Introduction
According to data from 2024, colorectal cancer (CRC)
is the second leading cause of cancer-related mortality worldwide
and is the third most frequently diagnosed cancer (1). The development of CRC is a complex,
multistep process influenced by a combination of environmental and
genetic factors (2). Key risk
factors include advanced age, family history, inflammatory bowel
disease, dietary habits and lifestyle factors such as smoking and
physical inactivity (3). The
incidence and mortality rates of CRC vary globally, with higher
rates typically observed in developed countries (4); these differences may be due to
lifestyle variables, such as diets heavy in fat and poor in fiber,
a lack of physical activity and greater screening rates that result
in more frequent diagnoses. Despite advances in screening programs
and the introduction of various therapeutic interventions,
including radiation, chemotherapy, surgery and targeted therapy,
CRC remains a significant public health issue. Patient prognosis
largely depends on the tumor stage and molecular profile of the
cancer (5). Although progress has
been made, many questions remain unanswered regarding the
understanding of the molecular mechanisms of CRC, particularly in
terms of early diagnosis and therapeutic strategies. For example,
aberrant activation of the Wnt/β-catenin signaling pathway is a
common molecular event in CRC, but its specific role in disease
progression has not been fully elucidated (6). The heterogeneity of CRC presents
challenges in achieving optimal clinical outcomes, emphasizing the
need for innovative diagnostic biomarkers, therapeutic strategies
and prognostic indicators to enhance patient management and
outcomes in CRC.
Lectin galactoside-binding soluble 4
(LGALS4), a member of the galactose lectin family, serves a
role in a variety of biological processes (7). In CRC, the expression level of
LGALS4 is closely associated with tumor progression,
invasiveness and drug resistance. It has been reported that
LGALS4 expression is high in normal colonic epithelial cells
and significantly lower in CRC tissues, suggesting that it may
function as a tumor suppressor (7).
Loss of function of LGALS4 has been associated with enhanced
proliferation, migration and invasion of tumor cells (7). At the molecular level, LGALS4
is able to induce cell cycle arrest by regulating the levels of
cell cycle-related proteins, such as Cyclin D1, p21 and p15,
thereby inhibiting the proliferation of tumor cells (8). In addition, LGALS4 can inhibit
tumor development by affecting the Wnt/β-catenin signaling pathway,
an aberrant activation pathway commonly found in CRC (9). Increased expression of LGALS4
levels reduces the expression level of β-catenin and upregulates
inhibitory factors of the Wnt signaling pathway, such as Ephrin B1,
which suppresses the proliferation and invasion of tumor cells. At
the clinical level, the expression level of LGALS4
correlates with the prognosis of patients with CRC and the decrease
in its expression may predict the severity of the disease and the
response to treatment (10). In
addition, the expression level of LGALS4 correlates with
tumor resistance to the chemotherapeutic agent oxaliplatin,
suggesting its potential for therapeutic application (10).
Glycolysis, a crucial metabolic pathway, produces
pyruvate from glucose conversion while producing ATP and NADH
(11). In CRC, glycolysis is
significantly upregulated, an occurrence known as the Warburg
effect, where cancer cells predominantly rely on aerobic glycolysis
even when oxygen is abundant (12).
This metabolic reprogramming supports rapid cell proliferation and
tumor growth, making glycolysis a focal point in CRC research. Zuo
et al (13) investigated the
impact of the long non-coding RNA maternally expressed gene 3
(MEG3) on glycolysis in CRC, reporting that MEG3 suppresses
glycolysis by promoting the ubiquitin-dependent degradation of
c-Myc, a key regulator of glycolytic genes. Overexpression of MEG3
significantly reduces glycolysis, glycolytic capacity and lactate
production in CRC cells; conversely, MEG3 knockdown produces the
opposite result. Additionally, MEG3 activation by vitamin D
suggests a potential therapeutic value in CRC treatment through
glycolysis modulation. Similarly, Zhu et al (14) reported that microRNA (miR)-146b-5p
enhances cell proliferation, glycolysis and invasiveness in CRC by
specifically targeting the pyruvate dehydrogenase E1 subunit b.
Overexpression of miR-146b-5p enhanced these processes, while
knockdown inhibited them, underscoring its oncogenic role. Zhu
et al (15) identified five
glycolysis-related genes (enolase 3, glypican 1, prolyl
4-hydrxylase subunit a 1, sperm associated antigen 4 and
stanniocalcin 2) that form a prognostic model, highlighting the
significant influence of aerobic glycolysis on CRC development.
These studies underscore the critical role of glycolysis and its
potential for CRC therapy, offering valuable insights into tumor
metabolism and potential therapeutic strategies.
Despite the established role of glycolysis in CRC
and the known tumor-suppressive function of LGALS4, the
precise mechanisms by which LGALS4 regulates glycolysis and
its potential as a therapeutic target in CRC remain understudied.
The present study aimed to address these gaps by investigating the
role of LGALS4 in modulating glycolysis and its interplay
with the β-catenin signaling pathway in CRC cells. These findings
introduced a novel perspective for CRC treatment, highlighting the
potential of LGALS4 as a therapeutic target and prognostic
indicator, which has not been extensively reported in previous
research.
Materials and methods
Data acquisition and analysis of
differentially expressed genes (DEGs)
COAD samples from the TCGA were obtained via the
Clinical Bioinformatics Assistant website (https://www.aclbi.com/static/index.html#/). The
GSE26571CRC microarray dataset was obtained from the GEO database
(https://www.ncbi.nlm.nih.gov/gds/).
The TCGA-COAD dataset comprised 455 tumor samples and 41 normal
control samples, while the GSE26571 dataset included 12 CRC samples
and 5 control samples. Probe IDs were transformed into gene symbols
and differential expression analysis was employed using the ‘Limma’
package (version 3.46.0) in R (version 4.1.2; Posit Software).
According to previous literature, genes with a fold change (FC)
>2 were identified as upregulated DEGs and those with an FC
<0.5 as downregulated DEGs (16,17).
Identification and enrichment analysis
of overlapping DEGs
The overlapping DEGs were subjected to Gene Ontology
(GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment
analyses using the Database for Annotation, Visualization and
Integrated Discovery. The GO analysis included the biological
process (BP), molecular function (MF) and cellular component (CC)
categories to comprehensively explore the functional roles of the
identified overlapping DEGs.
Construction of prognostic risk model
for overlapping DEGs
The Least Absolute Shrinkage and Selection Operator
(LASSO) Cox regression model was used to identify genes associated
with patient prognosis. The λ parameter in the LASSO model was used
to adjust the strength of selection of the variables in the model
to prevent overfitting. Smaller values of λ, which allowed more
variables to enter the model but may lead to excessive model
complexity and larger values of λ, which increased the penalization
strength and retained only the most significant variables,
simplified the model structure. The optimal λ value, λ.min=0.0382
was determined, through 10-fold cross-validation to ensure that the
model achieved the best balance between predictive accuracy and
simplicity. This process of optimizing λ enabled the model to
select only those genes with the strongest associations with
patient survival outcomes, which included ribosomal protein S17
(RPS17), G2/mitotic-specific Cyclin B1 (CCNB1),
karyopherin subunit a2 (KPNA2), TNF receptor associated
protein 1 (TRAP1), complement C8 chain (C8G),
LGALS4, lectin galactoside-binding soluble-2
(LGALS2), carbonic anhydrase 4 (CA4), and solute
carrier family 6 member 8 (SLC6A8). The risk score was
calculated using the following formula: Risk score=∑(coefficient ×
gene expression). Where the coefficient represented the coefficient
of each gene in the LASSO model and gene expression was the
expression level of the corresponding gene in the sample. The risk
score showed the effect of each gene on patient prognosis, with
positive coefficients indicating positive correlation and negative
coefficients indicating negative correlation. A model capable of
assessing the prognostic risk of patients was constructed, with
high risk scores predicting poorer prognostic outcomes. The score
of risk for each sample was calculated according to the following
formula: Risk score=(0.1219) × RPS17 + (−0.0582) ×
CCNB1 + (−0.0442) × KPNA2 + (−0.0137) × TRAP1
+ (0.0885) × C8G + (−0.1236) × LGALS4 + (−0.009) ×
LGALS2 + (−0.0063) × CA4 + (0.0012) × SLC6A8.
The COAD cohort from the TCGA database were stratified into low-
and high-risk groups according to the expression patterns of the
aforementioned genes. Kaplan-Meier (KM) analysis was employed to
ascertain the overall survival (OS) probability for the two risk
groups. The median survival time was calculated and the statistical
significance of survival differences between the groups was
evaluated using the log-rank test. To clarify relative risk, hazard
ratios (HRs) were calculated for the high-risk group. Additionally,
Receiver Operating Characteristic (ROC) curves were generated using
the timeROC package (https://CRAN.R-project.org/package=timeROC) and the
Area Under the Curve (AUC) values were calculated to evaluate the
predictive ability of the prognostic models for patient survival at
1, 3 and 5 years. Higher AUC values indicated stronger prognostic
prediction capabilities.
Construction of prognostic nomogram
and expression analysis of prognostic significant genes
Univariate and multivariate Cox regression analyses
were performed on signature genes and specific clinical predictive
variables, such as patient age and sex, using the ‘forestplot’
package (version 2.0.1; http://cran.r-project.org/web/packages/forestplot/vignettes/forestplot.html).
For each variable, the HRs, 95% CIs and P-values were calculated.
Key prognostic factors were identified based on variables with
P<0.05. Nomograms were created to predict the 1-, 3- and 5-year
survival probabilities using the rms software (version 3.6.1;
https://cran.r-project.org/web/packages/rms/index.html;
provided by R Foundation for Statistical Computing) (18). The consistency index (C-index) was
determined to assess the prediction accuracy of the model. The
performance of the model was then assessed by predicting survival
curves for 1, 3 and 5 years and by generating a calibration curve
to evaluate the accuracy of these predictions. The expression
levels of C8G, LGALS4 and RPS17 in different samples
were examined in the TCGA-COAD and GSE26571 datasets using the
SangerBox platform (version 3.0; http://vip.sangerbox.com/home.html).
Cell lines and culture
Based on previous literature, the LoVo, HCT-116 and
SW480 cell lines were selected and NCM460 cells were used as the
control cell line (19–21). LoVo, HCT-116 and SW480 cells were
obtained from the Cell Bank of the Chinese Academy of Sciences.
NCM460 cells were purchased from Jennio Biotech Co., Ltd. All cells
were cultured in DMEM, 1% penicillin-streptomycin and 10% FBS
(Gibco; Thermo Fisher Scientific, Inc.). Cells were maintained at
37°C with 5% CO2 and 20% O2.
Vector construction and
transfection
Total RNA was isolated from NCM460 cells using the
TRIzol® kit (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. cDNA was synthesized
using the PrimeScript RT kit (Takara Bio Inc.). cDNA was amplified
using the CloneAmp™ HiFi PCR Premix (Takara Bio, Inc.) to amplify
the corresponding LGALS4 fragment, which was cloned into the
pcDNA™3.1(+) vector (Thermo Fisher Scientific, Inc.).
The correct insertion of the gene was verified by DNA sequencing
and the recombinant plasmid was transformed and amplified in
Escherichia coli to prepare plasmid DNA for cell
transfection. For transfection, 1 µg plasmid DNA was used per well,
and transfection was performed at 37°C for 24 h. The LGALS4
overexpression plasmid was introduced into LoVo and HCT-116 cells
using Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) as the transfection reagent, and cells were
incubated for 48 h post-transfection before further
experimentation. Cells stably expressing LGALS4 were
screened by resistance. The selection method involved puromycin at
a concentration of 1 µg/ml for selection and 0.5 µg/ml puromycin
for maintenance. The amplification primer sequences used were as
follows: Forward (F), 5′-CTCGAGATGGCCTATGTCCCC-3′ and reverse (R),
5′-TCTAGATTAGATCTGGACATAGGACAAGG-3′.
Cell treatment
CRC cells were treated with various agents to
investigate their effects on glucose metabolism and cell viability.
Glucose was administered at concentrations of 10.0, 1.0 and 0.5 mM.
To inhibit glucose transport and hexokinase activity, cells were
treated with cytochalasin B (Cyto-B) at 20 µM and 3-bromopyruvate
(3-BrPA) at 10 µg/ml. Additionally, the 5-fluorouracil (5-FU)
anticancer drug was used at 50 µg/ml as an and XAV-939, a β-catenin
inhibitor, was applied at 10 µM to examine its impact on CRC cells.
All treatments were administered for 48 h.
Reverse transcription-quantitative PCR
(RT-qPCR)
TRNzol reagent (cat. no. DP424; Tiangen Biotech Co.,
Ltd.) was used to extract the total RNA from CRC cells following
the manufacturer's instructions. cDNA synthesis was performed using
a PrimeScript RT kit (Takara Bio Inc.). RT-qPCR was conducted on
the StepOnePlus Real-Time PCR System (Applied Biosystems; Thermo
Fisher Scientific, Inc.) using SYBR Green PCR Master Mix (Takara
Biotechnology Co., Ltd.). The thermocycling conditions were as
follows: Initial denaturation at 95°C for 10 min, followed by 40
cycles of denaturation at 95°C for 15 sec, annealing at 60°C for 30
sec and extension at 72°C for 30 sec. Gene expression levels were
quantified, normalized to GAPDH and calculated using the
2−ΔΔCq method (22).
Table I lists the primer sequences
for the genes investigated in the present study.
 | Table I.Primer sequences used for reverse
transcription-quantitative PCR. |
Table I.
