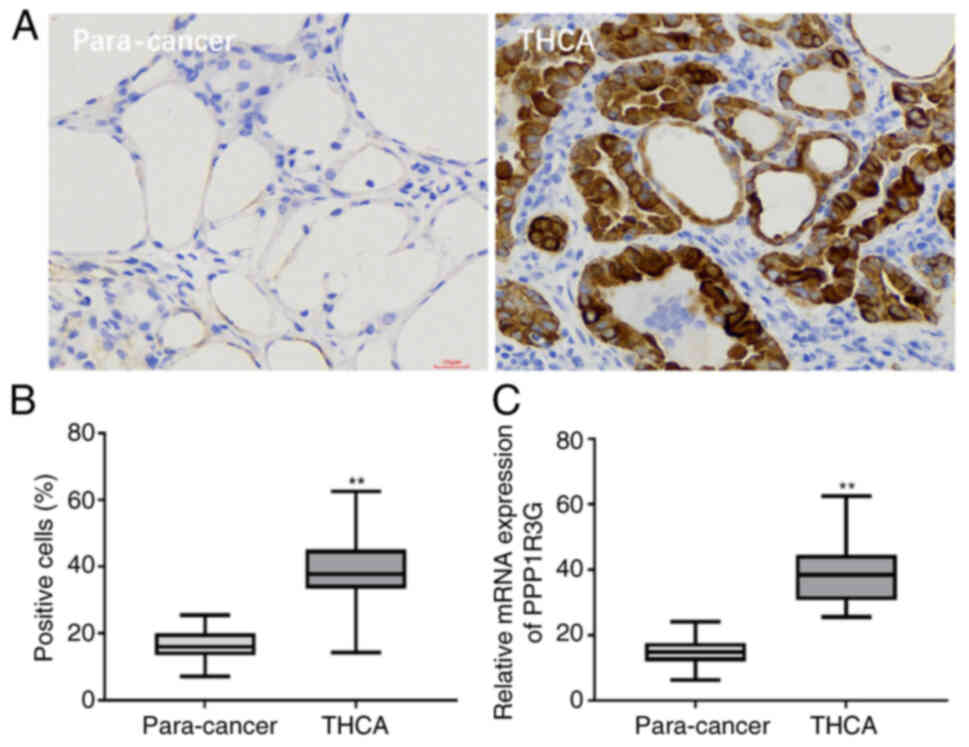
Immunohistochemistry and RT-qPCR were
performed to validate the expression of PPP1R3G in THCA
The expression of PPP1R3G in THCA and its
corresponding para-cancerous tissue was assessed using
immunohistochemistry, validating the data obtained from
bioinformatics analysis. The findings indicated a high expression
of PPP1R3G in 38/67 cases of THCA and in 19/67 cases of paired
para-cancer normal tissues (Fig.
3A). The detailed quantitative analysis of the expression
levels across all 67 cases was quantified based on the percentage
of positive cells (Fig. 3B). The
percentage of positive cells was 37.36 and 16.58 in the 67 cases of
THCA and the paired para-cancer normal tissues, respectively
[standard error of difference, 1.296; 95% confidence interval (CI),
−23.76 to −18.59]. The RT-qPCR results also demonstrated that the
RNA expression of PPP1R3G was expressed at significantly higher
levels in THCA tissues compared with those in the paired
para-cancer normal tissue (38.69 vs. 14.93%; difference between
means, 23.76±2.340; 95% CI, 19.02 to 28.50; Fig. 3C). Additionally, the prognostic
significance of clinical indicators was analyzed. The results
demonstrated a significant association between PPP1R3G expression
and clinical indicators in patients with THCA, including
pathological N stage, pathological stage, histological type and
extrathyroidal extension. However, no association was demonstrated
between PPP1R3G expression and pathological T stage or residual
tumor (Table I).
 | Table I.Association between PP1R3G and
clinical indicators of patients with thyroid carcinoma. |
Table I.
Association between PP1R3G and
clinical indicators of patients with thyroid carcinoma.
