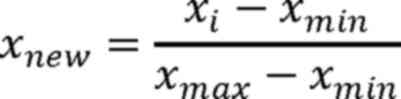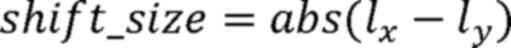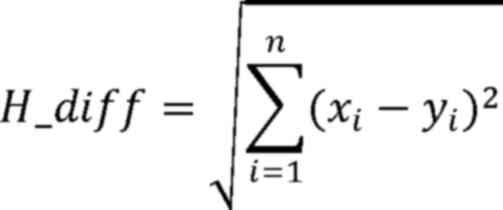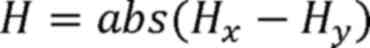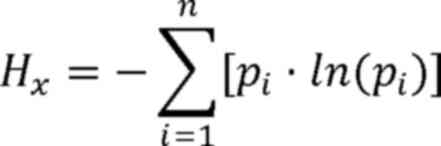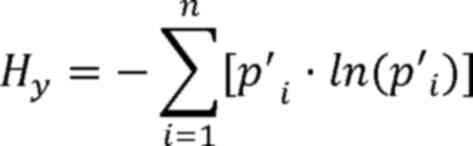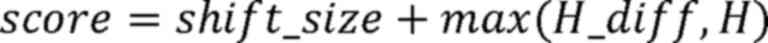Introduction
Microsatellites are short tandem repetitive DNA
sequences with repeating units of 1 to 6 bases that are spread
throughout the human genome. Notably, replication errors commonly
occur in microsatellites during cell division (1,2). For
the maintenance of homeostasis, the majority of replication errors
are recognized and repaired by the DNA mismatch repair (MMR)
system, which includes the mutL homolog 1 (MLH1), mutS Homolog 2
(MSH2), MSH6 and PMS1 homolog 2, mismatch repair system component
(PMS2) proteins. A deficiency in MMR (dMMR) leads to an increase in
microsatellite instability (MSI) (3). In such a state, MMR protein expression
levels reflect the status of MSI. Notably, MSI promotes
carcinogenesis and serves a major role in mechanisms underlying
malignant transformation (4), which
is a sensitive indicator of genetic instability in various types of
cancer, including endometrial and colorectal cancer (5–7).
At present, MSI is detected in clinical practice
using immunohistochemical (IHC) analysis of the impaired DNA MMR
proteins, and PCR is used for the analysis of microsatellite sites
(8). Notably, PCR-based
microsatellite analysis is the gold standard for MSI detection,
involving the examination of PCR product length in a limited set of
informative microsatellite sites (9). The Promega Corporation MSI analysis
system is one of the most widely used commercial PCR assays,
consisting of 5 mononucleotide markers for MSI detection, namely
BAT-25, BAT-26, NR-21, NR-24 and MONO-27 (10). Although MSI-PCR is widely used in
colorectal cancer and other gastrointestinal tumors, MMR-IHC is
recommended in endometrial cancer due to the relatively low
sensitivity of MSI-PCR (11,12).
Tumors with dMMR often exhibit high MSI (MSI-H) that is detected
using DNA-based testing (13);
however, results of a previous study reported a 1–10% discrepancy
between MMR protein and MSI status in numerous types of cancer
(11). In addition, previous
studies reported high levels of discrepancy between these factors
(6,14). Samples with MSI-H may exhibit MMR
proficiency (pMMR) as a result of MMR gene methylation (15) and MMR proteins may exhibit abnormal
functions with an expected antigen structure (16). Notably, MMR may exert effects on
factors other than the four common proteins, MLH1, MSH2, MSH6 and
PMS2, detected using IHC analysis (17). A previous study reported that
>20% of patients with endometrial cancer exhibit dMMR/MSI-H
status, and accurate identification of this type is crucial for
treatment optimization and the assessment of prognosis. Thus, the
use of IHC analysis alone in the detection of dMMR may lead to
inaccurate diagnoses of pMMR in patients with MSI-H (18). The development of a novel MSI
detection method with high levels of sensitivity is required.
Next-generation sequencing (NGS) is used for the
comprehensive analysis of genomic profiles and MSI status, and
simultaneous analysis may decrease the number of tissue samples
required and increase the efficiency of examination. NGS-based
algorithms demonstrate a comparable accuracy to PCR-based MSI
detection (19,20). Notably, existing algorithms, such as
MSIsensor (21) and MANTIS
(22), measure MSI levels using the
read-count distribution of microsatellites with different repeat
lengths. The aforementioned algorithms require the analysis of
>10 (or even ≥40) loci for accurate MSI evaluation (22). NGS-based microsatellite testing
selects mononucleotide repeats with stable repeat lengths among
samples with microsatellite stability (MSS) (9,23). At
present, various loci and numerous methods of MSI detection are
used in research, leading to low levels of reliability and a lack
of consistency.
In the present study, a novel algorithm was
developed for the detection of MSI status, using NGS for the
analysis of five mononucleotide repeats, namely BAT-25, BAT-26,
NR-21, NR-24 and MONO-27. Notably, the aforementioned loci are
often analyzed using MSI-PCR in clinical settings, with the ability
to represent the MSI status of a sample. NGS was integrated into
the algorithm to improve the sensitivity of the MSI detection,
which may lead to improved detection of pMMR in patients with
endometrial cancer and MSI-H.
Materials and methods
Patients
A total of 181 patients aged 37 to 86 years (median,
56) with endometrial cancer were retrospectively enrolled from the
First Affiliated Hospital of Wannan Medical College (Wuhu, China).
Inclusion criteria were as: i) Female; pathologically diagnosed as
endometrial cancer in the past 3 years; no other malignant tumors
nor serious chronic diseases; can be contacted and agree to
participate in the project and sign an informed consent form.
Exclusion criteria were set as: tissue sample retained in pathology
department was too small; tumor cells in the sample was less than
10%; patients lost contact or were unwilling to participate in the
research project.
These patients were diagnosed with endometrial
cancer from April 2021 to November 2022 and tissues were collected.
This was performed between November 2022 and June 2023. The present
study was approved by the Ethics Committee of the First Affiliated
Hospital of Wannan Medical College (approval no. 2022-110) and each
patient provided written informed consent for their clinical
information as well as their genomics data (from PCR and NGS) to be
reported in the journal. Tumor and matched adjacent non-tumor
tissues were collected from all patients and the MMR status was
verified using IHC analysis of MSH2, MSH6, PMS2 and MLH1 protein
expression levels. All IHC results were tested and reported by
pathologists in the pathology department of the hospital.
Antibodies including MLH1 (cat. no. ZM-0154, ZSGB-bio), MSH2 (clone
FE11, cat. no. ZA-0622, ZSGB-bio, China), MSH6 (clone EP49, cat.
no. ZA-0541, ZSGB-bio, China), and PMS2 (clone EP51, cat. no.
ZA-0542, ZSGB-bio, China), were stained using Dako's automated
staining system (LINK48, Dako, CA, USA) with 1:1,000 dilutions. All
staining procedures were performed according to the manufacturer's
recommendations and previous study (24).