Primer sequences used for reverse
transcription-quantitative PCR.
| Gene | Sequence
(5′-3′) |
|---|
| LGALS4 | F:
CCGGACATTGCCATCAACAG |
|
| R:
CAAAGCTCTTGCCTGTGGGA |
| BAX | F:
GAGCTAGGGTCAGAGGGTCA |
|
| R:
CCCCGATTCATCTACCCTGC |
| BCL2 | F:
ACCTACCCAGCCTCCGTTAT |
|
| R:
GAACTGGGGGAGGATTGTGG |
| CASP3 | F:
GCTGGATGCCGTCTAGAGTC |
|
| R:
ATGTGTGGATGATGCTGCCA |
| CASP9 | F:
ATTGCACAGCACGTTCACAC |
|
| R:
TATCCCATCCCAGGAAGGCA |
| CTNNB1 | F:
AAAGCGGCTGTTAGTCACTGG |
|
| R:
CGAGTCATTGCATACTGTCCAT |
| MYC | F:
CACCACCAGCAGCGACTCT |
|
| R:
GGCACCTCTTGAGGACCAGT |
| PKM | F:
GACTGCCTTCATTCAGACCCA |
|
| R:
GGGTGGTGAATCAATGTCCAG |
| SLC2A1 | F:
ATGCGGGAGAAGAAGGTCAC |
|
| R:
GTTGACGATACCGGAGCCAA |
| LDHA | F:
GGCCTGTGCCATCAGTATCT |
|
| R:
GGAGATCCATCATCTCTCCC |
| HK2 | F:
CCTGTGAATCGGAGAGGTCC |
|
| R:
ATTTTGGCGTCACAACTGCT |
| GAPDH | F:
ACCACAGTCCATGCCATCAC |
|
| R:
TCCACCACCCTGTTGCTGTA |
Western blot (WB) assay
Protease and phosphatase inhibitors (CoWin
Biosciences) were added to RIPA lysis buffer (Beijing Solarbio
Science & Technology Co., Ltd) to facilitate the preparation of
protein lysates from CRC cells. The BCA Protein Assay Kit (Beyotime
Institute of Biotechnology) was used to measure the protein
concentration. Proteins in equal quantities (20 µg/lane) were
separated using 10% SDS-PAGE and then transferred onto PVDF
membranes (Beyotime Institute of Biotechnology). The membranes were
blocked with 5% skim milk at room temperature for 1 h. After
blocking, the membranes were washed three times with Tris-buffered
saline containing 0.1% Tween-20. They were then incubated with
primary antibodies against LGALS4 (1:1,000; cat. no. ab175185),
Cyclin dependent kinase 1 (CDK1) (1:1,000; cat. no. ab265590),
Cyclin A2 (1:1,000; cat. no. ab227277), Cyclin B1 (1:2,000; cat.
no. ab181593), Bax (1:1,000; cat. no. ab32503), Bcl-2 (1:1,000;
cat. no. ab194583), caspase-9 (1:1,000; cat. no. ab32539),
caspase-3 (1:1,000; cat. no. ab90437), β-catenin (1:1,000; cat. no.
ab227499), c-myc (1:1,000; cat. no. ab32072), solute carrier family
2 member 1 (GLUT1) (1:5,000; cat. no. ab195021), pyruvate kinase M2
(PKM2) (1:1,000; cat. no. ab154816), lactate dehydrogenase A (LDHA)
(1:1,000; cat. no. ab300637) and hexokinase 2 (HK2) (1:1,000; cat.
no. ab209847) (all from Abcam) overnight at 4°C. After rinsing, the
membranes were incubated with secondary antibodies conjugated with
HRP (1:5,000; cat. no. D110011-0100; Sangon Biotech Co., Ltd) for 1
h at room temperature. GAPDH (1:5,000; cat. no. KC-5G4; Aksomics
Inc.) was used as an internal reference. Protein bands were
visualized using an ECL kit (Tiangen Biotech Co., Ltd.) and imaged
using a ChemiDoc system (Bio-Rad Laboratories, Inc.). Densitometric
analysis was performed using ImageJ software (version 2.0.0;
National Institutes of Health).
Cell counting kit-8 (CCK-8) assay
The CCK-8 assay kit (cat. no. KGA317s; Nanjing
KeyGen Biotech Co., Ltd) was used to evaluate cell viability. CRC
cells were seeded in 96-well plates at a density of
5×103 cells/well. Following transfection with either the
vector control or LGALS4 overexpression plasmid, CCK-8
reagent was added to each well and incubated for 2 h at 37°C. A
microplate reader (Shanghai Kehua Bio-Engineering Co., Ltd.) was
used to measure the absorbance of each sample at 450 nm after 0, 1,
2, 3, 4 and 5 days.
Cell invasion and migration
assays
Transwell assays were used to assess cell invasion
and migration. Transfected CRC cells (5×104 cells) were
suspended in serum-free DMEM in the upper chamber of the plate.
Subsequently, DMEM supplemented with 10% FBS was added to the lower
chamber of the plate. Cells were incubated at 37°C in a 5%
CO2 incubator for 24 h to allow migration. After the
incubation period, cells that had migrated to the underside of the
Transwell membrane were fixed using 4% paraformaldehyde at room
temperature for 15 min. Cells were then stained using DAPI for 10
min at room temperature to visualize their nuclei. Finally, the
number of migrating cells in the field of view were visualized
using an inverted fluorescence microscope. For the cell invasion
experiments, the top chamber of the Transwell was coated with
Matrigel (BD Biosciences) at 37°C for 20 min, as previously
reported (23).
Flow cytometry
For the assessment of cellular phenotypes via flow
cytometry, CRC cells were enzymatically dissociated using 0.25%
trypsin-EDTA (Thermo Fisher Scientific, Inc.) at 37°C for 5 min to
obtain a single-cell suspension. The cells were then lysed with 100
µl ice-cold RIPA buffer (containing protease and phosphatase
inhibitors) on ice for 30 min at 4°C to ensure complete cell lysis.
The cells were then subjected to dual staining with propidium
iodide (PI) for DNA content analysis and annexin V conjugated to
FITC (BD Biosciences) for the detection of apoptotic cells,
following the manufacturer's recommended protocols. Staining was
performed at room temperature for 15 min in the dark. This
procedure allowed for the discrimination of viable, early apoptotic
and necrotic cells based on their distinct fluorescence
characteristics. The PI staining also facilitated the evaluation of
the cell cycle distribution by reflecting the DNA content at
various stages (24–26). For cell cycle analysis, cells were
also treated with RNase A (10 µg/ml) for 15 min at 37°C to ensure
accurate DNA content analysis. The stained cells were analyzed
using a CytoFLEX flow cytometer (Beckman Coulter, Inc.) and the
resulting data were processed with FlowJo software (version 10.6.0;
FlowJo LLC)to quantify the apoptotic rate and to determine the cell
cycle phase distribution. The percentage of cells in the G1, S and
G2 phases were calculated and statistically compared across
different experimental conditions [transfection with either
LGALS4 overexpression plasmid or vector control, followed by
treatment with 5-FU (5.0 µM) or 0.5 mM glucose] to assess the
modulation of cell cycle progression by the treatments
administered.
Glucose uptake assay
2-[N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino]-2-deoxyglucose
(2-NBDG; cat. no. 186689-07-6; Anjiekai Biological Medicine
Technology) was used as a glucose tracer. Cells were seeded at
1×105 cells/well in 6-well plates in four replicates and
incubated overnight at 37°C and 5% CO2. The following
day, cells were subjected to glucose starvation for 4 h. Then, one
well was treated with DMEM containing glucose as a negative
control, while the other wells were incubated with 2-NBDG for 2 h
at 37°C. After the incubation, cells were collected by
trypsinization, followed by centrifugation at 4°C for 5 min at 300
× g, and rinsed twice with PBS. Subsequently, flow cytometry was
performed using a CytoFLEX flow cytometer). The geometric mean
fluorescence intensity of the cells was measured, with emission at
530 nm and excitation at 488 nm. Data analysis was conducted using
FlowJo software, version 10.6.0.
Colony formation assay
A colony formation assay was used to assess the
colony formation capacity of cells. Cells were seeded at a
concentration of 2×103 in 60 mm plates and cultured for
2 weeks at 37°C with 5% CO2, under varying glucose
concentrations (10.0, 1.0 and 0.5 mM). After fixation in methanol
at room temperature for 30 min, cells were stained using alkali
nitro tetrazolium blue chloride at room temperature for 20 min. A
chemiluminescence imager (Clinx) was used to image the cells, and
the colonies were counted manually. Colonies were defined as cell
clusters containing ≥50 cells. Each experiment was performed thrice
to ensure accuracy.
Lactate and ATP assays
To measure lactate production, 1×105
cells/well were seeded in 24-well plates and cultured at 37°C with
5% CO2 for 24 h. This process was repeated three times.
For ATP production measurement, 1×105 cells/well were
seeded in 96-well plates and incubated at 37°C with 5%
CO2 for 24 h in five replicates. Subsequently, the
medium in both assays was replaced with DMEM containing 1 mM
glucose and the cells were incubated overnight at 37°C with 5%
CO2. The following day, lactate in the culture media was
measured using a commercial lactate assay kit (cat. no. A109-2;
Nanjing Jiancheng Bioengineering Institute). The relative ATP
concentration was also measured using a commercial ATP assay kit
(cat. no. S0026; Beyotime Institute of Biotechnology). Both kits
were used according to the manufacturers' instructions.
Statistical analysis
Statistical analysis was conducted using R software.
Each experiment was carried out in triplicate and the results were
presented as the mean ± SD. For survival curve analysis,
Kaplan-Meier analysis was used to evaluate the overall survival,
and the statistical significance between survival curves was
assessed using the log-rank test. Differences between groups were
evaluated using a one-way ANOVA when there were more than two
experimental groups, followed by post-hoc analysis with Tukey's
test. When comparing only two groups, an unpaired Student's t-test
was applied P<0.05 was used to indicate a statistically
significant difference.
Results
Identification and enrichment analysis
of overlapping DEGs in TCGA-COAD and GSE26571 datasets
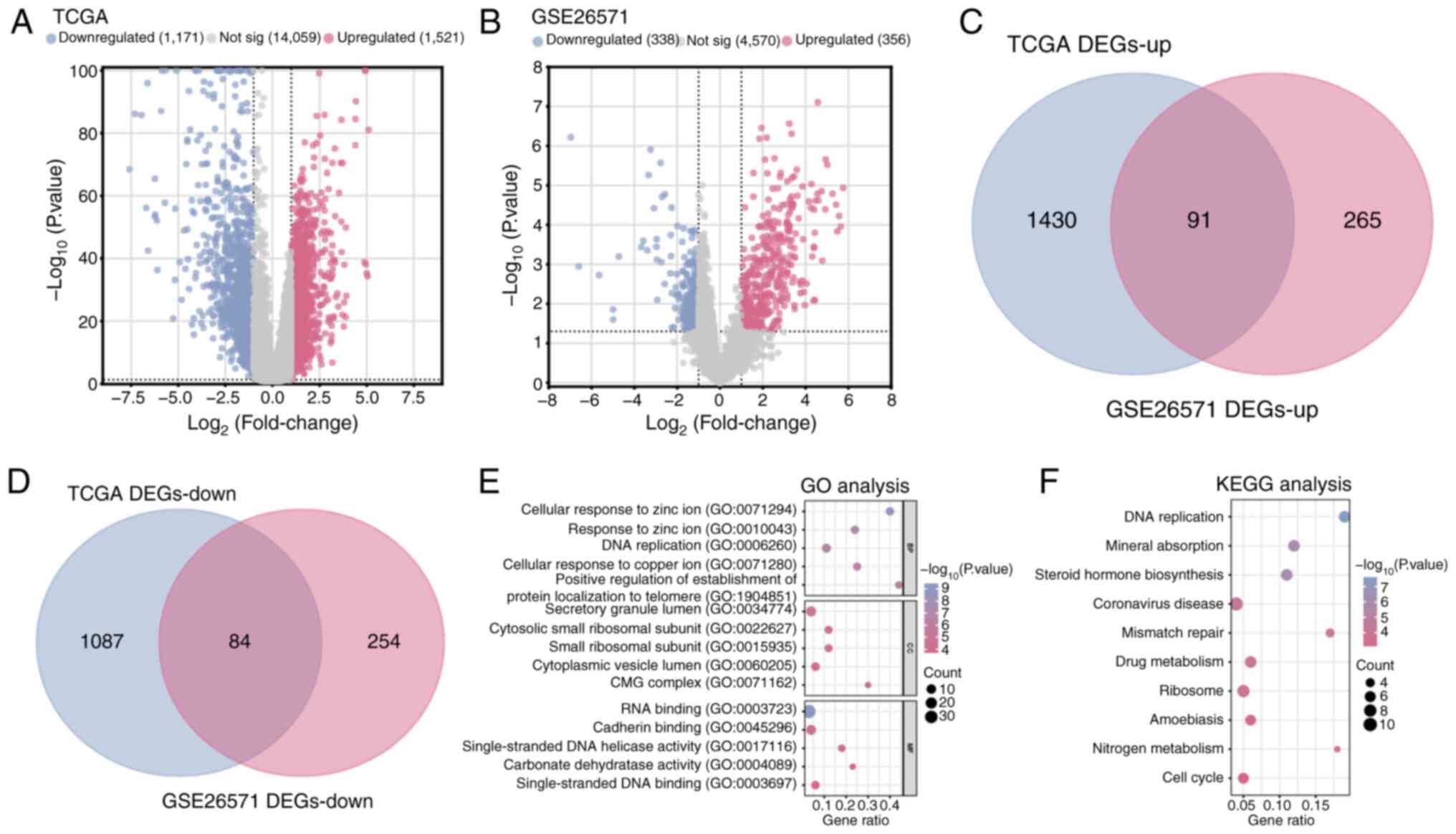
A total of 1,171 downregulated DEGs and 1,521
upregulated DEGs were identified in COAD samples compared with
normal samples in the TCGA database (Fig. 1A). From the GSE26571 dataset, 356
upregulated DEGs and 338 downregulated DEGs were identified in
colon cancer samples compared with normal samples (Fig. 1B). Subsequently, using
bioinformatics platforms, a cross-analysis of the upregulated and
downregulated DEGs from the TCGA-COAD dataset and the GSE26571
dataset identified a total of 175 overlapping DEGs (Fig. 1C and D). The enrichment analysis
demonstrated significant enrichment in terms related to ‘DNA
replication’ (GO: 0006260, BP), ‘response to zinc ion’ (GO:
0010043, BP), ‘CMG complex’ (GO: 0071162, CC), ‘small ribosomal
subunit’ (GO: 0015935, CC), ‘RNA binding’ (GO: 0003723, MF) and
‘cadherin binding’ (GO: 0045296, MF) (Fig. 1E), as well as pathways involved in
the ‘cell cycle’, ‘DNA replication’ and ‘ribosome’ (Fig. 1F).
Prognostic analysis of nine signature
genes in the risk model
A total of 175 overlapping DEGs (Table SI) were analyzed using the LASSO
Cox regression method. By employing cross-validation to identify
the optimal lambda value of 0.0382, nine genes (Table SII) were identified that served as
significant predictors of patient outcomes (Fig. 2A and B). These genes, identified
through their ability to discriminate between high-risk and
low-risk groups, were characterized by their expression patterns
that strongly associated with survival rates. The high-risk group,
as determined by the risk model, exhibited increased mortality and
diminished overall survival compared with the low-risk group, a
finding further supported by the Kaplan-Meier survival analysis
(Fig. 2C). A median survival time
of 5 years was identified for both groups, with a HR of 2.537,
indicating a pronounced impact on survival probability (Fig. 2D). Moreover, the model's predictive
accuracy was validated by the ROC curve, which demonstrated an area
under the curve of 0.696 at the 5 year mark, signifying a reliable
prediction of survival outcomes. This selection process and
subsequent analysis underscored the robustness of the risk model
and suggested a potential role for the identified genes in CRC
prognosis.