| Characteristic | Low expression of
PPP1R3G (n=29) | High expression of
PPP1R3G (n=38) | P-value |
|---|
| Pathological T
stage |
|
| 0.154 |
|
T1-T2 | 14 (20.9%) | 17 (25.3%) |
|
|
T3-T4 | 15 (22.4%) | 21 (31.3%) |
|
| Pathological N
stage |
|
| <0.001 |
| N0 | 16 (24.9%) | 14 (20.9%) |
|
| N1 | 13 (19.4%) | 24 (35.8%) |
|
| Pathological
stage |
|
| <0.001 |
| I–II | 18 (26.9%) | 10 (14.9%) |
|
|
III–IV | 11 (16.4%) | 28 (41.8%) |
|
| Histological
type |
|
| <0.001 |
|
Classical | 19 (28.4%) | 30 (44.8%) |
|
|
Follicular&Other&Tall
Cell | 10 (14.9%) | 8 (11.9%) |
|
| Residual tumor |
|
| 0.634 |
| R0 | 26 (38.8%) | 33 (49.3%) |
|
|
R1-R2 | 3 (4.5%) | 5 (7.5%) |
|
| Extrathyroidal
extension |
|
| 0.011 |
|
Yes | 6 (9.0%) | 13 (19.4%) |
|
| No | 23 (34.3%) | 25 (37.3%) |
|
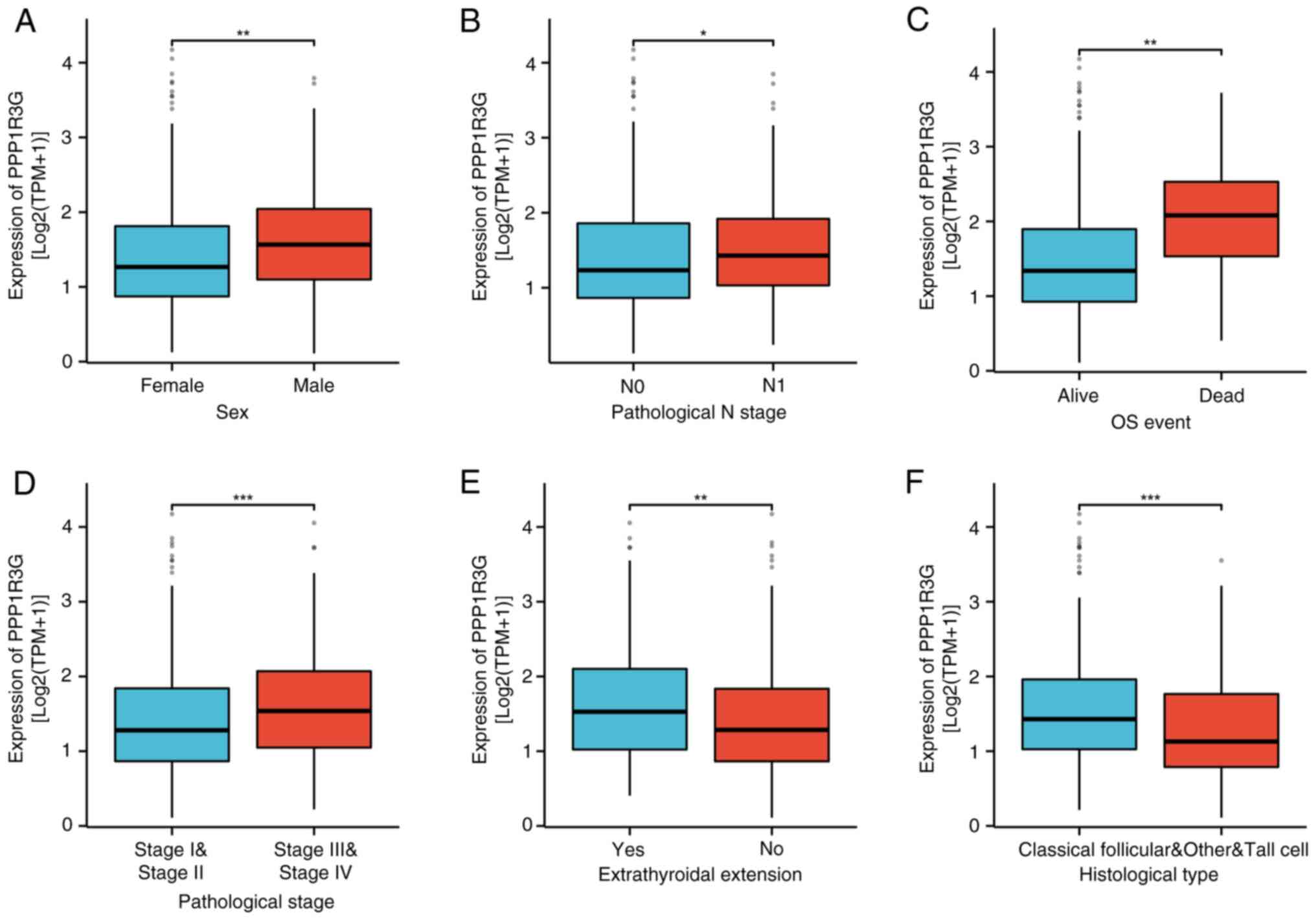
PPP1R3G is significantly associated
with histological grade, tumor location and TNM stage in THCA
An assessment of TCGA database and an analysis of
both the RNA-seq data and clinical information from TCGA THCA
project was performed. The results demonstrated that there was a
significant increase in PPP1R3G expression among male patients
(n=140) compared with female patients (n=375) (Fig. 4A). Additionally, the N1 stage group
(n=231) exhibited a significant increase compared with the N0 stage
group (n=234) (Fig. 4B), as well as
the OS event dead group (n=16) compared with the OS event alive
group (n=499; Fig. 4C) and the
stage III–IV group (n=171) compared with the stage I–II group
(n=342; Fig. 4D). Moreover, a
significantly higher expression level was observed in the
extra-thyroid extension group (n=154) compared with the
non-extension group (n=343) (Fig.
4E). Lastly, when compared with the non-classical
(Follicular&Other&Tall Cell) group (n=148), the classical
group (n=367) also exhibited a significantly elevated expression
level of PPP1R3G (Fig. 4F).
Furthermore, the findings demonstrated significant associations
between the expression of PPP1R3G and several clinical indicators
in patients with THCA, including pathological N stage, pathological
stage, sex, OS event, primary neoplasm focus type, histological
type, extrathyroidal extension and neoplasm location (Table II).
 | Table II.Association between PPP1R3G and
clinical indicators in thyroid carcinoma, assessed using The Cancer
Genome Atlas. |
Table II.