Surgically specimens were fixed in 10%
neutral-buffered formalin for 24–72 h at room temperature and
embedded in paraffin. DNA was extracted from 10-µm-thick sections
of formalin-fixed paraffin-embedded (FFPE) tumor tissue blocks
using the GeneRead DNA FFPE kit (Qiagen, GmbH), according to the
manufacturer's instructions. Samples were analyzed using MSI-PCR
and NGS. After extraction, DNA quality was evaluated by 1% agarose
gel electrophoresis and the concentration of all samples was
quantifed using the Qubit dsDNA HS Assay kit (Termo Fisher
Scientifc, Waltham, MA, USA) with a Qubit 3.0 Fluorometer.
Spike-in samples with synthetic
DNA
For each of the five microsatellite loci, namely
BAT-25, BAT-26, NR-21, NR-24 and MONO-27, four plasmids were
synthesized by Sangon Biotech (China). These included a wild-type
fragment and deletions of the wild-type, consisting of 1-, 2- and
3-bp deletions. Plasmids were utilized as spike-in fragments and
mixed into the DNA of noncancerous endometrial tissue at a ratio of
1:1.
MSI-PCR analysis
MSI-PCR analysis was performed using the MSI
Analysis System (Promega Corporation) (25) as previously described (26,27).
Briefly, the five microsatellite loci, namely BAT-25, BAT-26,
NR-21, NR-24 and MONO-27, and two pentanucleotide repeats PENTAC
and PENTAD, were amplified in a single multiplex 25 µl PCR
reaction. PENTAC and PENTAD were used as reference genes to detect
potential contamination. Fluorescently labeled primers used for
MSI-PCR analysis are supplied by Sangon Biotech and listed in
Table SI. The following
thermocycling conditions were used for the PCR: Initial
denaturation at 95°C for 11 min and 96°C for 1 min; 10 cycles of
94°C for 30 sec, ramp 68 sec to 58°C, hold for 30 sec, ramp 50 sec
to 70°C and hold for 1 min; 20 cycles at 90°C for 30 sec, ramp 60
sec to 58°C, hold for 30 sec, ramp 50 sec to 70°C and hold for 1
min; 60°C for 30 min; hold at 4°C. PCR products were analyzed using
a 3500 Genetic Analyzer (Thermo Fisher Scientific, Inc.).
GeneMapper 6.0 (Thermo Fisher Scientific, Inc.) was used to
determine the size differences between tumor samples and adjacent
tissues. A tumor was defined as exhibiting MSI-H if ≥2 markers were
unstable, and MSS was defined according to the presence of ≤1
unstable mononucleotide markers in the tumor sample. The term
‘unstable’ was used for markers with a shift of ≥2 bp, or if the
shoulder pattern extended the range of the smallest peak by ≥2 bp
in the tumor allele.
MSI detection using NGS
NGS was performed on a NextSeq 500 or Novaseq 6000
(Illumina, Inc.) using a custom amplicon-based gene panel that
comprised five microsatellite loci included in the Promega
Corporation MSI kit. Initially, libraries were generated with the
Hieff NGS™ OnePot Pro DNA Library Prep Kit (Shanghai Yeasen
Biotechnology Co., Ltd.) according to the manufacturer's protocol.
Briefly, 20 ng fragmented genomic DNA was used to amplify the
target regions and amplified products were purified (Table SII). Subsequent rounds of PCR were
carried out through the addition of sequencing adapters and
barcodes to amplicons. Following the purification of the library,
quantification of the DNA library was performed using Labchip GX
Touch (PerkinElmer). The libraries with 1 pM concentration were
then sequenced using the Novaseq 6000 NGS (Illumina, Inc.)
platforms and NovaSeq 6000 SP reagent kit (100 cycles; cat. no.
2002746; Illumina Inc.), according to the manufacturer's
instructions using 2X150 bp paired-end reads at an average depth of
5,000× for tissue.
A novel algorithm, MSIPeak, was developed in the
present study to determine the MSI status of all samples using NGS
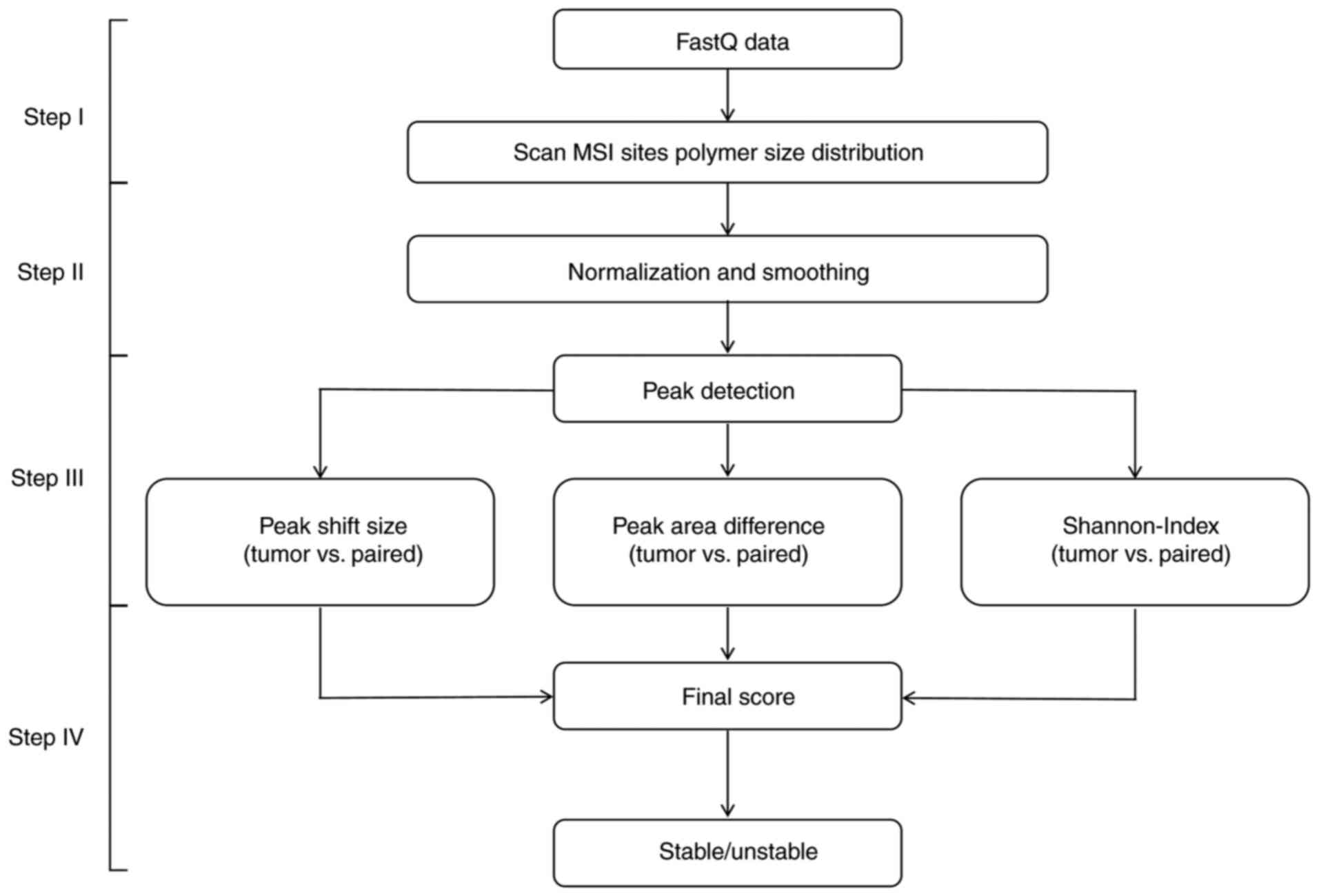
read-count distribution. MSIPeak program flow was divided into four
main steps, as follows (Fig.