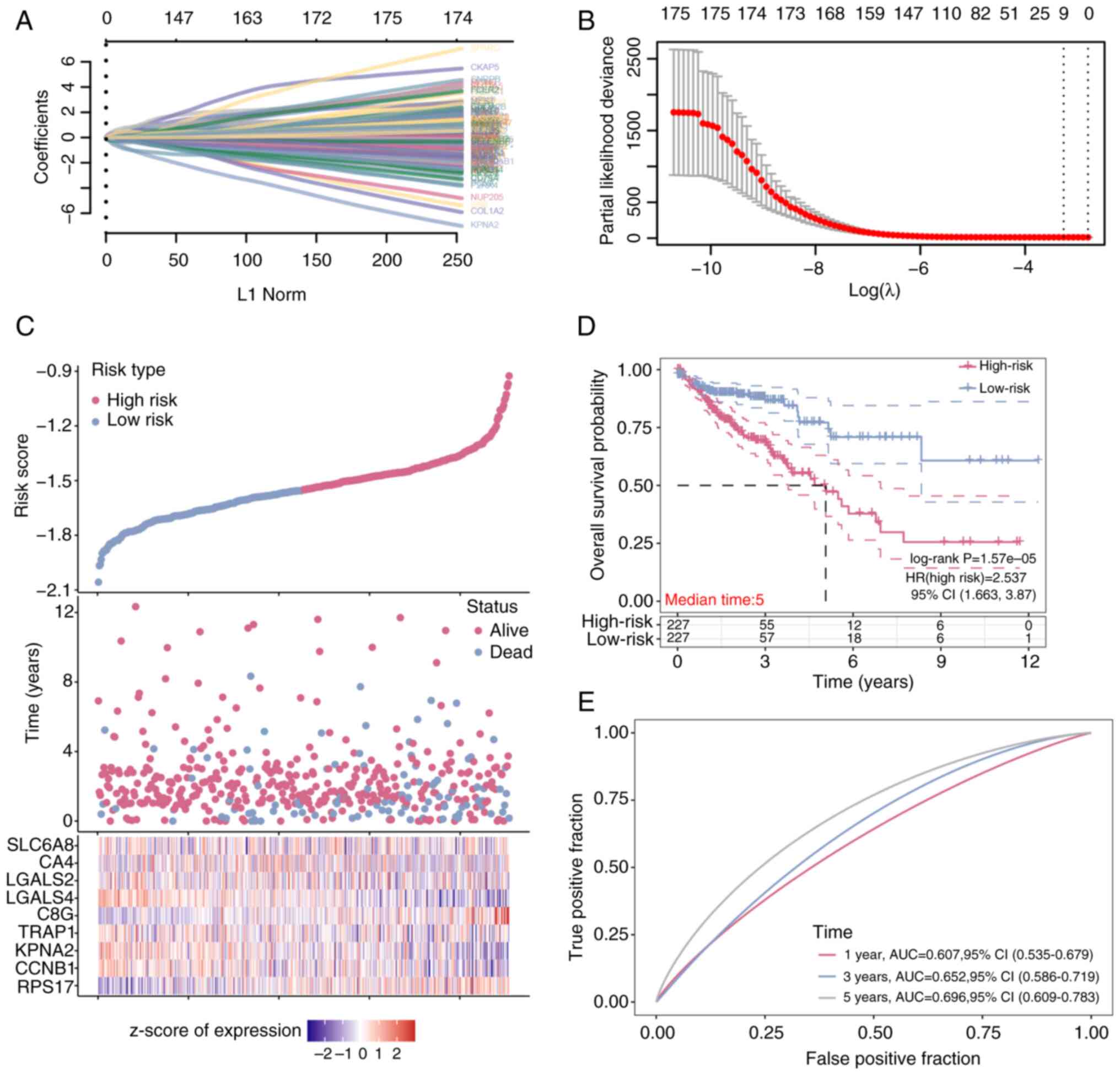 | Figure 2.Prognostic analysis of nine signature
genes in overlapping DEGs. (A) Least Absolute Shrinkage and
Selection Operator-Cox regression model analysis on 175 overlapping
differentially expressed genes. The absolute values of the
coefficients for the different genes were plotted against their
corresponding L1 regularization norm values. (B) The relationship
between 10-fold cross-validation partial likelihood deviation and
log(λ). (C) Risk model analysis of the selected sample data. (D)
Kaplan-Meier survival curve analysis of the high-risk group and the
low-risk group in the risk model (E) Receiver operating
characteristic curve analysis of the risk model at 1, 3 and 5 years
post-treatment. AUC, area under the curve; RPS17, ribosomal
protein S17; CCNB1, G2/mitotic-specific Cyclin-B1;
KPNA2, karyopherin subunit 2; TRAP1, TNF receptor
associated protein 1; C8G, complement C8 chain;
LGALS, lectin galactoside-binding soluble 4; CA4,
carbonic anhydrase 4; SLC6A8, solute carrier family 6 member
8. |
Nomogram analysis of key prognostic
variables and screening of hub genes
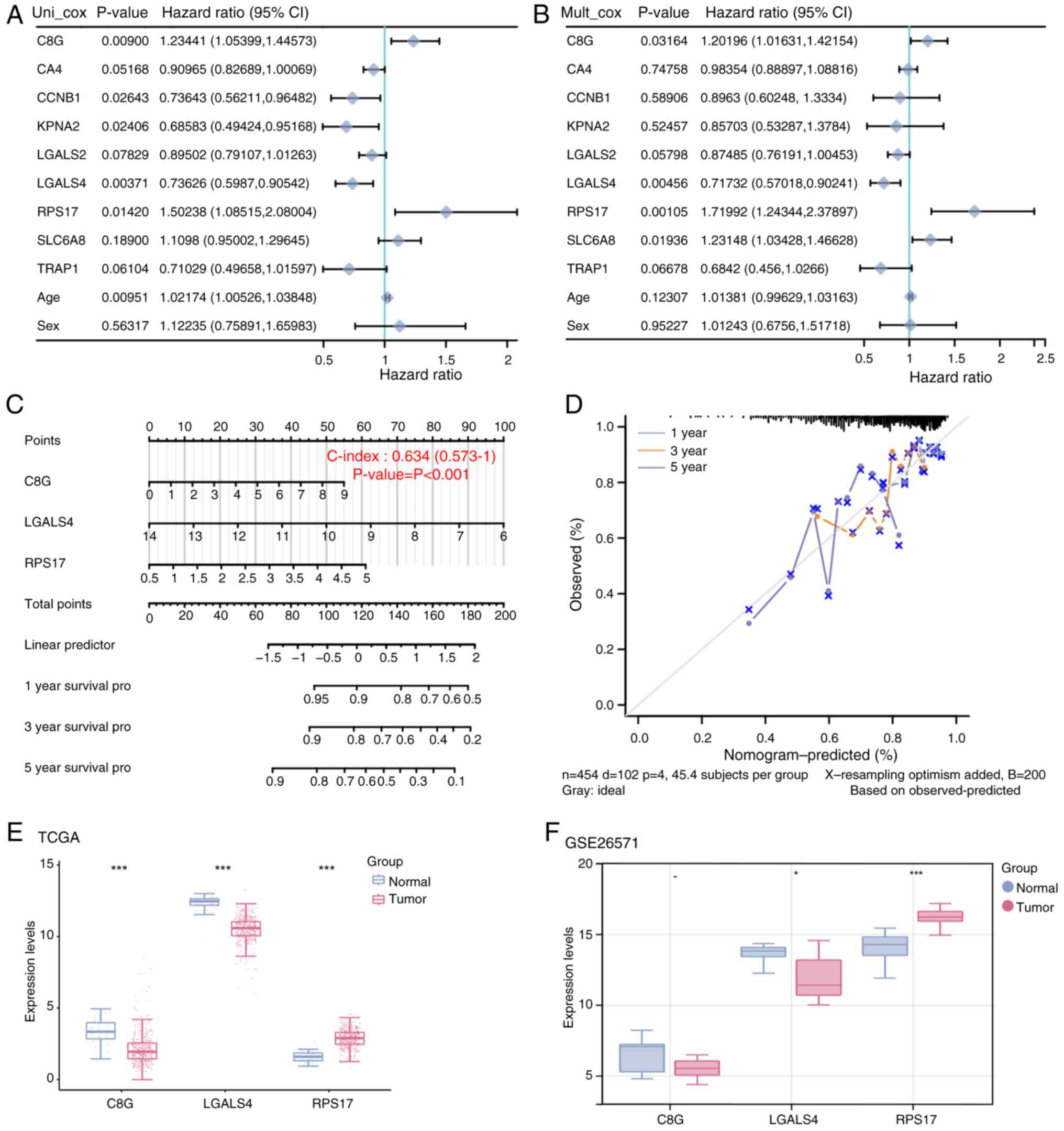
A total of three factors were identified as
statistically significant after analyzing 9 genes and 2 clinical
variables in the risk model: C8G, LGALS4 and RPS17
(Fig. 3A and B). Using these
insights, a predictive model with a C-index of 0.634 was
constructed (Fig. 3C). The
calibration curve showed the highest consistency with model
predictions at 1 year, followed by 3 and 5 years, indicating that
these variables have predictive power for patient survival
(Fig. 3D). The expression of the
three prognostic significant genes was assessed in both the
TCGA-COAD and GSE26571 datasets. These findings indicated that
C8G was significantly under-expressed in tumor samples in
the TCGA-COAD dataset, but was not significantly expressed in the
GSE26571 dataset. LGALS4 was notably under-expressed in the
tumor samples of both datasets, while RPS17 was
significantly over-expressed (Fig. 3E
and F). A previous study reported that LGALS4 is a
potential prognostic factor in CRC patients, but its role in CRC
glycolysis remains unclear (27).
Therefore, LGALS4 was selected as a hub gene for further
investigation.
Overexpression of LGALS4 inhibited the
proliferation, migration and invasion of CRC cells
The expression of LGALS4 was assessed in
normal cells (NCM460) and CRC cells (LoVo, HCT-116 and SW480) using
RT-qPCR and WB. These results demonstrated that LGALS4 was
significantly under-expressed in LoVo and HCT-116 cells compared
with normal cells (Fig. 4A-C).
Therefore, these two cell lines were selected for further
experiments. The RT-qPCR and WB analyses showed efficient
overexpression of LGALS4 in LoVo and HCT-116 cells (Fig. 4D-F). Based on the results of the
CCK-8 assay, on day 5 of cell culture, a significant decrease in
cell viability of cells overexpressing LGALS4 was observed
in the LoVo cell line, which was ~25% compared with the control
cells. For the HCT-116 cell line, the cell viability of cells
overexpressing LGALS4 also showed a significant decrease to
~50% compared with the control (Fig.
4G-H). These results suggested that LGALS4 may inhibit
cell proliferation by modulating the expression of cell
cycle-related proteins or their activities, which in turn inhibits
cell proliferation. The Transwell assays demonstrated that the
invasive and migratory abilities of LoVo cell lines were
significantly decreased upon LGALS4 overexpression compared
with control cells (Fig. 4I-L).
Specifically, the invasion capacity was decreased 4-fold and the
migration capacity was decreased 2.5-fold compared with the
control. A similar effect was demonstrated in the HCT-116 cell
line, in which the invasive capacity was reduced by 3-fold and the
migratory capacity by 2-fold compared with control cells. This
effect may stem from the effects of LGALS4 on cell adhesion,
reorganization of the cytoskeleton or degradation of the
extracellular matrix, which are all critical aspects of the cell
migration and invasion process (28).
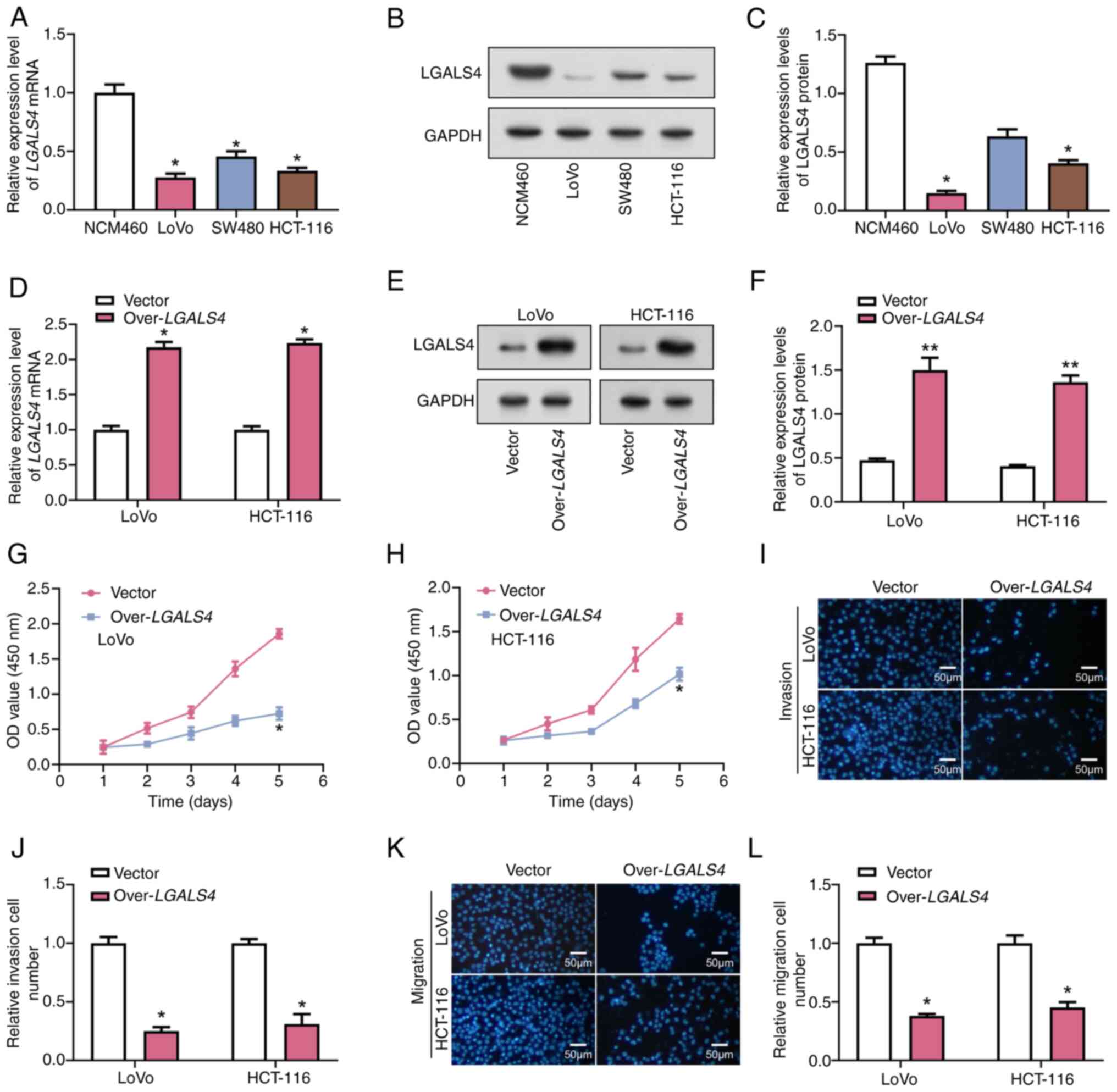 | Figure 4.Overexpression of LGALS4
inhibits the proliferation, migration and invasion of CRC cells.