Association between PPP1R3G and
clinical indicators in thyroid carcinoma, assessed using The Cancer
Genome Atlas.
|
Characteristics | Low expression of
PPP1R3G | High expression of
PPP1R3G | P-value |
|---|
| Pathological T
stage |
|
| 0.132 |
| T1-T2
(n=313) | 165 (32.4) | 147 (28.8) |
|
| T3-T4
(n=198) | 90 (17.6) | 108 (21.2) |
|
| Pathological N
stage |
|
| 0.016 |
| N0
(n=229) | 127 (27.5) | 102 (22.1) |
|
| N1
(n=233) | 103 (22.3) | 130 (28.1) |
|
| Pathological
stage |
|
| <0.001 |
| I–II
(n=340) | 188 (36.9) | 152 (29.8) |
|
| III–IV
(n=170) | 66 (12.9) | 104 (20.4) |
|
| Sex |
|
| 0.004 |
| Female
(n=373) | 201 (39.3) | 172 (33.6) |
|
| Male
(n=139) | 55 (10.7) | 84 (16.4) |
|
| Age, years |
|
| 0.093 |
| ≤45
(n=243) | 131 (25.6) | 112 (21.9) |
|
| >45
(n=269) | 125 (24.4) | 144 (28.1) |
|
| Histological
type |
|
| 0.002 |
|
Classical (n=366) | 167 (32.6) | 199 (38.9) |
|
|
Follicular&Other&Tall
Cell (n=146) | 89 (17.4) | 57 (11.1) |
|
| Residual tumor |
|
| 0.670 |
| R0
(n=392) | 201 (44.7) | 191 (42.4) |
|
| R1-R2
(n=58) | 28 (6.2) | 30 (6.7) |
|
| Extrathyroidal
extension |
|
| 0.012 |
| Yes
(n=154) | 64 (13.0) | 90 (18.2) |
|
| No
(n=340) | 183 (37.0) | 157 (31.8) |
|
| OS event |
|
| 0.011 |
| Alive
(n=496) | 253 (49.4) | 243 (47.5) |
|
| Dead
(n=16) | 3 (0.6) | 13 (2.5) |
|
| Primary neoplasm
focus type |
|
| 0.032 |
|
Multifocal (n=233) | 128 (25.5) | 105 (20.9) |
|
|
Unifocal (n=269) | 122 (24.3) | 147 (29.3) |
|
| Neoplasm
location |
|
| 0.010 |
| Left
lobe (n=178) | 80 (15.8) | 98 (19.4) |
|
|
Bilateral (n=88) | 47 (9.3) | 41 (8.1) |
|
| Isthmus
(n=22) | 5 (1.0) | 17 (3.4) |
|
| Right
lobe (n=218) | 121 (23.9) | 97 (19.2) |
|
| Thyroid gland
disorder history |
|
| 0.317 |
| Normal
(n=286) | 141 (31.1) | 145 (31.9) |
|
| Nodular
hyperplasia, lymphocytic thyroiditis and other (n=168)s | 91 (20.0) | 77 (17.0) |
|
Diagnostic predictive value for
patients with THCA is significantly enhanced by assessing the
expression level of PPP1R3G
A prognostic evaluation for THCA was performed by
analyzing the value of PPP1R3G. RNA-seq data and clinical data from
the STAR process of the THCA project were obtained from TCGA
database and were organized. Univariate Cox regression analysis
indicated that the occurrence of the pathological T4 stage and high
PPP1R3G expression were significant factors associated with
unfavorable outcomes in patients diagnosed with THCA. Furthermore,
through the use of multivariate Cox regression analysis, it was
determined that even when considering other variables, pathological
T4 stage and high PPP1R3G expression were still prognostic
indicators for an unfavorable outcome in patients diagnosed with
THCA (Table III).
 | Table III.Prognostic value of PPP1R3G in
patients with thyroid carcinoma, determined using both univariate
and multivariate Cox regression analyses. |
Table III.
Prognostic value of PPP1R3G in
patients with thyroid carcinoma, determined using both univariate
and multivariate Cox regression analyses.
|