1).
Step I: The sequencing data of each tumor
tissue and matched adjacent tissues were read in FastQ format
files. For each MSI locus, reads coverage information, including
reads count, was extracted from FastQ files.
Step II: Minimum-maximum normalization was
performed on the reads count of each microsatellite locus. Values
were scaled to the range (0,1) for subsequent data processing.
Here, i represents a single microsatellite locus,
xi represents the reads count prior to normalization,
xnew represents the reads count value following
normalization, and xmin and xmax represent
the minimum and maximum values of the reads count for each locus,
respectively.
Reads count values were smoothed using the sliding
window. Following normalization and smoothing, peak data of the
microsatellite loci of tumor and matched adjacent tissues were
analyzed. The local maximum values of each repeat were compared
with the values of neighboring points.
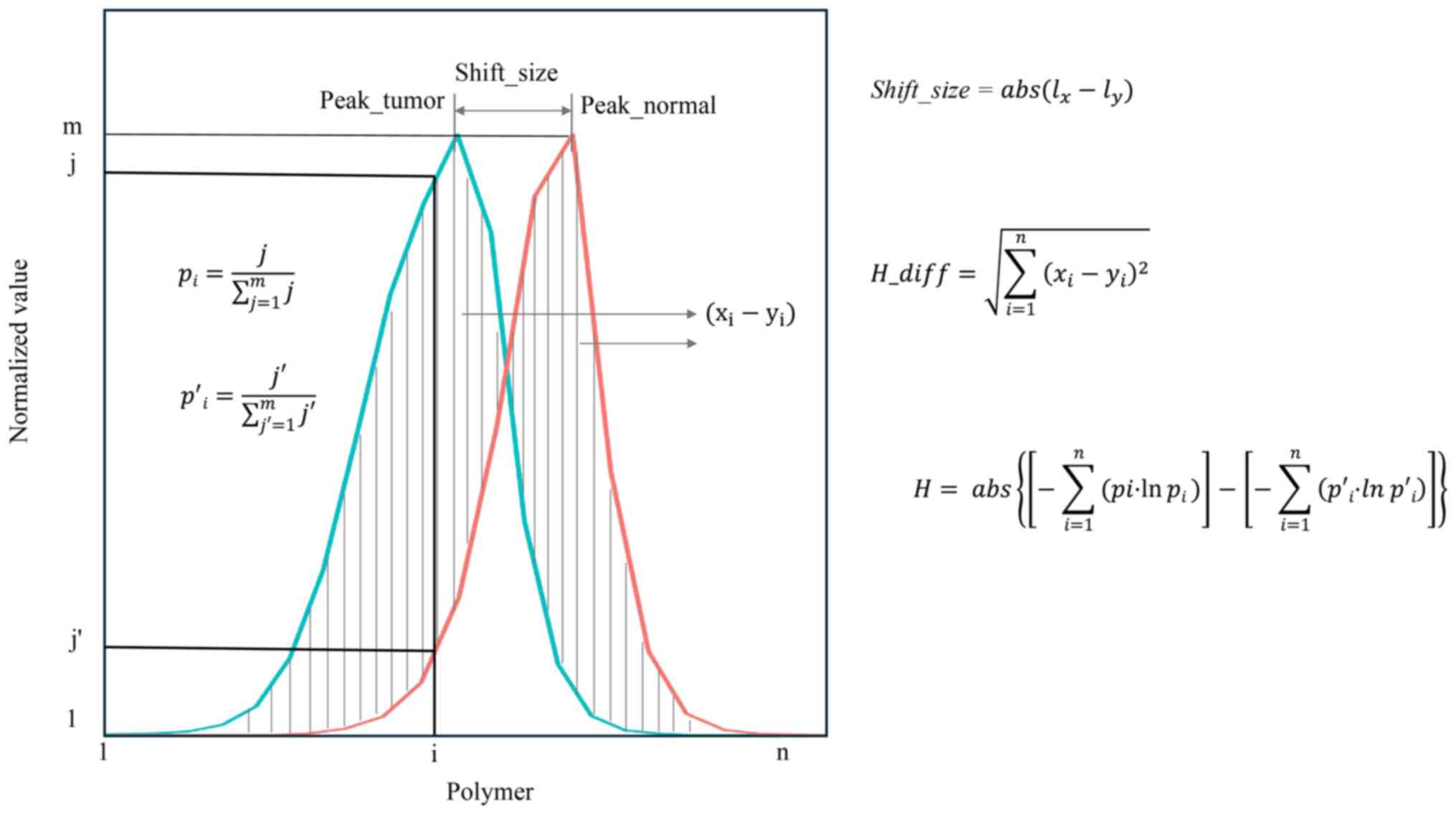
Step III: For each peak determined in the
tumor and adjacent tissues, peak shift size, peak area difference
and Shannon coefficient difference were calculated to score the MSI
status of each locus (Fig. 2):
Here, i represents a single microsatellite locus,
lx represents the peak value of the microsatellite locus
in the tumor sample and ly represents the peak value of
the microsatellite locus in the matched adjacent tissue.
Here, H_diff represents the area difference of each
microsatellite locus peak, × and y refer to the vectors of area
values of MSI loci in tumor and adjacent tissues, respectively, i
represents a single microsatellite locus, and xi and
yi represent the area values of the ith MSI locus in
tumor and adjacent tissues, respectively.
Here, H represents the Shannon coefficient
difference between tumor and adjacent tissues. Hx
represents the Shannon-Wiener diversity index of the tumor sample,
Hy represents the Shannon-Wiener diversity index of the
adjacent sample, i represents a single microsatellite locus,
pi represents the relative abundance of the ith
microsatellite locus in the tumor sample and p'i
represents the relative abundance of the ith microsatellite locus
in the adjacent sample.
Step IV: The final score for each MSI locus
was calculated using the following equation:
When the score was ≥1.10, the MSI status of this
locus was considered unstable. After the stability of all five
markers had been determined, the MSI status of the patient was
evaluated. Samples with two or more unstable markers were
considered MSI-H and samples with <2 unstable markers were
considered MSS.
Comparison of MSIPeak with MSIsensor
and MANTIS
Among previously published NGS-based MSI studies,
MSIsensor (21) and MANTIS
(22) were widely used analytical
methods (2,5,28,29).
The calculation principles of these two algorithms are markedly
different from MSIPeak (Table I).