(A) RT-qPCR was used to detect the expression levels of
LGALS4 in normal cells (NCM460) and CRC cells (LoVo, HCT-116
and SW480). (B) WB analysis was performed to detect the protein
expression levels of LGALS4 in normal cells (NCM460) and CRC cells
(LoVo, HCT-116, SW480). (C) Semi-quantification of the WB analysis
showing the protein expression levels of LGALS4 in normal cells
(NCM460) and CRC cells (LoVo, HCT-116, SW480). *P<0.05 vs.
NCM460. (D) RT-qPCR was used to assess the overexpression
efficiency of LGALS4 in LoVo and HCT-116 cells. (E) WB
analysis further confirmed the overexpression efficiency of LGALS4
in LoVo and HCT-116 cells. (F) Semi-quantification of the WB
analysis further confirming the overexpression efficiency of LGALS4
in LoVo and HCT-116 cells. The Cell Counting Kit-8 assay was used
to detect the effect of LGALS4 overexpression on the
viability of (G) LoVo and (H) HCT-116 cells. The Transwell assay
was used to detect the effect of LGALS4 overexpression on
the (I and J) invasion and (K and L) migration capacities of CRC
cells. *P<0.05, **P<0.01 vs. Vector. RT-qPCR, reverse
transcription-quantitative PCR; WB, western blot; LGALS4,
lectin galactoside-binding soluble 4; over, overexpression; CRC,
colorectal cancer. |
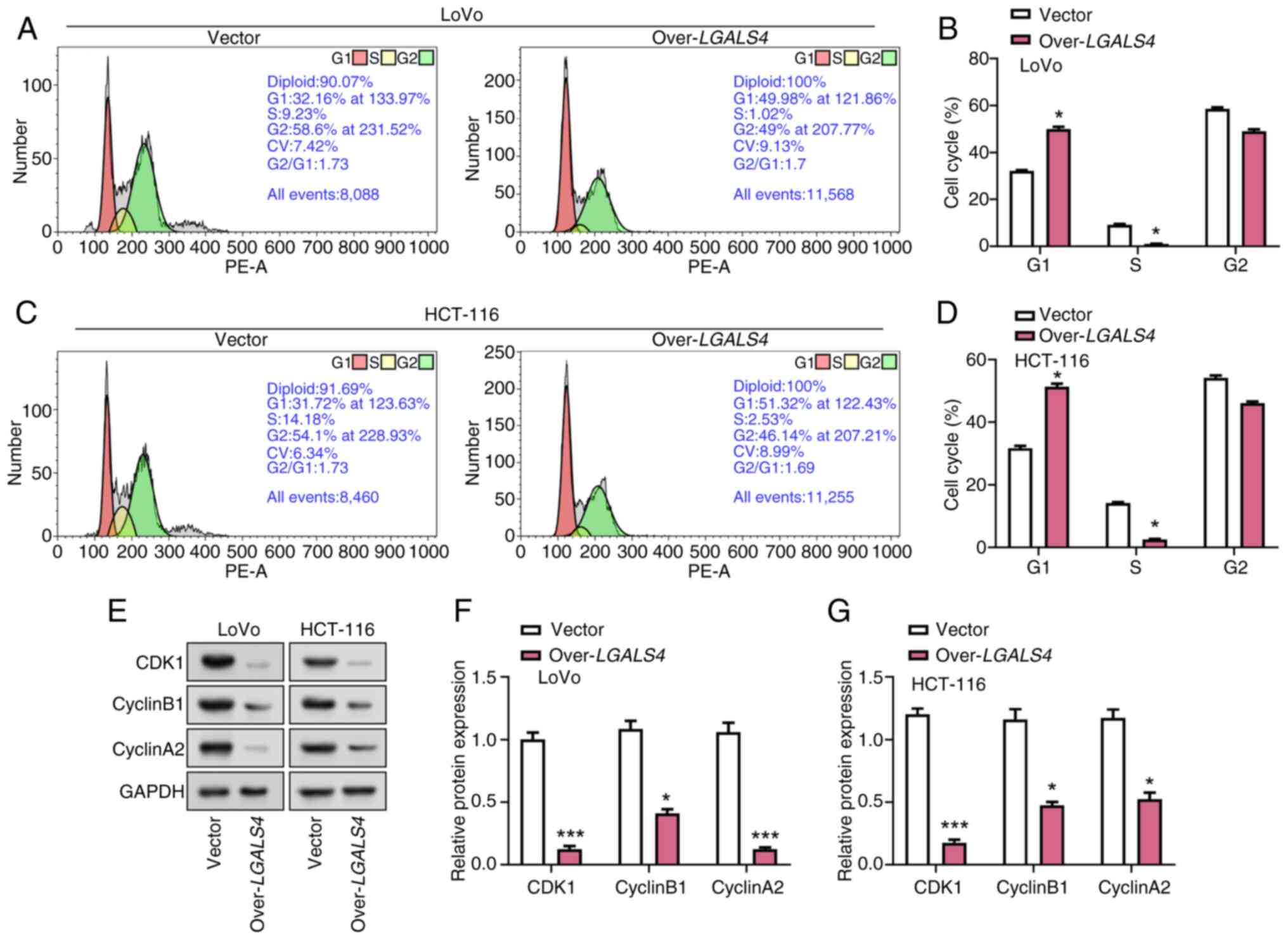
LGALS4 overexpression induces cell
cycle arrest in CRC cells
To explore the effect of LGALS4
overexpression on the cell cycle progression of CRC cells, flow
cytometric analysis was performed on LoVo and HCT-116 cells that
were transfected with either an LGALS4 overexpression vector
or a control vector. These results demonstrated a significant
effect of overexpression of LGALS4 on the cell cycle
distribution of LoVo and HCT-116 cells. Compared with control
cells, the proportion of cells overexpressing LGALS4 was
significantly higher in the G1 phase and significantly lower in the
S phase. In LoVo cells, overexpression of LGALS4 resulted in
a ~1.5-fold increase in the proportion of cells in G1 phase and a
~10-fold decrease in the proportion of cells in S phase compared
with the control cells. For HCT-116 cells, overexpression of
LGALS4 resulted in a ~1.8-fold increase in the proportion of
G1-phase cells and a ~15-fold decrease in the proportion of S-phase
cells compared with control cells (Fig.
5A-D). The expression levels of cell cycle-related proteins
Cyclin B1, CDK1 and Cyclin A2 were further analyzed as these
proteins serve a key role in driving the cell cycle transition from
G1 to S phase (29). These results
demonstrated that LGALS4 overexpression significantly
decreased the protein expression levels of CDK1, Cyclin B1 and
Cyclin A2 in CRC cells compared with the control group (Fig. 5E-G). These results suggested that
LGALS4 overexpression may cause a G1 phase cell cycle arrest
by downregulating the levels of cell cycle regulatory proteins.
Overexpression of LGALS4 promoted CRC
cell apoptosis
Flow cytometry was used to analyze the effect of
LGALS4 overexpression on the apoptosis of LoVo and HCT-116
cells. These results demonstrated that the apoptosis rate of CRC
overexpressing LGALS4 was significantly increased by
~2.5-fold compared with control cells (Fig. 6A-C). RT-qPCR results showed that
LGALS4 overexpression significantly increased the expression
levels of CASP3, BAX and CASP9 and decreased the
expression level of BCL2 (Fig.
6D and E). These results were also confirmed by WB assay
results (Fig. 6F-H).
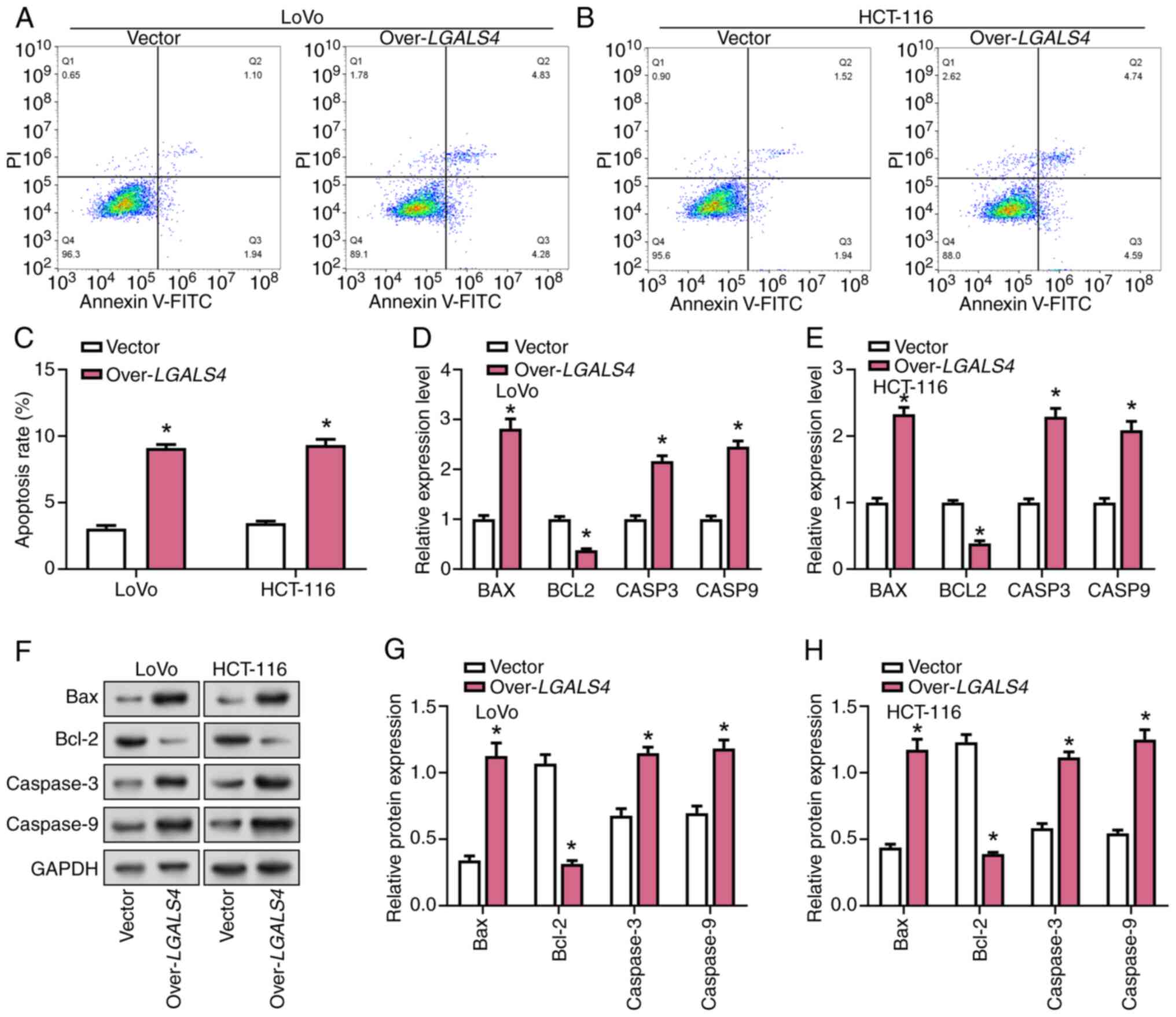 | Figure 6.Overexpression of LGALS4
promotes CRC cell apoptosis. Flow cytometric analysis showing the
effect of LGALS4 overexpression on apoptosis in (A) LoVo and
(B) HCT-116 cells. (C) Quantification of apoptosis in LoVo and
HCT-116 cells following LGALS4 overexpression. RT-qPCR
analysis showing the relative mRNA expression levels of
apoptosis-related genes (BAX, BCL2, CASP3, and CASP9)
in (D) LoVo and (E) HCT-116 cells following LGALS4
overexpression. (F) Western blot analysis showing the protein
expression of Bax, Bcl-2, caspase-3, and caspase-9 in LoVo and
HCT-116 cells following LGALS4 overexpression.
Semi-quantification of protein expression levels of Bax, Bcl-2,
caspase-3 and caspase-9 in (G) LoVo and (H) HCT-116 cells following
LGALS4 overexpression. *P<0.05 vs. vector. CRC,
colorectal cancer; RT-qPCR, reverse transcription-quantitative PCR;
LGALS4, lectin galactoside-binding soluble 4; CASP3,
caspase-3; CASP9, caspase-9. |
Overexpression of LGALS4 inhibited
aerobic glycolysis in CRC
Cancer cells typically rely on higher glucose
concentrations to maintain their rapid glycolytic processes. By
depriving glucose, the nutrient-limited conditions in the tumor
microenvironment can be mimicked and the adaptation and survival of
cancer cells to stressful conditions can be studied (30). Colony formation assays delineated
the glucose dependency of LGALS4-overexpressing cells,
demonstrating an inherent reliance on exogenous glucose for optimal
proliferation. Compared with control cells, LGALS4
overexpressing cells showed enhanced survival under glucose-scarce
conditions, particularly at a glucose concentration of 0.5 mM,
suggesting that they may have acquired metabolic adaptations to the
glucose-deficient microscopic environment (Fig. 7A-D). Subsequent flow cytometric
analysis, utilizing Annexin V-FITC staining, corroborated the
diminished apoptotic propensity of LGALS4-overexpressing CRC
cells subjected to glucose deprivation at 0.5 mM, underscoring
their enhanced survival kinetics relative to control cell
populations (Fig. 7E-G). Cyto-B and
3-BrPA target the initiation steps of the glycolytic pathway such
as glucose uptake and the first phosphorylation reaction,
respectively (31). The use of
these inhibitors could aid in the understanding of the metabolic
adaptations of LGALS4 overexpressing cells when the
glycolytic pathway is inhibited. Targeted inhibition of glucose
transport and hexokinase activity was performed using Cyto-B at a
concentration of 20 µM and 3-BrPA at a concentration of 10 µg/ml,
respectively. The cytotoxic effects of these inhibitors on
LGALS4-overexpressing cells were ascertained through CCK-8
assays following a 48 h incubation. These results demonstrated a
preservation of cell viability among LGALS4-overexpressing
cells, compared with control cells, suggesting the potential
presence of a recalibrated metabolic phenotype conferring
resistance to glycolytic inhibition (Fig. 7H and I). The increased survival of
cyto-B-treated control cells may be due to the fact that the
inhibitory effect of cyto-B on glucose transport did not completely
block the energy supply of the cells, and the cells may be
sustained by other metabolic pathways such as fat oxidation or
amino acid metabolism. In addition, the concentration of cyto-B may
not be sufficient to completely inhibit glucose uptake, or the
cells may be somewhat adapted to cyto-B treatment.