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|
|---|
|
Characteristics | n | Hazard ratio (95%
CI) | P-value | Hazard ratio (95%
CI) | P-value |
|---|
| Pathological T
stage | 510 |
|
|
|
|
| T1 | 143 | Reference |
| Reference |
|
| T2 | 169 | 1.010
(0.168–6.067) | 0.992 | 1.141
(0.190–6.852) | 0.886 |
| T3 | 175 | 1.602
(0.309–8.303) | 0.575 | 1.395
(0.269–7.244) | 0.692 |
| T4 | 23 | 11.521
(2.303–57.635) | 0.003 | 12.220
(2.437–61.285) | 0.002 |
| Pathological M
stage | 295 |
|
|
|
|
| M0 | 286 | Reference |
|
|
|
| M1 | 9 | 4.258
(0.909–19.952) | 0.066 |
|
|
| PPP1R3G | 512 |
|
|
|
|
|
Low | 256 | Reference |
| Reference |
|
|
High | 256 | 4.164
(1.186–14.623) | 0.026 | 4.371
(1.232–15.505) | 0.022 |
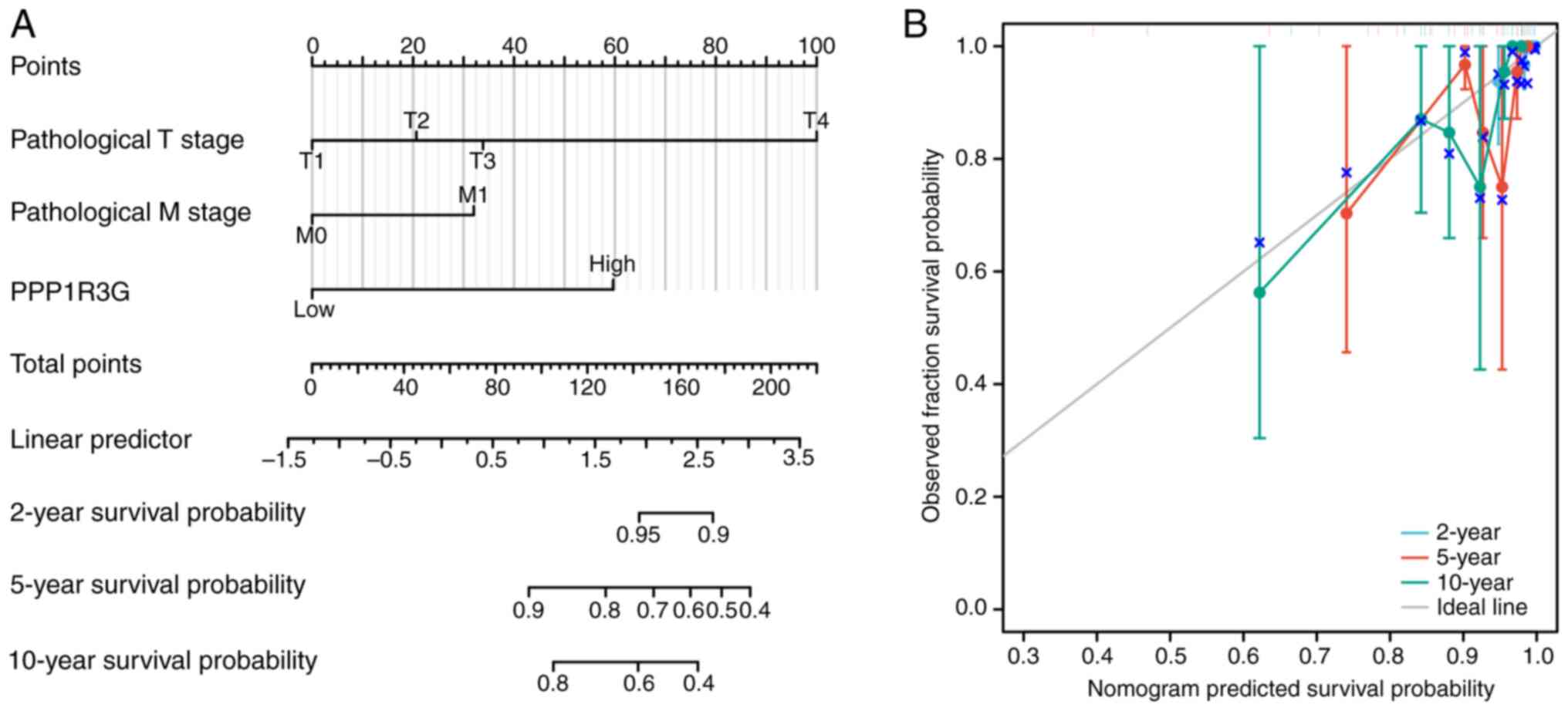
In addition, a nomogram curve was developed to
estimate the OS rates at 2, 5 and 10 years for patients with THCA
(n=507) by considering both pathological T4 stage and PPP1R3G
expression (Fig. 5A). The results
revealed that PPP1R3G could independently predict an unfavorable
prognosis in patients diagnosed with THCA. The combination of
pathological T4 stage and PPP1R3G expression was used to forecast
OS at intervals of 2, 5 and 10 years in patients with THCA. A line
with bias correction was created to approximate the intended curve
(Fig. 5B).
Involvement of PPP1R3G in THCA
assessed using GSEA
To elucidate potential gene regulatory networks
linked to PPP1R3G, RNA-seq data from TCGA database (n=571) was
used. The data were categorized into high- and low-expression
groups based on the median expression levels of the PPP1R3G
molecule. Using the DESeq2 package [1.36.0], the top 12
differentially expressed genes that co-expressed with PPP1R3G were
identified and were represented as a heatmap (Fig. 6A). Subsequently, GSEA was performed
and the results were visualized using the ggplot2 package.
Additionally, GO and KEGG analysis were used to classify the gene
list. The findings revealed significant enrichment of several
functional clusters such as ‘keratinization’, ‘keratinocyte
differentiation’, ‘epidermal cell differentiation’ and ‘skin
development’ in patients exhibiting high expression of PPP1R3G in
THCA (Fig. 6B). In addition, the
visualized outcomes of GSEA were presented. The analysis revealed
notable enrichment of several gene functional clusters,
encompassing ‘cell cycle checkpoints’, ‘mitotic G1 phase and G1-S
transition’, DNA replication, G2-M checkpoints, ‘synthesis of DNA’,
mitotic spindle checkpoint’, ‘retinoblastoma gene in cancer’,
‘mitotic metaphase and anaphase’ and ‘S phase’ in patients with
high PPP1R3G expression in THCA (Fig.
6C). Finally, genes were ranked in descending order based on
their average similarity to other genes. The top gene represents
the highest similarity with other genes, indicating that it has the
strongest association and is more likely to have a key role. The
analysis of the data suggested that small proline-rich protein
(SPRR)3 and SPRR1B may serve a crucial role in the functioning of
PPP1R3G (Fig. 6D).