Therefore, MSIPeak was compared with the MSIsensor and MANTIS
algorithms. MSISensor2 (30), an
upgraded version of MSIsensor, and Mantis were run according to
their manuscript, for the analysis of in-house whole-exome
sequencing (WES) data from 25 endometrial cancer samples. The WES
library was constructed using the commercial Hi-Exon 35 Panel and
supporting library construction kit (cat. no. P10016-96, Shanghai
HeYin Biotechnology Co., LTd.). A total of 50 ng fragmented genomic
DNA was used for a capture-based library (Table SIII) according to the
manufacturer's protocol of the library construction kit. After the
quantification of the DNA library by the Labchip GX Touch
(PerkinElmer), WES library with 1 pM concentration was performed on
the same Novaseq 6000 NGS (Illumina, Inc.) platforms and NovaSeq
6000 SP reagent kit (100 cycles; cat. no. 2002746; Illumina Inc.),
according to the manufacturer's instructions using 2×150 bp
paired-end reads at an average depth of 150×. To obtain clean
reads, FASTQ files from tumor tissue and white blood samples were
done by fastp (https://github.com/OpenGene/fastp, version 0.19.3).
Clean reads were mapped to the reference genome (hg38/GRCh38) by
Burrows-Wheeler aligner (BWA, https://github.com/lh3/bwa, version 0.7.12-r1039) and
perform alignment processing by SAMtools
(https://github.com/samtools/samtools, version 0.1.19–96b5f2294a).
The quality score was recalibrated using GATK
(https://github.com/broadinstitute/gatk, version 4.1.0.0) to
generate the final binary SAM (BAM) files used for subsequent
analyses. Lastly MSI status was detected using the MANTIS (version
v1.0.5) (22) and MSISensor2
(Version 0.1) (30).
 | Table I.Comparison of MSIPeak with the
published algorithms based on next-generation sequencing data. |
Table I.
Comparison of MSIPeak with the
published algorithms based on next-generation sequencing data.
| Parameters | MSIPeak | MSISensor | MANTIS |
|---|
| No. of loci | Five | Tens to
thousands | Dozens to
thousands |
| Origin of the
loci | Fixed | Genome-wide or
target screening | Genome wide or
target screening |
| Data
preprocessing | Obtainment of
coverage information of loci, and perform normalization and data
smoothing processing. | Calculation of the
coverage of each locus without mentioning normalization and data
smoothing steps. | Calculation of the
coverage of each locus and data normalization. |
| Comparison between
tumor and normal samples | Peak shift, Peak
area difference and Shannon coefficient. difference | Number of
repetitions and allele distribution for each locus. | The repeat length
distribution and stability level of each locus. |
| Scoring criteria of
each locus | The final score of
each MSI locus is obtained by the peak shift, peak area difference
and Shannon coefficient difference. Loci with a score ≥1.10 are
rated as unstable. | Calculation of the
proportion of unstable positioning points, and if the proportion
exceeds a threshold, it is rated as unstable. The threshold is
determined by the cumulative distribution of this indicator on a
set of samples. | The average L1 norm
of all loci is the MSI score of the sample. If the score exceeds
the threshold, it is rated as unstable. |
| Criteria of
MSI | ≥2 of 5 loci are
unstable | Default 20% | Default 0.4 |
Statistical analysis
The chi-square test was used to compare the
frequencies of MSI-H and MSS tumors identified through PCR and NGS,
with the dMMR and pMMR status determined by IHC. For chi-square
test analysis, P<0.001 was considered to indicate a
statistically significant difference in PCR and NGS in ability to
detect MSI-H. Cohen's κ was calculated to evaluate the level of
agreement between IHC-based and molecular-based methods, PCR and
NGS. A Cohen's κ of P<0.001 was considered to indicate a
statistically significant difference between methods. All data
presented in figures and tables are reported as percentages for
categorical comparisons. P<0.05 was considered to indicate a
statistically significant difference. The statistical analyses were
performed using R software (version 4.3.2; RStudio).
Results
MSI-PCR and MSI-NGS using synthetic
DNA samples
The present study demonstrated that spike-in DNA
sample profiles with 3-bp deletions displayed notable left-shifts,
which could be distinguished from those of the wild-type fragments
(Fig. 3A). Spike-in DNA sample
profiles with 2-bp deletions exhibited ambiguous extensions on the
left shoulder of BAT25 and NR24, compared with the wild-type
fragments. However, there were no notable shifts in BAT26, NR21 and
MONO27 with 2-bp deletions or in the five markers with 1-bp
deletions (Fig. 3A and B).
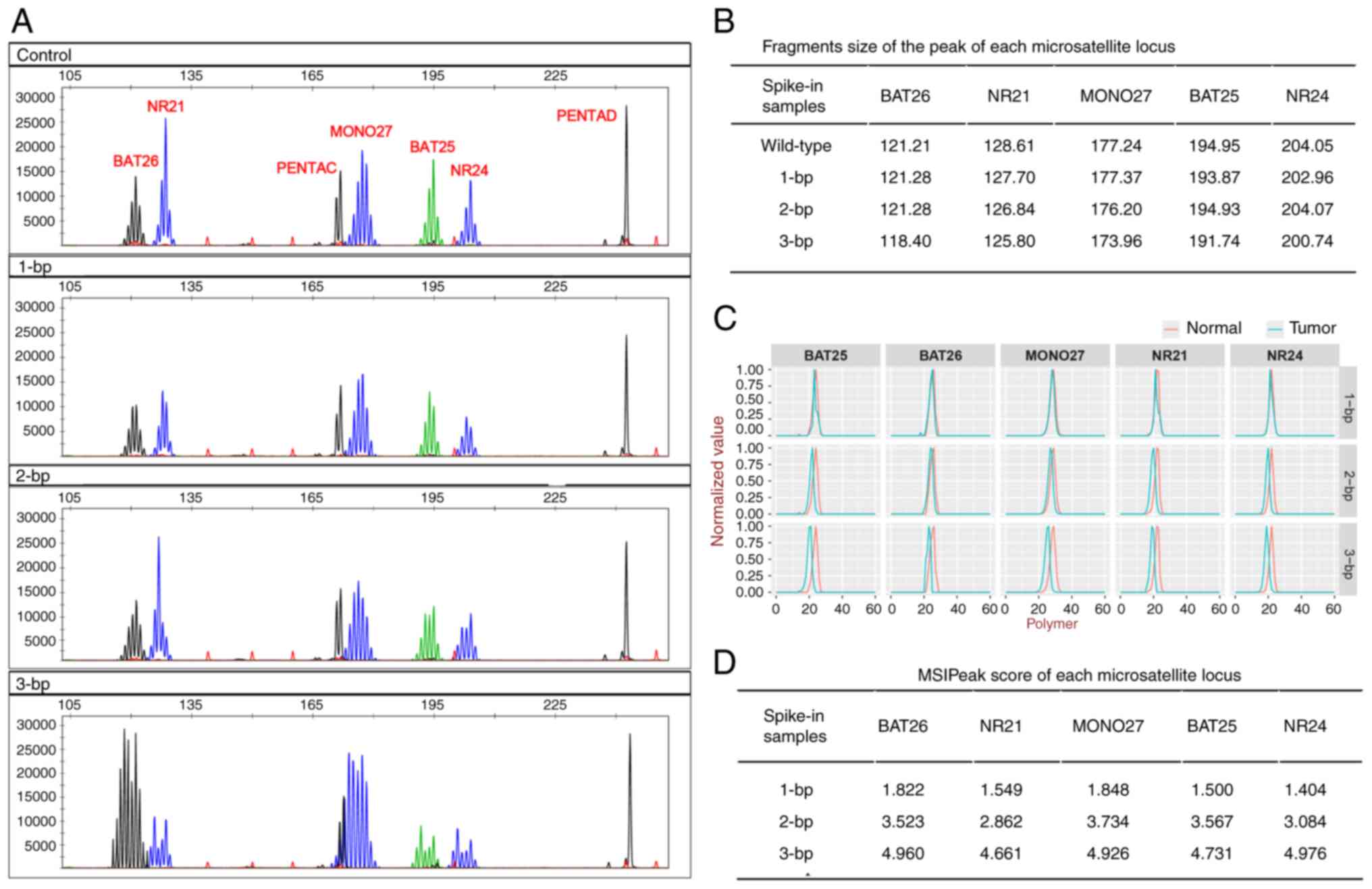 | Figure 3.MSI detection using spike-in DNA
samples. (A) MSI-PCR detection of spike-in DNA samples with
wild-type fragments (control), 1-, 2- and 3-bp deletions, from top
to bottom. The polymorphic pentanucleotide repeat markers PENTAC
and PENTAD were used as quality controls for sample authentication.