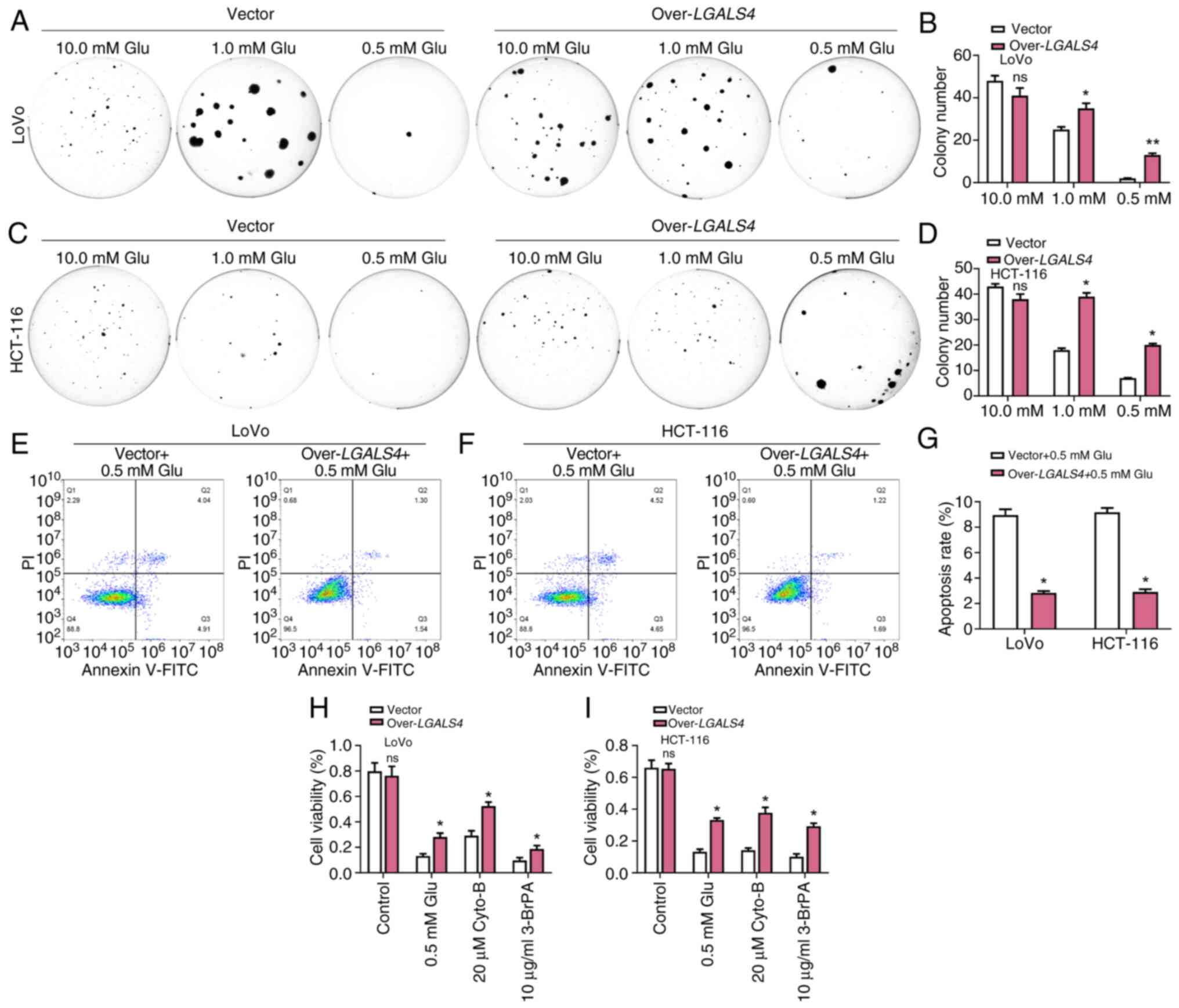 | Figure 7.Overexpression of LGALS4
inhibits aerobic glycolysis in CRC cells. (A) Colony formation
assays were performed on LoVo cells after overexpression of
LGALS4 and treatment with 10.0, 1.0 and 0.5 mM of Glu. (B)
Semi-quantification of colony formation in LoVo cells
overexpressing LGALS4. (C) Colony formation assay showing
the effect of LGALS4 overexpression on HCT-116 cells
following treatment with 10.0, 1.0 and 0.5 mM Glu. (D)
Semi-quantification of colony formation in HCT-116 cells
overexpressing LGALS4. Flow cytometric analysis showing the
apoptosis of (E) LoVo and (F) HCT-116 cells treated with 0.5 mM Glu
after LGALS4 overexpression. (G) Quantification of apoptosis
in LoVo and HCT-116 cells treated with 0.5 mM Glu after
LGALS4 overexpression. Quantification of cell viability in
(H) LoVo and (I) HCT-116 cells overexpressing LGALS4 or a
control vector under treatment with 0.5 mM Glu, 20 µM Cyto-B, and
10 µg/ml 3-BrPA. *P<0.05, **P<0.01 vs. vector. CRC,
colorectal cancer; Glu, glucose; Cyto-B, cytochalasin B; 3-BrPA,
3-Bromopyruvate; LGALS4, lectin galactoside-binding soluble
4; over, overexpression; ns, not significant. |
LGALS4 overexpression enhanced
5-FU-induced apoptosis and inhibited glucose metabolism in CRC
cells
5-FU is an anticancer drug used to treat various
types of cancer, including CRC. It typically inhibits DNA synthesis
by interfering with the biosynthesis of pyrimidine nucleotides
(32). Flow cytometry analysis
showed that overexpression of LGALS4 significantly increased
apoptosis of CRC cells compared with control cells, and the
pro-apoptotic effect was significantly increased when combined with
5-FU treatment compared with the control (Fig. 8A-C). WB analysis demonstrated
significantly increased protein expression levels of apoptosis
markers caspase-9, caspase-3 and Bax, and significantly decreased
protein expression levels of Bcl-2 in CRC cells overexpressing
LGALS4 compared with controls. The effects of LGALS4
overexpression on these markers were significantly enhanced by 5-FU
treatment (Fig. 8D-F).
Additionally, 2-NBDG uptake, ATP production and lactate levels were
measured in LoVo and HCT-116 cells. These results showed that
overexpression of LGALS4 significantly decreased ATP
production, lactate levels and glucose uptake in CRC cells compared
with the control (Fig. 8G-8K).
These results suggest that LGALS4 overexpression may enhance
5-FU-induced apoptosis in CRC cells and disrupt glucose metabolism,
further inhibiting cell viability.
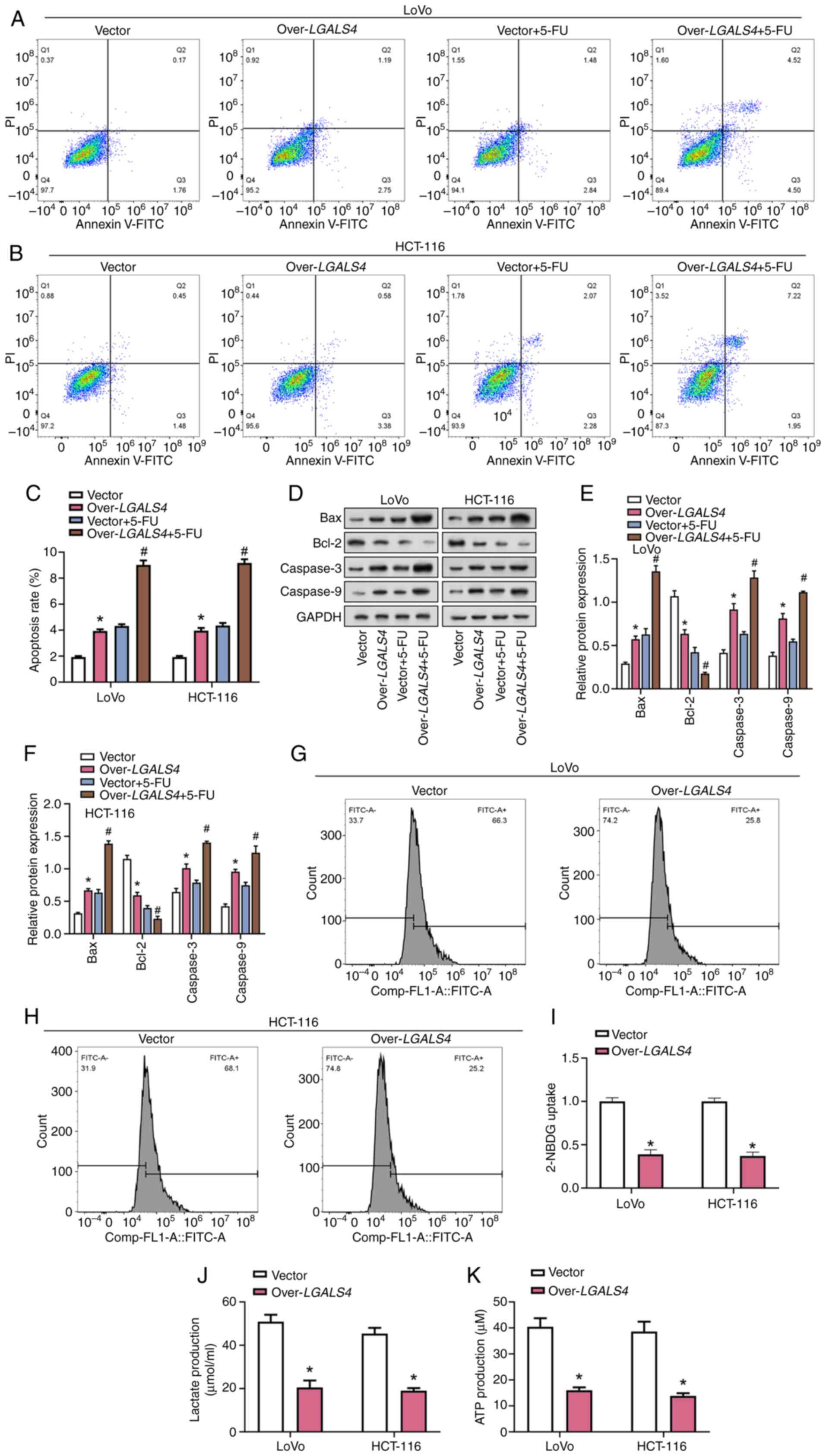 | Figure 8.LGALS4 overexpression enhances
5-FU-induced apoptosis and inhibits glycolysis in colorectal cancer
cells. Flow cytometric analysis was performed to assess apoptosis
in (A) LoVo and (B) HCT-116 cells following LGALS4
overexpression, with or without treatment with 50 µg/ml 5-FU. (C)
Quantification of apoptosis in LoVo and HCT-116 cells following
LGALS4 overexpression, with or without 50 µg/ml 5-FU
treatment. (D) Western blot analysis was performed to detect the
expression of apoptotic proteins (caspase-3, caspase-9, Bax and
Bcl-2) in LoVo and HCT-116 cells following LGALS4
overexpression, with or without 50 µg/ml 5-FU treatment.
Semi-quantification of the western blot analysis results for
apoptotic proteins (caspase-3, caspase-9, Bax and Bcl-2) in (E)
LoVo and (F) HCT-116 cells following LGALS4 overexpression,
with or without 50 µg/ml 5-FU treatment. *P<0.05 vs. vector;
#P<0.05 vs. vector + 5-FU. (G and H) Flow cytometry
was used to measure the uptake of 2-NBDG in (G) LoVo and (H)
HCT-116 cells following LGALS4 overexpression. (I)
Quantification of 2-NBDG uptake in LoVo and HCT-116 cells
transfected with Vector or over-LGALS4. (J) A lactate
detection kit was used to measure lactate release from LoVo and
HCT-116 cells following LGALS4 overexpression. (K) An ATP
detection kit was used to measure ATP production in LoVo and
HCT-116 cells following LGALS4 overexpression. *P<0.05
vs. vector. 5-FU, 5-Fluorouracil; LGALS4, lectin
galactoside-binding soluble 4; over, overexpression; 2-NBDG,
2-[N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino]-2-deoxyglucose. |
LGALS4 modulated β-catenin signaling
to inhibit glycolysis in CRC cells
To ascertain the impact of LGALS4
overexpression on glycolysis-related proteins in CRC cells,
conducted RT-qPCR analysis was performed on CRC cells that were
transfected with either an LGALS4 overexpression vector or a
control vector. These findings showed that overexpression of
LGALS4 significantly downregulated the expression levels of
key glycolysis-related factors, such as CTNNB1, MYC, solute
carrier family 2 member 1 (SLC2A1), pyruvate kinase M1/2
(PKM), HK2 and LDHA compared with control
cells (Fig. 9A and B). WB analysis
showed similar results, with a significant decrease in the
expression levels of these proteins following LGALS4
overexpression compared with the controls (Fig. 9C-E). HCT-116 and LoVo cells were
treated with the β-catenin inhibitor XAV-939, in addition to
inducing LGALS4 overexpression. These results demonstrated
that inhibition of β-catenin significantly enhanced the
downregulation of the glycolysis-related factors that were induced
by LGALS4 overexpression (Fig.
9F-J). These findings suggest that LGALS4 overexpression
may inhibit the expression of key glycolysis-related proteins in
CRC cells and this effect is further potentiated by β-catenin
inhibition. The Wnt/β-catenin signaling pathway serves a central
role in cell proliferation, migration and invasion and
LGALS4 potentially inhibits the malignant behavior of tumor
cells by inhibiting this signaling pathway and reducing the
expression of glycolysis-related genes.