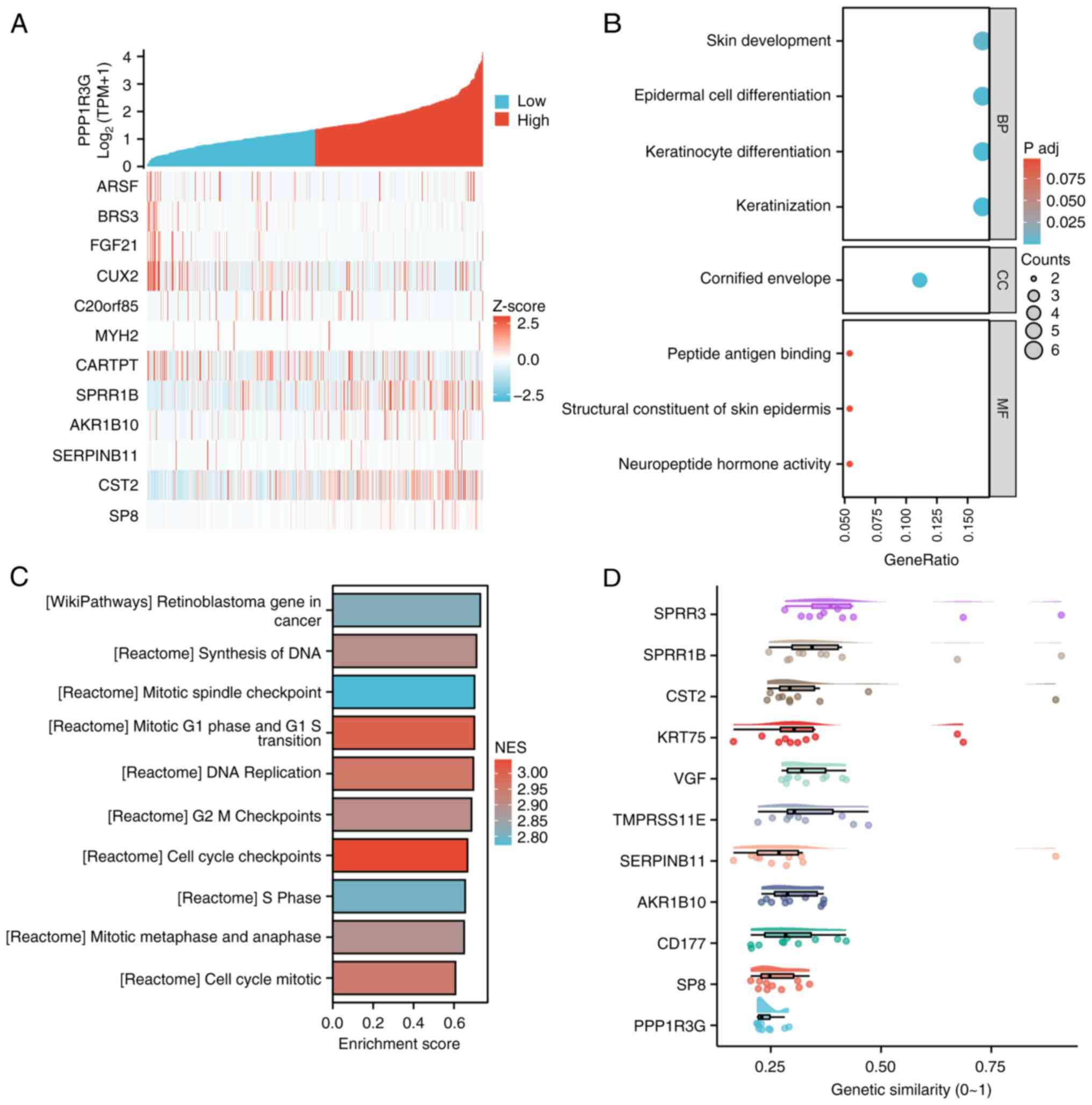 | Figure 6.Differentially expressed genes were
identified and subjected to GO/KEGG and GSEA analysis to cluster in
patients with THCA (n=571). (A) Heat map displaying the
co-expression of PPP1R3G mRNA with other genes. (B) Ranked list of
differentially expressed genes analyzed using GO/KEGG analysis to
facilitate clustering. (C) GSEA analysis performed on the gene list
for clustering purposes. (D) Key genes were identified by
constructing a gene interaction network and calculating their
importance based on network topology. PPP1R3G, protein phosphatase
1 regulatory subunit 3G; GO, Gene Ontology; KEGG, Kyoto
Encyclopedia of Genes and Genomes; GSEA, Gene Set Enrichment
Analysis; TPM, transcripts per million; BP, biological process; CC,
cellular component, MF, molecular function; NES, normalized
enrichment score. |
Involvement of PPP1R3G in THCA immune
invasion properties
To assess the immune invasion properties in patients
with THCA (n=571) and high PPP1R3G expression, correlation analysis
was performed. Immune-related genes that co-expressed with PPP1R3G
were identified and visually represented using a heat map (Fig. 7A). Additionally, the experimental
findings demonstrated an association between the PPP1R3G expression
level and the proportional representation of 24 distinct immune
cell populations in the tumor microenvironment (Fig. 7B). Notably, increased levels of
PPP1R3G were associated with enhanced infiltration of dendritic
cells (DCs), activated (a)DCs, eosinophils, immature (i)DCs,
macrophages, neutrophils, T helper (Th) type 1 cells and regulatory
T cells (Tregs). Furthermore, a negative association was observed
between elevated PPP1R3G expression and infiltrating
characteristics of Th type 17 cells.