(B) Fragment sizes of peaks for each microsatellite locus in
spike-in DNA samples. (C) MSI-NGS detection of spike-in DNA samples
with 1-, 2- and 3-bp deletions, displayed from top to bottom. The
red line represents wild-type samples and the blue line represents
spike-in samples with 1–3-bp deletions. Polymer, number of base
repeats size of each microsatellite locus; normalized-value,
minimum-maximum normalization value of each microsatellite locus at
each polymer. (D) MSIPeak score of each microsatellite locus in
spike-in DNA samples. MSI, microsatellite instability; bp, base
pair; NGS, next-generation sequencing. |
Results obtained using MSI-NGS are presented as
peaks, which were comparable with those obtained using MSI-PCR
(Fig. 3C). Peaks of the spike-in
DNA samples with 1-bp deletions exhibited subtle shifts compared
with those of the wild-type fragments. However, the score was more
than the threshold of 1.10 (Fig.
3D). Spike-in DNA samples with 1- or 2-bp deletions exhibited
shifts and scores that were indicative of MSI (Fig. 3C and D).
MMR/MSI detection in FFPE samples
A total of 39 endometrial cancer samples were
identified as dMMR and the remaining 142 samples were identified as
pMMR (Table II). Within the 39
dMMR samples, 16 were classified as MSI-H using PCR testing and 36
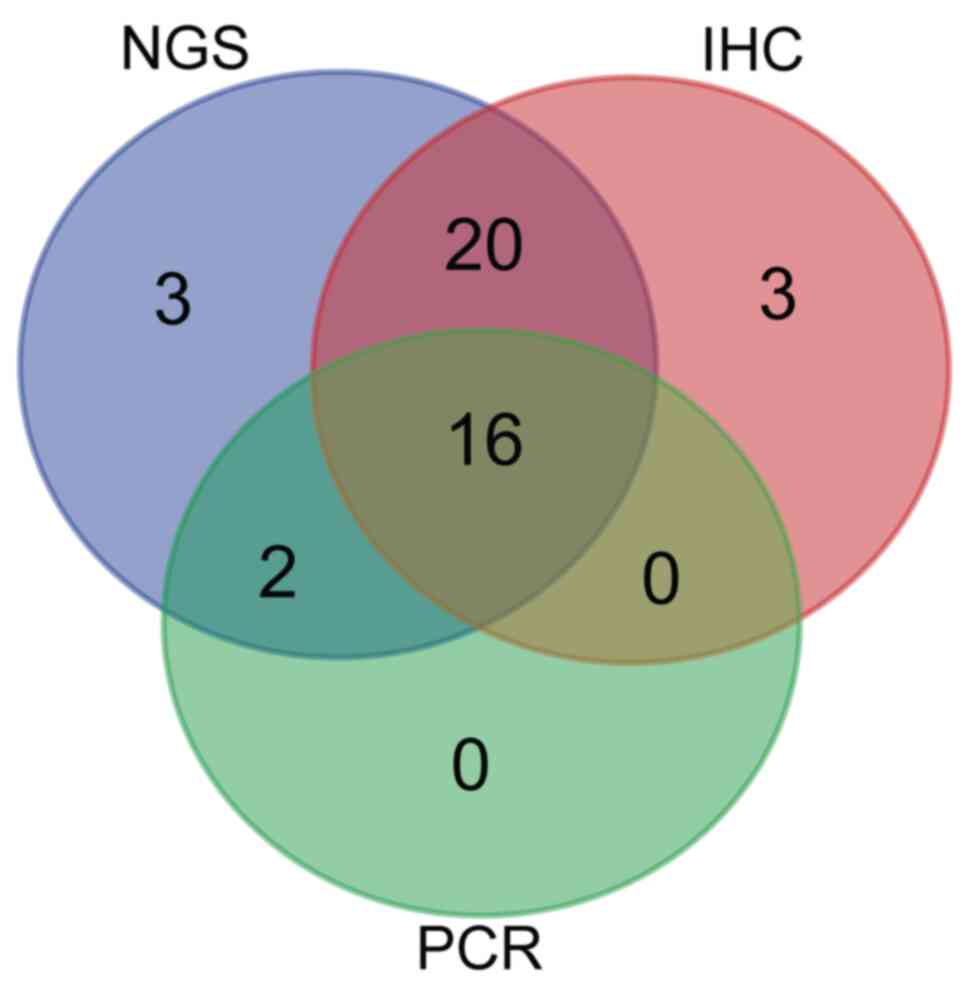
were identified as MSI-H using NGS (Table II). The concordance between IHC and
NGS was significantly higher compared with that between IHC and PCR
(Cohen's κ=0.492 vs. 0.872; P<0.001; Table II). All 16 dMMR/MSI-H samples
confirmed using IHC and PCR were also defined as MSI-H using NGS
(Fig. 4; Tables SIV and SV). In addition, a further 20 MSI-H
samples were identified using NGS alone, with the profiles
exhibiting minor shifts that did not meet the criteria for MSI-H
based on PCR analysis (data not shown).
 | Table II.Concordance between MMR-IHC and PCR-
or NGS-based methods. |
Table II.