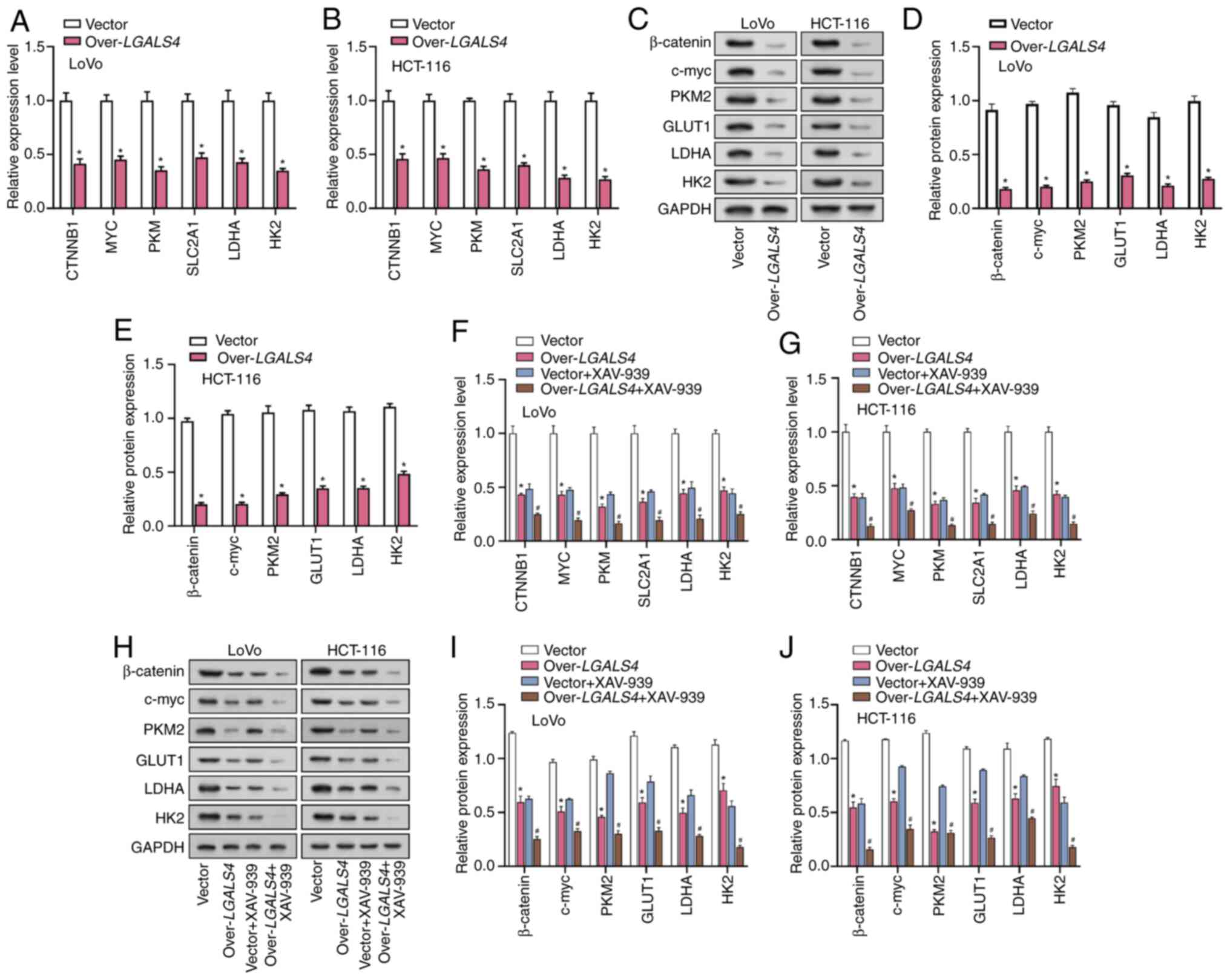 | Figure 9.LGALS4 affects β-catenin
signaling to inhibit glycolysis. RT-qPCR analysis of
glycolysis-related factors, including CTNNB1, MYC, PKM, SLC2A1,
LDHA, and HK2, in (A) LoVo and (B) HCT-116 cells
following LGALS4 overexpression. (C) WB analysis showing the
protein expression of glycolysis-related factors in LoVo and
HCT-116 cells following LGALS4 overexpression.
Semi-quantification of the WB analysis results for
glycolysis-related factors (β-catenin, c-myc, PKM2, GLUT1, LDHA and
HK2) in (D) LoVo and (E) HCT-116 cells following LGALS4
overexpression. *P<0.05 vs. vector. RT-qPCR analysis of
glycolysis-related factors in (F) LoVo and (G) HCT-116 cells
treated with or without the β-catenin inhibitor XAV-939 following
LGALS4 overexpression. (H) WB analysis showing the protein
expression of glycolysis-related factors in LoVo and HCT-116 cells
treated with or without the β-catenin inhibitor XAV-939 following
LGALS4 overexpression. Semi-quantification of the WB
analysis results for glycolysis-related factors in (I) LoVo and (J)
HCT-116 cells treated with or without the β-catenin inhibitor
XAV-939 following LGALS4 overexpression. *P<0.05 vs.
vector; #P<0.05 vs. vector + XAV-939. RT-qPCR,
reverse transcription-quantitative PCR; WB, western blot;
LGALS4, lectin galactoside-binding soluble 4; CTNNB1,
catenin beta 1; MYC, Myc proto-oncogene; PKM,
pyruvate kinase M1/M2; SLC2A1, solute carrier family 2
member 1; CRC, colorectal cancer; PKM2, pyruvate kinase type M2;
GLUT1, solute carrier family 2 member 1; LDHA, lactate
dehydrogenase A; HK2, hexokinase 2; over,
overexpression. |
Discussion
CRC is a multifaceted disease characterized by
genetic and molecular alterations that drive its progression and
impact patient outcomes (33). To
better understand the molecular underpinnings of CRC, the present
study performed a bioinformatics analysis of DEGs using the
TCGA-COAD and GSE26571 datasets. This analysis showed overlapping
DEGs, primarily involved in processes such as DNA replication, cell
cycle and ribosome function. Genes involved in DNA replication and
cell cycle pathways may be associated with rapid proliferation and
tumor development in CRC. CRC is closely associated with aberrant
DNA replication, as evidenced by the prevalence of DNA polymerase
e, catalytic subunit mutations such as p.S297F in colorectal and
endometrial carcinomas (34).
Furthermore, Zurlo et al (35) demonstrated that Cladosporol A
induces G1-phase cell cycle arrest in CRC cells, particularly HT-29
cells, by upregulating the expression of p21(waf1/cip1),
which reduces Cyclin levels and inhibits CDK activity. Zinc is a
cofactor for many enzymes and transcription factors and is
essential for cell proliferation and differentiation (36). Genes responsive to zinc ion levels
may be involved in regulating cellular adaptation to changes in the
microenvironment (37). Ribosomes
are sites of protein synthesis and their biosynthesis is closely
linked to the metabolic requirements of cell growth and tumor cells
(38). Calcineurin is a cell
adhesion molecule involved in cell-cell interactions and
maintenance of tissue structure. Changes in the expression of its
related genes may affect the invasiveness and metastatic ability of
tumor cells (39). Another study by
Zurlo et al (40) reported
that Cladosporol A inhibits CRC proliferation by enhancing the
expression of p21(waf1/cip1) through Sp1-peroxisome
proliferator activated receptor γ interaction. This compound also
induces β-catenin degradation, thereby suppressing the
β-catenin/T-cell factor (TCF) pathway and promoting E-cadherin
expression, which impedes cell cycle progression. Prognostic
analysis of overlapping DEGs identified significant prognostic
genes, including C8G, LGALS4 and RPS17. Expression
analysis demonstrated significantly decreased expression levels of
C8G and LGALS4 in tumor samples from the TCGA-COAD
and GSE26571 datasets. Yu et al (41) identified RPS17 as a hub gene
in the co-expression network of differentially expressed genes in
CRC with microsatellite instability, suggesting its significant
role in the ribosome pathway's involvement in CRC.
LGALS4 is a protein encoded by the LGALS4
gene in humans. It belongs to the galectin family and has the
ability to bind and recognize β-galactoside sugars (42). LGALS4 is primarily expressed
in the gastrointestinal tract, where it serves essential roles in
cell-cell adhesion, epithelial differentiation and mucosal immunity
(43). Its involvement in various
physiological processes, including intestinal homeostasis,
inflammation and cancer progression has previously been reported.
Watanabe et al (44)
reported that elevated levels of circulating Galectin-4 in
patients with CRC correlate with disease progression, suggesting
its potential as a follow-up marker post-surgery. Galectin-1
may be useful for patient screening and Galectin-4 can
complement CEA/CA19-9 in enhancing CRC monitoring. Additionally,
Zhou et al (45)
demonstrated that surface profiles of CRC cells and
tumor-infiltrating lymphocytes from surgical samples align with
prognostic categories and minimal antigenic panels, including
Galectin-4, providing potential predictors for disease
relapse and patient survival. Furthermore, Satelli et al
(7) reported that Galectin-4
acts as a potential tumor suppressor in CRC, with its
downregulation observed in adenomas and invasive carcinomas.
Overexpression induces cell cycle arrest, reduces migration and
sensitizes cells to apoptosis, suggesting its significance in CRC
biology through interaction with Wnt signaling proteins and
downregulation of Wnt target genes. In the present study,
overexpression of LGALS4 limited the capacity of CRC cells
to migrate, proliferate and invade and disrupted CRC cell cycle
distribution. Meanwhile, overexpression of LGALS4 promoted
apoptosis in CRC cells. This suggests that LGALS4 could
potentially serve as both a therapeutic target and a potential
prognostic marker, underscoring its importance in cancer research
and clinical applications.
Glucose is a primary energy source for cells and a
simple sugar. It is vital for cellular metabolism and is
transported into cells through glucose transporters. Once inside
the cell, glucose undergoes glycolysis, a metabolic process known
as the energy investment phase and the energy payback phase, which
produces ATP and NADH by converting glucose to pyruvate (46). Glycolysis, which takes place in the
cytoplasm, consists of 10 enzyme-catalyzed stages. ATP is consumed
during the energy investment phase to phosphorylate glucose and its
intermediates. The energy payback phase involves the production of
NADH and ATP. Glycolysis is an anaerobic process, meaning it does
not require oxygen and is essential for energy production under
both aerobic and anaerobic conditions (47). This pathway is crucial for cells
with high energy demands, such as muscle and cancer cells. Zhou
et al (48) reported that
Dioscin inhibits aerobic glycolysis in CRC cells by
degrading S-phase kinase-associated protein 2 via Cadherin
1, consequently reducing CRC proliferation. Additionally, Zhou
et al (49) showed that
increased expression of PKM2 in CRC promotes aerobic glycolysis,
cell proliferation and migration, suggesting PKM2 may act as a
promising therapeutic target for CRC. Moreover, Zhao et al
(50) reported that Sam68, an
RNA-binding protein, promotes aerobic glycolysis in CRC by
regulating the alternative splicing of PKM2, enhancing glycolysis
and cell proliferation. This underscores the potential of Sam68 as
a target for therapy in CRC. In the present study, overexpression
of LGALS4 significantly enhanced the survival of CRC cells
under low glucose conditions. Although this phenomenon appears
contradictory to the effect of LGALS4 downregulating key
factors of glycolysis, this may potentially be due to the
reprogramming of metabolic pathways in CRC cells facilitated by
LGALS4. LGALS4 may have maintained the metabolic demand and
energy supply of cells under low-glucose conditions either by
activating non-glycolytic energy-generating pathways or by
enhancing the efficiency of cellular utilization of nutrients. In
addition, LGALS4-overexpressing CRC cells exhibited relative
resistance to glucose deprivation and glycolytic inhibition, which
may be related to its effects on cell cycle and apoptotic pathways,
making the cells more tolerant to metabolic stress. LGALS4
may enhance cellular adaptation during glycolytic inhibition by
regulating signaling pathways related to cell survival, such as the
β-catenin signaling pathway. The decreased expression level of
LGALS4 observed in CRC tissues may be related to its role as
a tumor suppressor. Downregulation of LGALS4 expression in a
variety of cancers correlates with a better prognosis, whereas in
the present study, the downregulation of LGALS4 may be
related to metabolic reprogramming of the tumor cells to adapt to
rapid proliferation and to evade immune surveillance. The present
study showed that LGALS4-overexpressing CRC cells exhibited
increased survival under glucose deprivation conditions and showed
tolerance to glycolytic inhibition compared with negative controls.
Flow cytometry results confirmed that LGALS4 significantly
reduced apoptosis induced by glucose deprivation. These findings
suggest that therapeutic pathways targeting aerobic glycolysis may
provide new strategies for CRC treatment in the future.
5-FU is an anticancer drug used to treat various
cancers, such as breast cancer, hepatocellular carcinoma (HCC) and
CRC (51). A previous study by Zou
et al (52) reported that
5-FU induces cytotoxicity and apoptosis in cancer cells through a
ROS-mediated mitochondrial pathway and allicin enhances the
antitumor activity of 5-FU by increasing apoptosis and reducing
mitochondrial membrane potential in HCC cells. Similarly, Zou et
al (53) showed that insulin
pretreatment enhances the anticancer effects of 5-FU in esophageal
and colonic cancer cells by increasing 5-FU uptake, promoting
apoptosis and upregulating the expression of cleaved caspase-3,
thereby inhibiting cell proliferation more effectively.
Additionally, Zuo et al (54) showed that 5-FU inhibits cell
proliferation and induces apoptosis in HepG2 liver cancer cells,
similar to the impacts of chikusetsusaponin IV and V, which also
promote cell cycle arrest and enhance apoptotic protein activation,
underscoring their potential in cancer treatment. The present study
demonstrated that overexpression of LGALS4 promoted
apoptosis and inhibited aerobic glycolysis in CRC cells. Based on
this finding, it was hypothesized that 5-FU may have a synergistic
effect with LGALS4 overexpression by further enhancing the
inhibitory effect on tumor cells. Specifically, LGALS4
overexpression in CRC cells enhances the apoptotic response to 5-FU
treatment and induces metabolic changes, reducing glycolytic
activity and energy production. This may reduce the metabolism and
elimination of 5-FU, thereby accumulating 5-FU in tumor cells and
enhancing its anticancer effect. 2-NBDG, a glucose analog, is used
to measure glucose uptake (55).
Glycolysis, a metabolic process, transforms glucose into energy and
lactate (56). Lactate release is a
product of glycolysis and ATP, the main energy source within the
cell, is produced through glycolysis (57). Therefore, inhibition of glycolysis
by 5-FU will affect ATP production, leading to energy deficiency,
while reducing lactate release and 2-NBDG uptake (58). Zhou et al (59) reported that Gefitinib causes A549
and H1975 non-small cell lung cancer cells to undergo programmed
cell death and inhibit glycolysis, as evidenced by reduced glucose
uptake, lowered ATP levels and increased apoptosis rates.
Additionally, Zu et al (60)
showed that by linking glucose metabolism with lipid synthesis, ATP
citrate lyase (ACL) catalyzes the conversion of citrate to
acetyl-CoA and oxaloacetate by utilizing ATP and CoA. ACL,
overexpressed in various types of cancer, including CRC, serves as
a possible target for therapeutic cancer by disrupting
glycolysis-driven lipogenesis. Moreover, Zuo et al (61) reported that miR-4443 downregulates
TRIM14, suppressing energy metabolism and metastasis in
papillary thyroid carcinoma (PTC). miR-4443 inhibits ATP production
and aerobic glycolysis by targeting TRIM14, indicating its
role in PTC progression and energy regulation. In the present
study, this was evidenced by increased apoptotic markers, decreased
2-NBDG uptake and decreased lactate and ATP levels. These results
suggested that LGALS4 may exert anti-CRC effects by
inhibiting glycolysis.