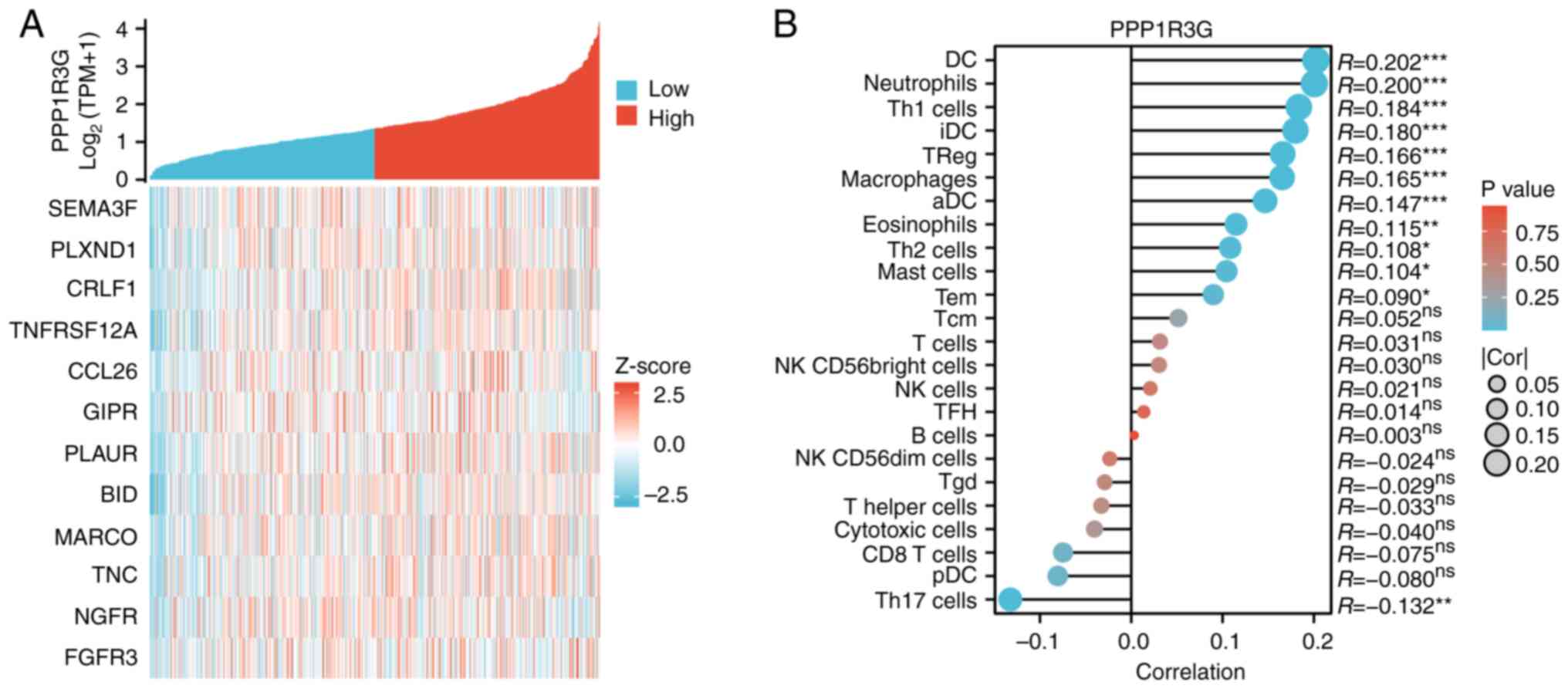 | Figure 7.Correlation between infiltrated immune
cells and the PPP1R3G expression level in patients with THCA
(n=571). (A) Correlation analysis of PPP1R3G and immune checkpoint
molecule expression. (B) Relationship between the expression levels
of PPP1R3G and infiltrated immune cells assessed in clinical
samples obtained from patients diagnosed with THCA. THCA, thyroid
carcinoma; PPP1R3G, protein phosphatase 1 regulatory subunit 3G;
SEMA3F, sema domain, Immunoglobulin Domain (Ig), Short Basic
Domain, Secreted, (Semaphorin) 3F; PLXND1, plexin D1; CRLF1,
Cytokine Receptor Like Factor 1; TNFRSF12A, TNF Receptor
Superfamily Member 12A; CCL26, C-C Motif Chemokine ligand 26; GIPR,
Gastric Inhibitory Polypeptide Receptor; PLAUR, Plasminogen
Activator, Urokinase Receptor; BID, BH3 Interacting Domain Death
Agonist; MARCO, Macrophage Receptor With Collagenous Structure;
TNC, Tenascin C; NGFR, Nerve Growth Factor Receptor; FGFR3,
Fibroblast Growth Factor Receptor 3; TPM, transcripts per million;
DC, dendritic cells; Th, T helper; Treg, regulatory T cells; Tcm,
central memory T cells; Tem, effector memory T cell; aDC, activated
dendritic cells; iDC, immature DC; pDC, plasmacytoid dendritic
cell; Treg, regulatory T cell; TFH, Follicular helper T cell; Tgd,
γ/δ T cell; NK, natural killer. |
Discussion
The understanding of the mechanism involved in the
spread of cancer to distant sites and the discovery of possible
targets for treating tumors are essential and foundational
components of gene therapy (22).
The current study used bioinformatics analyses of an RNA-seq
dataset derived from clinical samples in TCGA to perform a
comprehensive and detailed assessment of PPP1R3G expression. The
aim was to evaluate its association with clinicopathological
characteristics, survival outcomes and functional involvement in
the development of THCA. The results suggested that there was a
significant increase in the expression of PPP1R3G in tumor samples,
indicating its potential role in regulating the progression of
THCA. These findings propose that PPP1R3G could potentially be used
as a biomarker for early detection and prognosis prediction in
patients with THCA, particularly within specific stages of the
disease, pathological classifications and metastatic subgroups.
Additionally, the immunohistochemical analysis provided further
evidence supporting the gene sequencing findings, as it revealed an
increase in PPP1R3G expression in the cytoplasm and cellular
membrane of THCA tissues when compared with adjacent non-cancerous
tissues. Additionally, there was a significant correlation between
PPP1R3G expression and clinical indicators in patients with THCA,
including pathological N stage, pathological stage, histological
type and extrathyroidal extension. Therefore, PPP1R3G may represent
a promising target for the development of diagnostic strategies in
patients with THCA.