Concordance between MMR-IHC and PCR-
or NGS-based methods.
| A, Tumors with dMMR
identified by IHC technology |
|---|
|
|---|
| Type | PCR, n (%) | NGS, n (%) | Chi-square
P-value |
|---|
| MSI-H | 16 (8.84) | 36 (19.89) | <0.001 |
| MSS | 23 (12.71) | 3 (1.66) | 0.444 |
|
| B, Tumors with
pMMR identified by IHC technology |
|
| Type | PCR, n
(%) | NGS, n
(%) | Chi-square
P-value |
|
| MSI-H | 2 (1.10) | 5 (2.76) | <0.001 |
| MSS | 140 (77.35) | 137 (75.69) | 0.251 |
Comparison with MSISensor2 and
MANTIS
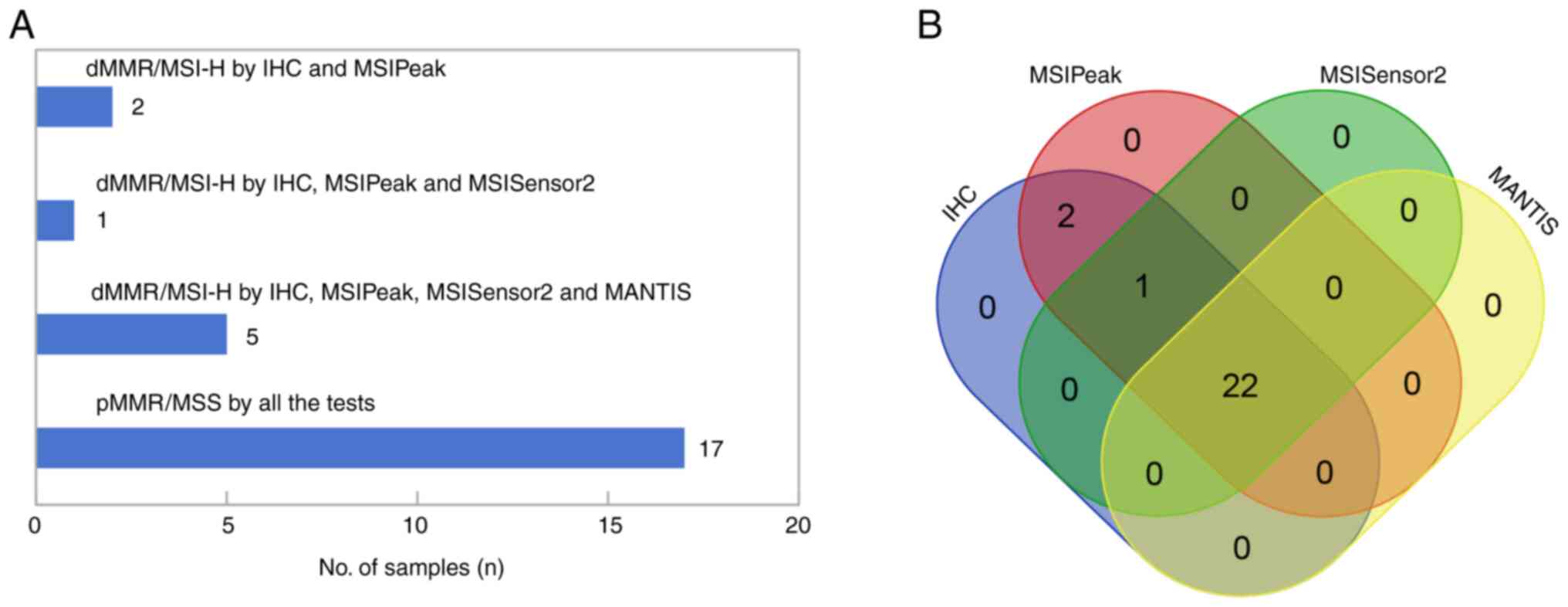
Among the 25 samples with available WES data, 8 dMMR
and 17 pMMR cases were identified using IHC analysis. MSIPeak,
MSISensor2 and MANTIS consistently classified the 17 pMMR samples
as MSS (Fig. 5A). However, MSIPeak
was the only algorithm to identify all 8 dMMR samples as MSI-H,
while MSISensor2 and MANTIS classified 2 and 3 dMMR samples as MSS,
respectively (Fig. 5A and B;
Table SVI).
Discussion
In endometrial cancer, MMR detection using IHC
analysis has been recommended over MSI detection using PCR.
Notably, results obtained using MSI and MMR exhibited low levels of
concordance in gynecologic tumors compared with gastrointestinal
tumors (11). The subtle leftward
shifts in endometrial cancer were 1–3 bp, whereas those observed in
colorectal cancer were >6 bp (31,32).
The results of the present study obtained using MSI-PCR showed that
synthetic DNA fragments with 1–2 bp differences displayed ambiguous
shifts that could not be distinguished from the matched adjacent
tissue samples. In addition, results obtained using MSI-PCR
demonstrated that numerous samples could not be classified as MSI
based on the shifts of their peaks. These ambiguous shifts imply
that endometrial cancer samples with 1–2 bp shifts cannot be
differentiated from MSS samples, contributing to the low
concordance between MSI and MMR in endometrial cancer.
Limitations of IHC analysis for the detection of MMR
(15–18,33)
have led to the requirement for detecting specific microsatellite
repeats. Thus, numerous NGS-based MSI detection methods have been
introduced (20,34–37),
and these have detected a greater number of microsatellite markers
compared with the 5 to 6 markers detected using PCR. Microsatellite
markers analyzed using NGS technology have varied among studies and
have only been demonstrated in specific cohorts or tumor types.
However, the five markers in the Promega Corporation system
(8) have been widely used in
clinical practice for a number of tumor types (3–7).
Notably, these markers are used to represent the status of MMR
proteins. In the present study, all samples defined as MSI-H using
MSI-PCR were also defined as MSI-H using MSI-NGS. These results
suggested that the novel algorithm developed in the present study
exhibits the capability to identify relatively large shifts in
endometrial cancer samples. Samples with IHC analysis-verified dMMR
and PCR-verified MSS were further categorized into two groups
according to shift using NGS combined with MSIPeak. These results
suggest that the novel algorithm may exhibit potential in the
identification of samples with sublet shifts.
MSIPeak uses the same markers as PCR, but levels of
sensitivity are improved compared with PCR. Potential reasons
include that, first, the interpretation of PCR results relies on
the analysis of capillary electrophoresis patterns, which has a
certain degree of subjectivity. Independent investigators may
interpret electrophoresis patterns differently, potentially
resulting in poor reproducibility of the results. In cases where
endometrial MSI is offset by 1–3 bp, errors may occur (32). By contrast, MSIPeak performs
minimum-maximum normalization and data smoothing during the data
preprocessing, which may reduce the impact of sequencing depth and
data fluctuations on MSI detection. Furthermore, MSIPeak analyzes
the differences in peak values between tumor samples and matched
adjacent tissues from multiple dimensions, including the peak
shift, peak area difference and Shannon coefficient difference
(38,39). Thus, the MSI status was
comprehensively evaluated to provide more accurate detection
results.
MSIPeak sorts loci from small to large based on the
distribution of different ploymer repetitions at each locus.
Subsequently, the peak of each locus is identified, and shift size,
area difference and Shannon-Wiener diversity index are evaluated in
tumor and adjacent tissues (38,39).