The β-catenin signaling pathway is essential for a
number of cellular functions, including cell proliferation,
differentiation and migration. β-catenin is phosphorylated by a
destruction complex when Wnt ligands are not present, leading to
its degradation (62). Wnt
activation inhibits the destruction complex, enabling β-catenin to
accumulate and move into the nucleus (63). There, it interacts with
TCF/lymphoid enhancer-binding factor (LEF)
transcription factors to activate target gene expression, such as
Cyclin D1 and c-Myc, promoting cell growth and survival.
Dysregulation of this pathway is associated with various diseases,
including cancer, making it an important target for therapeutic
intervention (64). Zou et
al (65) reported that elevated
circular (circ) RNA circ_0068464 levels in CRC contribute to cell
migration, proliferation and activation of the Wnt/β-catenin
signaling pathway. Its interaction with miR-383 further exacerbates
CRC progression, suggesting therapeutic potential in targeting this
pathway. Similarly, a study by Zou et al (66) showed that increased circCASK in CRC
promotes tumor growth and invasion by upregulating six homeobox 1
expression. Forkhead box c2 transcriptionally induces circCASK
expression, thereby activating the Wnt/β-catenin signaling pathway
and accelerating the development of CRC. In addition, there is a
close connection between β-catenin and glycolysis. Zhou et
al (67) reported that
Dihydrolipoamide S-acetyltransferase (DLAT), a
glycolysis-related gene, is overexpressed in HCC, contributing to
poor prognosis. Its downregulation inhibits PI3K/Akt and
Wnt/β-catenin signaling pathways, highlighting the role of
DLAT as a potential therapeutic target in HCC. Additionally,
Zhou et al (68) reported
that cryptotanshinone suppresses breast cancer cell migration,
invasion and proliferation by targeting glycolysis-related
proteins, such as PKM2. This suggests a potential connection
between β-catenin and glycolysis in breast cancer, suggesting PKM2
may be a promising therapeutic avenue. The interaction between
LGALS4 and the β-catenin signaling pathway may impact the
metabolic properties of CRC cells. Overexpression of LGALS4
may interfere with the intranuclear accumulation of β-catenin or
its interaction with TCF/LEF and inhibit the
transcription of genes downstream of the Wnt signaling pathway such
as c-Myc, PKM2 and GLUT1. In addition, LGALS4 may regulate
the expression of the metabolic enzymes LDHA and HK
and activate the AMPK signaling pathway by altering intracellular
ATP levels or AMP/ATP ratios, thereby inhibiting metabolic enzyme
expression. The effect of LGALS4 on the expression or
function of the glucose transporter protein GLUT1 may reduce
glucose uptake, thereby affecting glucose-dependent metabolic
pathways. As a tumor suppressor protein, overexpression of
LGALS4 inhibits tumor cell growth and metabolism through a
variety of mechanisms, including downregulation of genes closely
related to tumor metabolism. LGALS4-induced cell cycle
arrest and promotion of apoptosis may also indirectly affect the
expression of metabolism-related genes as these cellular processes
are closely related to the metabolic status of cells. The RT-qPCR
and WB analyses in the present study showed that LGALS4
overexpression significantly reduced the levels of
glycolysis-related proteins in CRC cells. When CRC cells were
subjected to 10 µM β-catenin inhibitor XAV-939 for 72 h, the
expression levels of glycolysis-related proteins further decreased.
This indicated that LGALS4 may affect β-catenin signaling to
inhibit glycolysis, thereby potentially inhibiting CRC
development.
The present study investigated the role of
LGALS4 in CRC glycolysis. Although LGALS4 has been
identified as a potential prognostic factor for patients with CRC,
its specific impact in tumor glycolysis has not been fully
elucidated. Through analysis of the TCGA-COAD and GSE26571
databases, it was demonstrated that LGALS4 expression was
significantly downregulated in CRC tissues and strongly correlated
with patient survival, suggesting a potentially important role in
CRC development. Glycolysis is a major energy source for cancer
cells and its aberrant activation is tightly linked to rapid
proliferation and invasiveness of tumors. It was hypothesized that
LGALS4 may affect the metabolic properties of CRC cells by
regulating the glycolytic pathway. The present preliminary data
suggested that LGALS4 overexpression inhibited glycolytic
activity in CRC cells and that this effect may be associated with
changes in the β-catenin signaling pathway. Given the role of
β-catenin in the regulation of glycolytic gene expression, it could
be suggested that LGALS4 may regulate glycolysis through this
signaling pathway. Therefore, the function of LGALS4 in CRC
glycolysis and its associated molecular mechanism were
analyzed.
In summary, overexpression of LGALS4 exerted
a multifaceted inhibitory effect in CRC cells, significantly
affecting key biological properties of tumor cells. Firstly, it
inhibited the glycolytic process, reducing the cell's dependence on
glucose and decreasing lactate production and ATP generation,
thereby directly limiting the cell's energy supply. Second,
LGALS4 caused cell cycle arrest in the G1 phase, which
prevented normal cell cycle progression by decreasing the
expression levels of cell cycle-related proteins such as CDK1,
Cyclin B1 and Cyclin A2. In addition, LGALS4 overexpression
promoted apoptosis, which was closely related to the changes in the
expression of apoptosis-related proteins such as Bax, Bcl-2,
caspase-3 and caspase-9, increasing the rate of apoptosis.
Meanwhile, LGALS4 affected the β-catenin protein signaling
pathway, decreasing the expression of glycolysis-related factors
such as β-catenin protein, c-Myc, GLUT1, PKM2, HK2 and LDHA. A
previous study has shown that the β-catenin protein signaling
pathway is a signaling pathway that serves a central role in cell
proliferation, migration and invasion (69). LGALS4 may also cause
metabolic reprogramming, which further reduced aerobic glycolysis
in CRC cells and inhibited the rapid proliferation and invasiveness
of tumor cells. Notably, LGALS4 overexpressing cells showed
increased sensitivity to the chemotherapeutic drug 5-FU, which may
be achieved by enhancing 5-FU-induced apoptosis. LGALS4
reduced 2-NBDG uptake and decreased ATP production and lactate
release, suggesting a potential direct effect on cellular energy
metabolism.
The present study suggested that LGALS4 may
provide a new potential target for CRC therapy. As a protein whose
expression is downregulated in CRC and is associated with patient
survival, LGALS4 has the ability to regulate glycolysis and
promote apoptosis in tumor cells, which provides a scientific basis
for the development of new therapeutic approaches. By inhibiting
glycolysis, LGALS4 is able to reduce the energy supply of
tumor cells, directly targeting their metabolic needs for rapid
proliferation. In addition, LGALS4 overexpression promoted
apoptosis, providing a new therapeutic avenue for inducing tumor
cell death. More importantly, LGALS4 increased the
sensitivity of tumor cells to chemotherapeutic agents such as 5-FU,
which may help to improve the efficacy of existing therapeutic
regimens, particularly in drug-resistant tumors. Meanwhile, the
regulatory effect of LGALS4 on the β-linker protein
signaling pathway provided a new perspective on the control of
tumor cell proliferation and invasion. These properties not only
demonstrate the potential of LGALS4 in individualized
medicine, but also highlight its advantages in overcoming existing
therapeutic limitations. With further research, LGALS4 may
be a key factor in improving treatment outcomes for patients with
CRC.
Although the present study demonstrated the
potential role of LGALS4 in CRC in in vitro
experiments and bioinformatics analyses, there were a number of
limitations. First, the findings need to be further validated by
in vivo models to ensure the accuracy and reliability of the
biological effects. Second, the sample size and population
representation may limit the generalizability of the findings. In
addition, the long-term effects and specific molecular mechanisms
of LGALS4 need to be explored in depth. To address these
limitations, future studies should conduct in vivo
experiments in animal models to assess the actual therapeutic
potential and safety of LGALS4. The mechanism of
LGALS4 downregulation of the expression of factors such as
β-catenin, c-Myc, PKM2, GLUT1, LDHA and HK is currently unknown.
Techniques such as chromatin immunoprecipitation sequencing and RNA
sequencing should be used to investigate how LGALS4 affects
the transcriptional activity of the β-catenin signaling pathway and
its associated target genes. Through methods such as
immunoprecipitation and mass spectrometry, the direct or indirect
interactions between LGALS4 and proteins such as β-catenin and
c-Myc can be explored in addition to how these interactions affect
their functions. In addition, metabolomics approaches should be
used to analyze the metabolic changes in LGALS4
overexpressing cells to understand how LGALS4 regulates
cellular metabolic pathways, particularly glycolytic processes. By
observing the effects of LGALS4 overexpression on the cell
cycle and apoptosis, how these cellular events are linked to the
regulation of the expression of metabolism-related genes could be
determined. In the present study, apoptosis and cell cycle
distribution of CRC cells were quantified using PI staining
combined with flow cytometry. Although this method provided
valuable information on cell cycle status and apoptosis rates, it
also has its inherent limitations. For example, PI staining may not
be able to distinguish between the various stages of the cell cycle
and there may be some bias in the assessment of cell survival
status. In addition, due to the limitations of the present study
conditions, 5-bromo-2-deoxyuridine (BRDU) staining was not
performed to further validate cell proliferation. However, BRDU
staining should be performed in future studies to complement the
results of PI staining and provide additional validation of the
findings of the present study. By combining these a more
comprehensive understanding of the biology of CRC cells could be
expected and these results could potentially provide a more solid
experimental basis for future studies.
The present study highlighted the role of LGALS4 in
CRC and its potential as a therapeutic target. Bioinformatics
analysis demonstrated that LGALS4 was significantly
downregulated in CRC and was associated with patient survival.
Overexpression of LGALS4 resulted in a significant
upregulation of caspase-3 and caspase-9, a phenomenon that may be
achieved through multiple mechanisms. First, LGALS4
overexpression promoted apoptosis in CRC cells. Caspase-3 and
caspase-9 act as key executors in the apoptotic pathway and
caspase-9 acts as an initiating caspase to activate effector
caspase-3, thereby triggering apoptosis. Second, LGALS4
overexpression inhibited aerobic glycolysis in CRC cells and
affected the β-catenin signaling pathway, which serves a crucial
role in cell survival and apoptosis. By decreasing the activity of
the β-catenin signaling pathway, LGALS4 may promote the
upregulation of caspase-3 and caspase-9, which in turn drives the
apoptotic process. In addition, G1-phase cell cycle arrest caused
by LGALS4 overexpression may have triggered a cellular
stress response that activated apoptotic pathways including caspase
family proteins. LGALS4 may also directly or indirectly
regulate the expression of caspase-3 and caspase-9, which acted as
post-transcriptional modifiers affecting the stability or
translational efficiency of specific genes. Furthermore, possible
intracellular feedback mechanisms may upregulate the expression of
apoptosis-related genes upon detection of survival stress or
abnormal signals to remove damaged cells. Taken together, the
upregulation of caspase-3 and caspase-9 levels in CRC cells by
LGALS4 overexpression may be due to its direct effect on
apoptotic pathways and its potential inhibitory effect on the
β-catenin signaling pathway, which exerts an antitumor effect in
CRC.
The present study demonstrated the important role
of LGALS4 in CRC and its value as a potential future
therapeutic target. These findings showed that LGALS4
expression was downregulated in CRC tissues and correlated with
poor patient prognosis, suggesting its role as a tumor suppressor.
Functionally, overexpression of LGALS4 inhibited glycolysis
and inhibited cell cycle progression in CRC cells, leading to G1
phase arrest while promoting apoptosis. In addition, the regulatory
effect of LGALS4 on the β-catenin signaling pathway may have
an inhibitory effect on the proliferation, migration and invasion
of tumor cells. Notably, LGALS4 overexpressed cells
increased higher sensitivity to the commonly used chemotherapeutic
drug 5-FU, which potentially provides a novel research avenue to
improve the efficacy of chemotherapy. The present study also
highlighted the impact of LGALS4 in the metabolic
reprogramming of tumors, indicating potential new perspectives for
metabolically targeted therapies. These results further the current
understanding of the molecular mechanisms of CRC and provide
directions for future individualized treatment strategies and the
development of new drugs, which may improve the treatment outcome
of patients with CRC.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgments
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
SL, KY and JY were responsible for conception and
design of the study. CX, KY, ZQ and YC were responsible for data
acquisition. SL, LY and BS were responsible for data analysis and
interpretation. SL, TZ, JX and YC were responsible for statistical
analysis. SL and BS drafted the manuscript. SL and KY confirm the
authenticity of all the raw data. All authors read and approved the
final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Roshandel G, Ghasemi-Kebria F and
Malekzadeh R: Colorectal cancer: Epidemiology, risk factors, and
prevention. Cancers. 16:15302024. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Parmar S and Easwaran H: Genetic and
epigenetic dependencies in colorectal cancer development.
Gastroenterol Report. 10:goac0352022. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yu J, Feng Q, Kim JH and Zhu Y: Combined
effect of healthy lifestyle factors and risks of colorectal
adenoma, colorectal cancer, and colorectal cancer mortality:
Systematic review and meta-analysis. Front Oncol. 12:8270192022.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wong MC, Huang J, Lok V, Wang J, Fung F,
Ding H and Zheng ZJ: Differences in incidence and mortality trends
of colorectal cancer worldwide based on sex, age, and anatomic
location. Clin Gastroenterol Hepatol. 19:955–966.e61. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Biller LH and Schrag D: Diagnosis and
treatment of metastatic colorectal cancer: A review. JAMA.
325:669–685. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhao H, Ming T, Tang S, Ren S, Yang H, Liu
M, Tao Q and Xu H: Wnt signaling in colorectal cancer: Pathogenic
role and therapeutic target. Mol Cancer. 21:1442022. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Satelli A, Rao PS, Thirumala S and Rao US:
Galectin-4 functions as a tumor suppressor of human colorectal
cancer. Int J Cancer. 129:799–809. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Michalak M, Golde V, Helm D, Kaltner H,
Gebert J and Kopitz J: Combining Recombinase-mediated cassette
exchange strategy with quantitative proteomic and Phosphoproteomic
analyses to inspect intracellular functions of the tumor suppressor
galectin-4 in colorectal cancer cells. Int J Mol Sci. 23:64142022.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang GL, Pan LL, Huang T and Wang JH: The
transcriptome difference between colorectal tumor and normal
tissues revealed by single-cell sequencing. J Cancer. 10:5883–5890.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cheraghi-Shavi T, Jalal R and Minuchehr Z:
TGM2, HMGA2, FXYD3, and LGALS4 genes as biomarkers in acquired
oxaliplatin resistance of human colorectal cancer: A systems
biology approach. PLoS One. 18:e02895352023. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chandel NS: Glycolysis. Cold Spring Harb
Perspect Biol. 13:a0405352021. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jaiswara PK, Gupta VK, Rawat SG, Tiwari
RK, Sonker P, Maurya RP and Kumar A: Targeting of Aerobic
Glycolysis: An Emerging Therapeutic Approach Against Colon Cancer.