To date, the precise role and underlying mechanism
of PPP1R3G in THCA tumorigenesis, development and metastasis remain
elusive. The present study suggests that PPP1R3G could be a useful
molecular marker for predicting the prognosis of patients with
THCA. PPP1R3G expression was a significant prognostic factor for
adverse outcomes, as demonstrated by both univariate and
multivariate Cox regression analyses, even when accounting for
confounding variables. These findings suggest that detecting
PPP1R3G expression may help identify patients at high risk of THCA
progression and poor survival rates, making it a potentially
valuable clinical tool. Additionally, RNA-seq data analysis was
performed to identify genes and functional gene clusters associated
with high PPP1R3G expression in patients with THCA. The GSEA
revealed a significant enrichment in several functional clusters
related to ‘keratinization’, ‘keratinocyte differentiation’,
‘epidermal cell differentiation’ and ‘skin development’ among
patients with high expression of PPP1R3G. GSEA demonstrated a
notable enrichment in several gene functional clusters associated
with ‘cell cycle checkpoints’, ‘mitotic G1 phase and G1-S
transition’, DNA replication, G2-M checkpoints, ‘synthesis of DNA’,
mitotic spindle checkpoint’, ‘retinoblastoma gene in cancer’,
‘mitotic metaphase and anaphase’ and ‘S phase’ among patients
exhibiting elevated PPP1R3G expression. The results of the current
study have the potential to offer innovative perspectives for the
development of therapies aimed at addressing patients with
high-risk THCA. In addition, the gene encodes envelope proteins of
keratinocytes known as SPRR3 and SPRR1B (23,24),
which have been predicted to serve a vital role in the functioning
of PPP1R3G. SPRR1B is associated with cervical intraepithelial
neoplasia (25), whilst SPRR3 has
been linked with esophageal cancer (26). The present data revealed that the
PPP1R3G may serve a role in THCA keratinization. However, the
impact of keratinization in THCA tumorigenesis has not been
reported before. Furthermore, the present study used RNA-seq
analysis to demonstrate unique gene expression patterns in
infiltrating immune cells. The current study findings demonstrated
association between PPP1R3G expression and increased infiltration
of DCs, aDCs, eosinophils, iDCs, macrophages, neutrophils, Th1
cells and Tregs in patients with THCA. Additionally, it was
demonstrated that elevated PPP1R3G expression was inversely
correlated with the presence of Th17 cells. Immunotherapy has shown
encouraging results in different types of cancer, including THCA
(27). Hence, additional research
is necessary to assess the potential influence of PPP1R3G on the
infiltration of immune cells in patients diagnosed with THCA.
Furthermore, it would be valuable to explore whether patients with
elevated levels of PPP1R3G expression could potentially experience
advantages from immunotherapy interventions.
Ultimately, in the current study, a regulatory
network associated with PPP1R3G was successfully established. In
general, PPP1R3G has the potential to serve as an unfavorable
prognostic indicator for THCA and exhibits an association with
immune cell infiltration in tumors. However, the present study had
certain limitations. Firstly, to confirm the prognostic importance
of PPP1R3G, a higher number of patients with THCA would be
necessary for validation. Moreover, whilst the presence of PPP1R3G
in patients with THCA has been verified through
immunohistochemistry and RT-qPCR, the absence of western blotting
experiments remains a limitation of the present study.
Additionally, due to constraints within the database used for the
present study, further exploration into the relationship between
PPP1R3G and tumor immunocytes was not feasible. In addition, the
cellular function of PPP1R3G needs to be further validated through
in vitro cellular and molecular experiments, which is
crucial for establishing its functional significance.
The findings of the current study indicated that
PPP1R3G was expressed at high levels in patients with THCA, as
determined through bioinformatics analysis and immunohistochemical
staining. Further analyses revealed an association between elevated
PPP1R3G expression and the pathological and clinical
characteristics of patients with THCA. Additionally, significant
upregulation patterns of PPP1R3G were demonstrated in several
functional signaling pathways, along with unique gene signatures
associated with immune infiltrating cells. These findings suggest
that PPP1R3G may serve as a valuable diagnostic and prognostic
marker for managing THCA.
Acknowledgements
Not applicable.
Funding
The financial assistance for the present study was supported by
the Natural Science Foundation of Shandong Province (grant no.
ZR2021QH032) and the Medical and Health Science and Technology
Project of Shandong Province (grant no. 202304070941).
Availability of data and materials
The datasets generated in the present study may be
requested from the corresponding author.
Authors' contributions
XZ, HL, HW and PW were involved in performing a
portion of the experiments and shaped the design of the study. XZ
and PW confirm the authenticity of all the raw data. XZ, HL, HW,
PW, SD, ZS and LL contributed to conceptualizing and designing the
study, acquiring, analyzing and interpreting data, drafting or
critically revising important intellectual content within the
article, and granting final approval for publication. All authors
have read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Zibo Central Hospital (Zibo, China; approval no.
IEC-form-030-2.0, 2024-147). The research program strictly followed
the scientific and ethical guidelines stated in the Declaration of
Helsinki, and written informed consent was obtained from all
patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Park J, Kang IK, Bae JS, Kim JS and Kim K:
Clinical significance of the lymph node ratio of the second
operation to predict Re-recurrence in thyroid carcinoma. Cancers
(Basel). 15:6242023. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Dunlap Q and Davies L: Differentiated
Thyroid Cancer Incidence. Surgery of the Thyroid and Parathyroid
Glands. Randolph GW: 3rd Edition. Elsevier; Philadelphia, PA: pp.