Results of the present study demonstrated that MSIPeak exhibited
higher levels of accuracy compared with alternate NGS-based
algorithms, such as MSISensor2 and MANTIS. However, further
analyses using a larger number of samples are required to verify
the results. Notably, MSISensor2 and MANTIS are designed to be
performed using microsatellite loci across the entire genome or
exon ranges (21,22,30).
Loci derived from different batches may vary, which may affect the
results. In addition, MSISensor2 and MANTIS have been extensively
applied in the context of colorectal cancer (2,5,28,29);
however, these algorithms are not widely used in endometrial
cancer. Certain parameters and/or thresholds of these algorithms
may require further refinement for effective MSI detection in
endometrial cancer.
MSIPeak demonstrated high levels of reproducibility
and adaptability for MSI detection in endometrial cancer. The five
common loci detected using MSIPeak are small in size, and these can
be integrated into other NGS sequencing panels, using associated
amplicons for amplicon-based panels or associated probes for
capture-based panels. Notably, this integration does not require
specifically designed sequencing panels, and is inexpensive
compared with WES or whole genome sequencing. Thus, NGS-based MSI
detection exhibits potential in patient diagnosis, with high levels
of flexibility and cost effectiveness.
In conclusion, a novel algorithm was developed for
the detection of MSI in the present study, namely MSIPeak. This
algorithm was designed to detect only five commonly used
microsatellite loci, allowing it to be easily integrated into
existing NGS panels, which could thereby lead to potential
reductions in experimental costs. Results obtained using MSIPeak
are presented in peak form for intuitive and convenient
identification, which is comparable with MSI-PCR. However, MSIPeak
demonstrated higher levels of accuracy and objectivity compared
with PCR. In addition, MSIPeak may exhibit potential in detecting
MSI in endometrial cancer. Further investigations with increased
sample sizes are required to validate the present results and to
explore the utility of this algorithm in other types of cancer,
such as colorectal cancer. Future investigations should focus on
refining and developing a widely applicable NGS-based MSI detection
algorithm that could be effectively used across various types of
cancer.
Supplementary Material
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Key Project of Wannan
Medical College (grant no. WK2022ZF02), Natural Science Foundation
of Anhui Provincial Education Department (grant no. 2023AH051779),
The First Affiliated Hospital/Yijishan Hospital of Wannan Medical
College (grant nos. KY27530533, YR202214, CX2023018 and GF2019G19)
and Anhui New Era Education Quality Project (Postgraduate
Education; grant no. 2022zyxwjxalk158).
Availability of data and materials
The data generated in the present study may be
found in The National Center for Biotechnology Information Sequence
Read Archive repository under accession number PRJNA1100268 or at
the following URL: https://www.ncbi.nlm.nih.gov/sra/PRJNA1100268.
Authors' contributions
BZ, YW, LD, XT, WS, WZ and YL acquired the data. BZ
and YW carried out the molecular experiments and drafted the
manuscript. LD analyzed and interpreted the data. XT and WS
performed histological examination of the tissue samples. WZ and YL
designed the research aims and acquired financial support for the
project. BZ and YL confirm the authenticity of all the raw data.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the First Affiliated Hospital of Wannan Medical
College (approval no. 2022-110). Each patient provided written
informed consent to participate.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
MSI-H
|
high microsatellite instability
|
|
MSS
|
microsatellite stability
|
|
PCR
|
polymerase chain reaction
|
|
NGS
|
next-generation sequencing
|
|
IHC
|
immunohistochemistry
|
|
MMR
|
mismatch repair
|
|
dMMR
|
mismatch repair deficiency
|
|
pMMR
|
MMR proficiency
|
|
FFPE
|
formalin-fixed paraffin-embedded
|
|
WES
|
whole-exome sequencing
|
|
WGS
|
whole-genome sequencing
|
|
Bp
|
base pair
|
References
|
1
|
Lynch HT, Snyder CL, Shaw TG, Heinen CD
and Hitchins MP: Milestones of lynch syndrome: 1895–2015. Nat Rev
Cancer. 15:181–194. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Li K, Luo H, Huang L, Luo H and Zhu X:
Microsatellite instability: A review of what the oncologist should
know. Cancer Cell Int. 20:162020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sinicrope FA and Sargent DJ: Molecular
pathways: Microsatellite instability in colorectal cancer:
prognostic, predictive, and therapeutic implications. Clin Cancer
Res. 18:1506–1512. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Aaltonen LA, Peltomäki P, Leach FS,
Sistonen P, Pylkkänen L, Mecklin JP, Järvinen H, Powell SM, Jen J,
Hamilton SR, et al: Clues to the pathogenesis of familial
colorectal cancer. Science. 260:812–816. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bonneville R, Krook MA, Kautto EA, Miya J,
Wing MR, Chen HZ, Reeser JW, Yu L and Roychowdhury S: Landscape of
microsatellite instability across 39 cancer types. JCO Precis
Oncol. 2017.PO.17.00073. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nádorvári ML, Kenessey I, Kiss A, Barbai
T, Kulka J, Rásó E and Tímár J: Comparison of standard mismatch
repair deficiency and microsatellite instability tests in a large
cancer series. J Transl Med. 22:1502024. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chung Y, Nam SK, Chang HE, Lee C, Kang GH,
Lee HS and Park KU: Evaluation of an eight marker-panel including
long mononucleotide repeat markers to detect microsatellite
instability in colorectal, gastric, and endometrial cancers. BMC
Cancer. 23:11002023. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
McConechy MK, Talhouk A, Li-Chang HH,
Leung S, Huntsman DG, Gilks CB and McAlpine JN: Detection of DNA
mismatch repair (MMR) deficiencies by immunohistochemistry can
effectively diagnose the microsatellite instability (MSI) phenotype
in endometrial carcinomas. Gynecol Oncol. 137:306–310. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Boyarskikh U, Kechin A, Khrapov E,
Fedyanin M, Raskin G, Mukhina M, Kravtsova E, Tsukanov A, Achkasov
S and Filipenko M: Detecting microsatellite instability in
endometrial, colon, and stomach cancers using targeted NGS. Cancers
(Basel). 15:50652023. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Murphy KM, Zhang S, Geiger T, Hafez MJ,
Bacher J, Berg KD and Eshleman JR: Comparison of the microsatellite
instability analysis system and the Bethesda panel for the
determination of microsatellite instability in colorectal cancers.