Colon Cancer Diagnosis and Therapy. Vol 2. Springer; pp. 225–244.
2021, View Article : Google Scholar
|
|
13
|
Zuo S, Wu L, Wang Y and Yuan X: Long
non-coding RNA MEG3 activated by vitamin D suppresses glycolysis in
colorectal cancer via promoting c-Myc degradation. Front Oncol.
10:2742020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhu Y, Wu G, Yan W, Zhan H and Sun P:
miR-146b-5p regulates cell growth, invasion, and metabolism by
targeting PDHB in colorectal cancer. Am J Cancer Res.
7:11362017.PubMed/NCBI
|
|
15
|
Zhu J, Wang S, Bai H, Wang K, Hao J, Zhang
J and Li J: Identification of five glycolysis-related gene
signature and risk score model for colorectal cancer. Front Oncol.
11:5888112021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xiong Y, Lei J, Zhao J, Lu Q, Feng Y, Qiao
T, Xin S, Han Y and Jiang T: A gene-based survival score for lung
adenocarcinoma by multiple transcriptional datasets analysis. BMC
Cancer. 20:10462020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
He R, Zhang M, He L, Huang J, Man C, Wang
X, Lang Y and Fan Y: Integrated analysis of Necroptosis-related
genes for prognosis, immune microenvironment infiltration, and drug
sensitivity in colon cancer. Front Med (Lausanne). 9:8452712022.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hou S, Li L, Hou H, Zhou T and Zhou H:
Establishment of nomogram to predict overall survival and
cancer-specific survival of local tumor resection in patients with
colorectal cancer liver metastasis with unresectable metastases: A
large population-based analysis. Discover Oncol. 15:3152024.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen S, Su T, Zhang Y, Lee A, He J, Ge Q
and Wang L, Si J, Zhuo W and Wang L: Fusobacterium nucleatum
promotes colorectal cancer metastasis by modulating KRT7-AS/KRT7.
Gut Microbes. 11:511–525. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xu J, Xiao Y, Liu B, Pan S, Liu Q, Shan Y,
Li S, Qi Y, Huang Y and Jia L: Exosomal MALAT1 sponges miR-26a/26b
to promote the invasion and metastasis of colorectal cancer via
FUT4 enhanced fucosylation and PI3K/Akt pathway. J Exp Clin Cancer
Res. 39:542020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tang Q, Chen J, Di Z, Yuan W, Zhou Z, Liu
Z, Han S, Liu Y, Ying G, Shu X and Di M: TM4SF1 promotes EMT and
cancer stemness via the Wnt/β-catenin/SOX2 pathway in colorectal
cancer. J Exp Clin Cancer Res. 39:2322020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Aslan M, Hsu EC, Liu S and Stoyanova T:
Quantifying the invasion and migration ability of cancer cells with
a 3D Matrigel drop invasion assay. Biol Methods Protoc.
6:bpab0142021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mao Z, Xiao H, Shen P, Yang Y, Xue J, Yang
Y, Shang Y, Zhang L, Li X, Zhang Y, et al: KRAS(G12D) can be
targeted by potent inhibitors via formation of salt bridge. Cell
Discov. 8:52022. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen H, Chen H, Zhang J, Wang Y, Simoneau
A, Yang H, Levine AS, Zou L, Chen Z and Lan L: cGAS suppresses
genomic instability as a decelerator of replication forks. Sci Adv.
6:eabb89412020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu Y, Zhang X, Wang W, Liu T, Ren J, Chen
S, Lu T, Tie Y, Yuan X, Mo F, et al: Ammonia promotes the
proliferation of bone marrow-derived mesenchymal stem cells by
regulating the Akt/mTOR/S6k pathway. Bone Res. 10:572022.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Acharjee A, Agarwal P, Nash K, Bano S,
Rahman T and Gkoutos GV: Immune infiltration and prognostic and
diagnostic use of LGALS4 in colon adenocarcinoma and bladder
urothelial carcinoma. Am J Transl Res. 13:113532021.PubMed/NCBI
|
|
28
|
Principe M, Borgoni S, Cascione M,
Chattaragada MS, Ferri-Borgogno S, Capello M, Bulfamante S,
Chapelle J, Di Modugno F, Defilippi P, et al: Alpha-enolase (ENO1)
controls alpha v/beta 3 integrin expression and regulates
pancreatic cancer adhesion, invasion, and metastasis. J Hematol
Oncol. 10:162017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ding L, Cao J, Lin W, Chen H, Xiong X, Ao
H, Yu M, Lin J and Cui Q: The roles of cyclin-dependent kinases in
cell-cycle progression and therapeutic strategies in human breast
cancer. Int J Mol Sci. 21:19602020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sun L, Suo C, Li ST, Zhang H and Gao P:
Metabolic reprogramming for cancer cells and their
microenvironment: Beyond the Warburg Effect. Biochim Biophys Acta
Rev Cancer. 1870:51–66. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fang Y, Shen ZY, Zhan YZ, Feng XC, Chen
KL, Li YS, Deng HJ, Pan SM, Wu DH and Ding Y: CD36 inhibits
β-catenin/c-myc-mediated glycolysis through ubiquitination of GPC4
to repress colorectal tumorigenesis. Nat Commun. 10:39812019.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang M, Song H, Yang S, Zhang Y, Tian Y,
Wang Y and Liu D: Deciphering the antibacterial mechanisms of
5-fluorouracil in Escherichia coli through biochemical and
transcriptomic analyses. Antibiotics. 13:5282024. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Malki A, ElRuz RA, Gupta I, Allouch A,
Vranic S and Al Moustafa AE: Molecular mechanisms of colon cancer
progression and metastasis: Recent insights and advancements. Int J
Mol Sci. 22:1302020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zou Y, Liu FY, Liu H, Wang F, Li W, Huang
MZ, Huang Y, Yuan XQ, Xu XY, Huang OP and He M: Frequent POLE1 p.
S297F mutation in Chinese patients with ovarian endometrioid
carcinoma. Mutat Res. 761:49–52. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zurlo D, Leone C, Assante G, Salzano S,
Renzone G, Scaloni A, Foresta C, Colantuoni V and Lupo A:
Cladosporol a stimulates G1-phase arrest of the cell cycle by
up-regulation of p21waf1/cip1 expression in human colon carcinoma
HT-29 cells. Mol Carcinog. 52:1–17. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Patil R, Sontakke T, Biradar A and Nalage
D: Zinc: An essential trace element for human health and beyond.
Food Health. 5:132023. View Article : Google Scholar
|
|
37
|
Haase H and Rink L: Zinc signals and
immune function. Biofactors. 40:27–40. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Pelletier J, Thomas G and Volarević S:
Ribosome biogenesis in cancer: New players and therapeutic avenues.
Nat Rev Cancer. 18:51–63. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Villalobo A: Ca2+ signaling and Src
functions in tumor cells. Biomolecules. 13:17392023. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zurlo D, Assante G, Moricca S, Colantuoni
V and Lupo A: Cladosporol A, a new peroxisome
proliferator-activated receptor γ (PPARγ) ligand, inhibits
colorectal cancer cells proliferation through β-catenin/TCF pathway
inactivation. Biochim Biophys Acta. 1840:2361–2372. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yu C, Hong H, Zhang S, Zong Y, Ma J, Lu A,
Sun J and Zheng M: Identification of key genes and pathways
involved in microsatellite instability in colorectal cancer. Mol
Med Rep. 19:2065–2076. 2019.PubMed/NCBI
|
|
42
|
Günther J and Galuska SP: A brief history
of galectin evolution. Front Immunol. 14:11473562023. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ferlizza E, Solmi R, Miglio R, Nardi E,
Mattei G, Sgarzi M and Lauriola M: Colorectal cancer screening:
Assessment of CEACAM6, LGALS4, TSPAN8 and COL1A2 as blood markers
in faecal immunochemical test negative subjects. J Adv Res.
24:99–107. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Watanabe M, Takemasa I, Kaneko N, Yokoyama
Y, Matsuo EI, Iwasa S, Mori M, Matsuura N, Monden M and Nishimura
O: Clinical significance of circulating galectins as colorectal
cancer markers. Oncol Rep. 25:1217–1226. 2011.PubMed/NCBI
|
|
45
|
Zhou J, Belov L, Chapuis P, Chan C,
Armstrong N, Kaufman KL, Solomon MJ, Clarke SJ and Christopherson
RI: Surface profiles of live colorectal cancer cells and tumor
infiltrating lymphocytes from surgical samples correspond to
prognostic categories. J Immunol Methods. 416:59–68. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Passarella S, Schurr A and Portincasa P:
Mitochondrial transport in glycolysis and gluconeogenesis:
Achievements and perspectives. Int J Mol Sci. 22:126202021.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Gupta R and Gupta N, Gupta R and Gupta N:
Glycolysis and gluconeogenesis. Fundamentals of bacterial
physiology and metabolism. 267–287. 2021. View Article : Google Scholar
|
|
48
|
Zhou L, Yu X, Li M, Gong G, Liu W, Li T,
Zuo H, Li W, Gao F and Liu H: Cdh1-mediated Skp2 degradation by
dioscin reprogrammes aerobic glycolysis and inhibits colorectal
cancer cells growth. EBioMedicine. 51:1025702020. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhou CF, Li XB, Sun H, Zhang B, Han YS,
Jiang Y, Zhuang QL, Fang J and Wu GH: Pyruvate kinase type M2 is
upregulated in colorectal cancer and promotes proliferation and
migration of colon cancer cells. IUBMB Life. 64:775–782. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhao J, Li J, Hassan W, Xu D, Wang X and
Huang Z: Sam68 promotes aerobic glycolysis in colorectal cancer by
regulating PKM2 alternative splicing. Ann Transl Med. 8:4592020.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Sethy C and Kundu CN: 5-Fluorouracil
(5-FU) resistance and the new strategy to enhance the sensitivity
against cancer: Implication of DNA repair inhibition. Biomed
Pharmacother. 137:1112852021. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zou X, Liang J, Sun J, Hu X, Lei L, Wu D
and Liu L: Allicin sensitizes hepatocellular cancer cells to
anti-tumor activity of 5-fluorouracil through ROS-mediated
mitochondrial pathway. J Pharmacol Sci. 131:233–240. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zou K, JU JH and Xie H: Pretreatment with
insulin enhances anticancer functions of 5-fluorou-racil in human
esophageal and colonic cancer cells. Acta Pharmacol Sin.
28:721–730. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Zuo T, Zhang Z, Jiang P, Zhang R, Ni D,
Yuan Y and Zhang S: Characterization of chikusetsusaponin IV and V
induced apoptosis in HepG2 cancer cells. Mol Biol Rep.
49:4247–4255. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Raeisi E and Mir LM: 2-NBDG, a fluorescent
analogue of glucose, as a marker for detecting cell
electropermeabilization in vitro. J Membr Biol. 245:633–642. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Lin HC, Chen YJ, Wei YH, Lin HA, Chen CC,
Liu TF, Hsieh YL, Huang KY, Lin KH, Wang HH and Chen LC: Lactic
acid fermentation is required for NLRP3 inflammasome activation.
Front Immunol. 12:6303802021. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Akram M: Mini-review on glycolysis and
cancer. J Cancer Educ. 28:454–457. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Mondal P, Tiwary N, Sengupta A, Dhang S,
Roy S and Das C: Epigenetic reprogramming of the glucose metabolic
pathways by the chromatin effectors during cancer. Subcell Biochem.
100:269–336. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Zhou Q, Li J, Pang J, Fan F, Li S and Liu
H: Gefitinib inhibits glycolysis and induces programmed cell death
in non-small cell lung cancer cells. Nan Fang Yi Ke Da Xue Xue Bao.
40:884–892. 2020.(In Chinese). PubMed/NCBI
|
|
60
|
Zu XY, Zhang QH, Liu JH, Cao RX, Zhong J,
Yi GH and Pizzorno G: ATP citrate lyase inhibitors as novel cancer
therapeutic agents. Recent Pat Anticancer Drug Discov. 7:154–167.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Zuo Xm, Sun Hw, Fang H, Wu Y, Shi Q and Yu
YF: miR-4443 targets TRIM14 to suppress metastasis and energy
metabolism of papillary thyroid carcinoma (PTC) in vitro. Cell Biol
Int. 45:1917–1925. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Shah K and Kazi JU:
Phosphorylation-dependent regulation of WNT/Beta-catenin signaling.
Front Oncol. 12:8587822022. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Jung YS and Park JI: Wnt signaling in
cancer: Therapeutic targeting of Wnt signaling beyond β-catenin and
the destruction complex. Exp Mol Med. 52:183–191. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
van Zuylen WJ, Rawlinson WD and Ford CE:
The Wnt pathway: A key network in cell signalling dysregulated by
viruses. Rev Med Virol. 26:340–355. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Zou Y, Liu L, Meng J and Dai M: Circular
RNA circ_0068464 combined with microRNA-383 regulates Wnt/β-catenin
pathway to promote the progression of colorectal cancer.
Bioengineered. 13:5113–5125. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Zou J, Huang Y, Chen Y, Wu Z, Xie H, Zhou
H and Xing C: FOXC2-induced circCASK aggravates colorectal cancer
progression by upregulating SIX1 expression. IUBMB Life.
75:659–672. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Zhou Y, Gu H, Shao B, Zhang S, Pall H,
Peixoto RD, Mok SRS and Zhu G: Glycolysis-related gene
dihydrolipoamide acetyltransferase promotes poor prognosis in
hepatocellular carcinoma through the Wnt/β-catenin and PI3K/Akt
signaling pathways. Ann Transl Med. 10:12402022. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Zhou J, Su CM, Chen HA, Du S, Li CW, Wu H,
Tsai SH and Yeh YT: Cryptanshinone inhibits the glycolysis and
inhibits cell migration through PKM2/β-catenin axis in breast
cancer. Onco Targets Ther. 13:8629–8639. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Wenxuan L, Liu L, Zhang L, Qiu Z, Wu Z and
Deng W: Role of gonadally synthesized steroid hormones in the
colorectal cancer microenvironment. Front Oncol. 13:13238262023.
View Article : Google Scholar : PubMed/NCBI
|