174–180. 2021, View Article : Google Scholar
|
|
3
|
Alzahrani AS: The risk of expanding risk
stratification in thyroid cancer. J Clin Endocrinol Metab.
108:e1147–e1148. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nabhan F, Dedhia PH and Ringel MD: Thyroid
cancer, recent advances in diagnosis and therapy. Int J Cancer.
149:984–992. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lieberman L and Worden F: Novel
therapeutics for advanced differentiated thyroid cancer. Endocrinol
Metab Clin North Am. 51:367–378. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Haymart MR: Progress and challenges in
thyroid cancer management. Endocr Pract. 27:1260–1263. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Grimm D: Recent advances in thyroid cancer
research. Int J Mol Sci. 23:46312022. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Santarpia L, El-Naggar AK, Cote GJ, Myers
JN and Sherman SI: PI3K/Akt and Ras/Raf-MAPK pathway mutations in
anaplastic thyroid cancer. JCEM. 93:278–284. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nikiforov YE and Nikiforova MN: Molecular
genetics and diagnosis of thyroid cancer. Nat Rev Endocrinol.
7:569–580. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gulfidan G, Soylu M, Demirel D, Erdonmez
HBC, Beklen H, Sarica PO, Arga KY and Turanli B: Systems biomarkers
for papillary thyroid cancer prognosis and treatment through
multi-omics networks. Arch Bioch Biophysics. 715:1090852021.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Beck AC, Rajan A, Landers S, Kelley S and
Weigel RJ, Bellizzi AM, Lal G, Sugg SL, Howe JR, Chan CH and Weigel
RJ: Expression of cancer stem cell markers in tall cell variant
papillary thyroid cancer identifies a molecular profile predictive
of recurrence in classic papillary thyroid cancer. Surgery.
171:245–251. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Du J, Xiang Y, Liu H, Liu S, Kumar A, Xing
C and Wang Z: RIPK1 dephosphorylation and kinase activation by
PPP1R3G/PP1γ promote apoptosis and necroptosis. Nat Commun.
12:70672021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang Y, Xu D, Huang H, Chen S, Wang L,
Zhu L, Jiang X, Ruan X, Luo X, Cao P, et al: Regulation of glucose
homeostasis and lipid metabolism by PPP1R3G-mediated hepatic
glycogenesis. Mol Endocrinol. 28:116–126. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Du J, Xiang Y, Liu H, Liu S, Kumar A, Xing
C and Wang Z: RIPK1 dephosphorylation and kinase activation by
PPP1R3G/PP1γ promote apoptosis and necroptosis. Nat Commun.
12:70672021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Israelian L, Nitschke S, Wang P, Zhao X,
Perri AM, Lee JPY, Verhalen B, Nitschke F and Minassian BA: Ppp1r3d
deficiency preferentially inhibits neuronal and cardiac Lafora body
formation in a mouse model of the fatal epilepsy Lafora disease. J
Neurochem. 157:1897–1910. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang Y, Gu J, Wang L, Zhao Z, Pan Y and
Chen Y: Ablation of PPP1R3G reduces glycogen deposition and
mitigates high-fat diet induced obesity. Mol Cell Endocrinol.
439:133–140. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhuo X, Chen L, Lai Z, Liu J, Li S, Hu A
and Lin Y: Protein phosphatase 1 regulatory subunit 3G (PPP1R3G)
correlates with poor prognosis and immune infiltration in lung
adenocarcinoma. Bioengineered. 12:8336–8346. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Dobin A and Gingeras TR: Mapping RNA-seq
Reads with STAR. Curr Protoc Bioinformatics. 51:112015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kim K, Kim JH, Park IS, Rho YS, Kwon GH
and Lee DJ: The updated AJCC/TNM staging system for papillary
thyroid cancer (8th Edition): From the perspective of genomic
analysis. World J Surg. 42:3624–3631. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang X, Meng X, Wang P, Luan C and Wang
H: Bioinformatics analysis for the identification of
Sprouty-related EVH1 domain-containing protein 3 expression and its
clinical significance in thyroid carcinoma. Sci Rep. 14:45492024.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shibata MA and Taniguchi K: Metastasis
Inhibition. Int J Mol Sci. 24:71232023. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hu X, Peng N, Qi F, Li J, Shi L and Chen
R: Cigarette smoke upregulates SPRR3 by favoring c-Jun/Fra1
heterodimerization in human bronchial epithelial cells. Future
Oncol. 14:2599–2613. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sasahira T, Kurihara-Shimomura M,
Shimomura H, Bosserhoff AK and Kirita T: Identification of oral
squamous cell carcinoma markers MUC2 and SPRR1B downstream of
TANGO. J Cancer Res Clin Oncol. 147:1659–1672. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Song Y, Pan H, Yang L, Fan Y, Zhang H, Pan
M and Zhang Y: DGUOK-AS1 promotes cervical squamous cell carcinoma
progression by suppressing miR-499a-5p that targets SPRR1B in
vitro. Biochem Biophys Res Commun. 585:177–184. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
de AST, Souza-Santos PT, de Oliveira DS,
Bernardo V, Lima SC, Rapozo DC, Kruel CD, Faria PA, Ribeiro Pinto
LF and Albano RM: Quantitative evaluation of SPRR3 expression in
esophageal squamous cell carcinoma by qPCR and its potential use as
a biomarker. Exp Mol Pathol. 91:584–589. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Naoum GE, Morkos M, Kim B and Arafat W:
Novel targeted therapies and immunotherapy for advanced thyroid
cancers. Mol Cancer. 17:512018. View Article : Google Scholar : PubMed/NCBI
|