J Mol Diagn. 8:305–311. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bartley AN, Mills AM, Konnick E, Overman
M, Ventura CB, Souter L, Colasacco C, Stadler ZK, Kerr S, Howitt
BE, et al: Mismatch repair and microsatellite instability testing
for immune checkpoint inhibitor therapy: Guideline from the college
of American pathologists in collaboration with the association for
molecular pathology and fight colorectal cancer. Arch Pathol Lab
Med. 146:1194–1210. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dedeurwaerdere F, Claes KB, Van Dorpe J,
Rottiers I, Van der Meulen J, Breyne J, Swaerts K and Martens G:
Comparison of microsatellite instability detection by
immunohistochemistry and molecular techniques in colorectal and
endometrial cancer. Sci Rep. 11:128802021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lindor NM, Burgart LJ, Leontovich O,
Goldberg RM, Cunningham JM, Sargent DJ, Walsh-Vockley C, Petersen
GM, Walsh MD, Leggett BA, et al: Immunohistochemistry versus
microsatellite instability testing in phenotyping colorectal
tumors. J Clin Oncol. 20:1043–1048. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lorenzi M, Amonkar M, Zhang J, Mehta S and
Liaw KL: Epidemiology of microsatellite instability high (MSI-H)
and deficient mismatch repair (dMMR) in solid tumors: A structured
literature review. J Oncol. 2020:18079292020. View Article : Google Scholar
|
|
15
|
McCarthy AJ, Capo-Chichi JM, Spence T,
Grenier S, Stockley T, Kamel-Reid S, Serra S, Sabatini P and Chetty
R: Heterogenous loss of mismatch repair (MMR) protein expression: A
challenge for immunohistochemical interpretation and microsatellite
instability (MSI) evaluation. J Pathol Clin Res. 5:115–129. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Stelloo E, Jansen AML, Osse EM, Nout RA,
Creutzberg CL, Ruano D, Church DN, Morreau H, Smit VTHBM, van Wezel
T and Bosse T: Practical guidance for mismatch repair-deficiency
testing in endometrial cancer. Ann Oncol. 28:96–102. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shia J: Immunohistochemistry versus
microsatellite instability testing for screening colorectal cancer
patients at risk for hereditary nonpolyposis colorectal cancer
syndrome. Part I. The utility of immunohistochemistry. J Mol Diagn.
10:293–300. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hechtman JF, Rana S, Middha S, Stadler ZK,
Latham A, Benayed R, Soslow R, Ladanyi M, Yaeger R, Zehir A and
Shia J: Retained mismatch repair protein expression occurs in
approximately 6% of microsatellite instability-high cancers and is
associated with missense mutations in mismatch repair genes. Mod
Pathol. 33:871–879. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhou T, Chen L, Guo J, Zhang M, Zhang Y,
Cao S, Lou F and Wang H: MSIFinder: A python package for detecting
MSI status using random forest classifier. BMC Bioinformatics.
22:1852021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Salipante SJ, Scroggins SM, Hampel HL,
Turner EH and Pritchard CC: Microsatellite instability detection by
next generation sequencing. Clin Chem. 60:1192–1199. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Niu B, Ye K, Zhang Q, Lu C, Xie M,
McLellan MD, Wendl MC and Ding L: MSIsensor: Microsatellite
instability detection using paired tumor-normal sequence data.
Bioinformatics. 30:1015–1016. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kautto EA, Bonneville R, Miya J, Yu L,
Krook MA, Reeser JW and Roychowdhury S: Performance evaluation for
rapid detection of pan-cancer microsatellite instability with
MANTIS. Oncotarget. 8:7452–7463. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhu L, Huang Y, Fang X, Liu C, Deng W,
Zhong C, Xu J, Xu D and Yuan Y: A novel and reliable method to
detect microsatellite instability in colorectal cancer by
next-generation sequencing. J Mol Diagn. 20:225–231. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yoshida H, Takigawa W, Kobayashi-Kato M,
Nishikawa T, Shiraishi K and Ishikawa M: Mismatch repair protein
expression in endometrial cancer: Assessing concordance and
unveiling pitfalls in two different immunohistochemistry assays. J
Pers Med. 13:12602023. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bacher JW, Flanagan LA, Smalley RL, Nassif
NA, Burgart LJ, Halberg RB, Megid WM and Thibodeau SN: Development
of a fluorescent multiplex assay for detection of MSI-high tumors.
Dis Markers. 20:237–250. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nakagomi T, Goto T, Hirotsu Y, Shikata D,
Yokoyama Y, Higuchi R, Amemiya K, Okimoto K, Oyama T, Mochizuki H
and Omata M: New therapeutic targets for pulmonary sarcomatoid
carcinomas based on their genomic and phylogenetic profiles.
Oncotarget. 9:10635–10649. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Takaoka S, Hirotsu Y, Ohyama H, Mochizuki
H, Amemiya K, Oyama T, Ashizawa H, Yoshimura D, Nakagomi K, Hosoda
K, et al: Molecular subtype switching in early-stage gastric
cancers with multiple occurrences. J Gastroenterol. 54:674–686.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Johansen AFB, Kassentoft CG, Knudsen M,
Laursen MB, Madsen AH, Iversen LH, Sunesen KG, Rasmussen MH and
Andersen CL: Validation of computational determination of
microsatellite status using whole exome sequencing data from
colorectal cancer patients. BMC Cancer. 19:9712019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yu F, Makrigiorgos A, Leong KW and
Makrigiorgos GM: Sensitive detection of microsatellite instability
in tissues and liquid biopsies: Recent developments and updates.
Comput Struct Biotechnol J. 19:4931–4940. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jia P, Yang X, Guo L, Liu B, Lin J, Liang
H, Sun J, Zhang C and Ye K: MSIsensor-pro: Fast, accurate, and
matched-normal-sample-free detection of microsatellite instability.
Genomics Proteomics Bioinformatics. 18:65–71. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang Y, Shi C, Eisenberg R and
Vnencak-Jones CL: Differences in microsatellite instability
profiles between endometrioid and colorectal cancers: A potential
cause for false-negative results? J Mol Diagn. 19:57–64. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wu X, Snir O, Rottmann D, Wong S, Buza N
and Hui P: Minimal microsatellite shift in microsatellite
instability high endometrial cancer: A significant pitfall in
diagnostic interpretation. Mod Pathol. 32:650–658. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tan WCC, Nerurkar SN, Cai HY, Ng HHM, Wu
D, Wee YTF, Lim JCT, Yeong J and Lim TKH: Overview of multiplex
immunohistochemistry/immunofluorescence techniques in the era of
cancer immunotherapy. Cancer Commun (Lond). 40:135–153. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ali AS and Alalem LS: Next-generation
sequencing and immunohistochemistry approaches for microsatellite
instability detection in endometrial cancer. Cell Mol Biol
(Noisy-le-grand). 69:237–242. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Bonneville R, Krook MA, Chen HZ, Smith A,
Samorodnitsky E, Wing MR, Reeser JW and Roychowdhury S: Detection
of microsatellite instability biomarkers via next-generation
sequencing. Methods Mol Biol. 2055:119–132. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Evrard C, Cortes U, Ndiaye B, Bonnemort J,
Martel M, Aguillon R, Tougeron D and Karayan-Tapon L: An innovative
and accurate next-generation sequencing-based microsatellite
instability detection method for colorectal and endometrial tumors.
Lab Invest. 104:1002972024. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Pang J, Gindin T, Mansukhani M, Fernandes
H and Hsiao S: Microsatellite instability detection using a large
next-generation sequencing cancer panel across diverse tumour
types. J Clin Pathol. 73:83–89. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Felinger A: 8 Peak detection. Data Handl
Sci Technol. 21:183–190. 1998.
|
|
39
|
Spellerberg IF and Fedor PJ: A tribute to
claude shannon (1916–2001) and a plea for more rigorous use of
species richness, species diversity and the ‘shannon-wiener’ index.
Glob Ecol Biogeogr. 12:177–179. 2003. View Article : Google Scholar
|