Introduction
Colorectal cancer (CRC) ranks third in incidence and
second in mortality of all cancer types worldwide (1). The incidence of CRC in the whole
population of China has gradually increased in recent years
(2). Some early stage CRC cases are
suitable for endoscopic treatment; however, surgery is the primary
and cornerstone treatment for curative purposes. Rectal cancer is
more complex due to the intricate anatomy of the pelvis (3). Total mesorectal excision (TME) is the
standard approach for treating rectal cancer. Furthermore,
neoadjuvant radiotherapy followed by TME and adjuvant chemotherapy
is the standard treatment for locally advanced rectal cancer (LARC;
staging, T3-4/N+M0). The combined treatment has led to a marked
reduction in the rate of local recurrence in the past 10 years
(4,5). Tumor downsizing and pathological
complete response (CR) following neoadjuvant chemoradiotherapy
(nCRT) has been observed in 15–20% of patients with LARC (4). However, no survival benefits were
observed in patients with rectal cancer treated with preoperative
radiation following TME by several long-term follow-up analyses
(5–7). Therefore, exploring an alternative
approach or identifying an nCRT response or survival predictor is
necessary and urgent. It has been reported that total neoadjuvant
therapy (TNT) could increase the CR rate to 30% and improve life
quality by anus preservation. nCRT combined with immunotherapy is
also a promising approach for treating patients with rectal cancer
(5). Regardless of whether nCRT,
TNT or combined immunotherapy is used, a biomarker that can predict
radiation response before or after nCRT remains necessary.
CKLF-like MARVEL transmembrane domain member 4
(CMTM4) is a member of the chemokine-like factor (CKLF)
superfamily, of which there are 9 members: CKLF and CMTM 1–8. The
MARVEL domain in CMTM4 is responsible for vesical trafficking and
membrane linking (8). To date, the
expression of CMTM4 has been found to be lower in tumor tissues
compared with adjacent normal tissues in CRC and clear cell renal
carcinoma (9,10). CMTM4 serves as a tumor suppressor
and has been shown to inhibit cell proliferation and migration
through the AKT, ERK1/2 and signal transducer and activator of
transcription (STAT) 3 pathways in CRC cell lines (9.10). It has
also been demonstrated that CMTM4 is upregulated in human head and
neck squamous cell carcinoma (HNSCC) and indicates poorer survival
and lymphatic metastasis by regulating epithelial-mesenchymal
transition (EMT) and programmed death ligand 1 (PDL1) expression
(11). The expression and
prognostic role of CMTM4 in hepatocellular carcinoma (HCC) proposed
by several groups from previous studies has been contradictory.
Chui et al (12) and Zhou
et al (13) reported that
upregulation of CMTM4 in patients with HCC was related to poor
survival. Studies from Tan et al (14) and Bei et al (15) reported that high expression of CMTM4
was associated with a good survival in patients with HCC. Studies
have shown that CMTM4/6 stabilizes PDL1 expression by inhibiting
protease or lysosome-mediated degradation (16,17).
Therefore, targeting the CMTM4/6-PDL1 pathway could be a new avenue
to enhance antitumor effectiveness of current PD-L1/PD-1 blocking
therapies. CMTM6, which shares 55% sequence homology with CMTM4,
has been reported to be involved in immunosuppressive
microenvironments in glioma, renal carcinoma and CRC (18). Data from The Cancer Genome Atlas
(TCGA) database showed that CMTM4 expression is negatively
correlated with cytotoxic, dendritic, T and CD8+ T cells
(13,14). The immune-related function of CMTM4
has rarely been reported. Furthermore, the role of CMTM4 in rectal
cancer, particularly in LARC treated with nCRT, remains
unclear.
In the present study, 228 patients with LARC were
retrospectively enrolled, including 178 with paired
pre-/post-operative tissues and 50 pathological complete response
(pCR) patients with preoperative tissues. The aim of the present
study was to reveal the predictive role of CMTM4 in patients with
LARC and its underlying biological mechanism in LARC and colon
cancer.
Materials and methods
Patient selection
Data were retrospectively collected from 228
patients with LARC who received intensity-modulated radiation
therapy (IMRT) with concurrent capecitabine treatment followed by
surgery at Peking University Cancer Hospital (Beijing, China)
between December 2008 and June 2015. The IMRT regimen consisted of
22 fractions of 2.3 Gy (gross tumor volume) and 1.9 Gy (clinical
target volume), which has been described previously (19). Surgery was recommended ≥8 weeks
after the completion of radiation. Adjuvant chemotherapy was also
routinely recommended to the patients. Each enrolled patient
satisfied the following criteria: i) Cancerous lesion located
within 10 cm from the anal verge; ii) cancer staged as T3-4 or any
T and N+ by endorectal ultrasonography (7th AJCC cancer staging
system) (20), pelvic magnetic
resonance imaging (MRI) or computed tomography; iii) presence of
distant metastases excluded by imaging examinations; iv)
preoperative radiotherapy of 50.6 Gy/22 fractions; and v) radical
surgery following TME. Patients with the following characteristics
were excluded from the present study: i) Previous chemotherapy or
pelvic radiation; ii) previous history (within 5 years) of
malignant tumor; and iii) presence of unresectable cancer. All
patients signed the consent forms before treatment. The patient
information and samples were gathered from the hospital surgical
database. The blocks of formalin-fixed paraffin-embedded (FFPE)
samples from each patient in this surgical database were stored at
The Department of Pathology at Peking University Cancer Hospital.
The slices of FFPE samples were stored at −80°C for long term and
−20°C for short term storage. The present retrospective study was
approved by The Ethics Committee of Peking University Cancer
Hospital (approval no. 2021KT93). Informed consent for inclusion in
the present study was waived by The Ethics Committee due to the
retrospective nature of the present study.
Immunohistochemistry (IHC)
FFPE tissue sections (5 µM) were prepared and
stained with hematoxylin (5 min) and eosin (2 min) at room
temperature for histological evaluation. For IHC, the sections were
deparaffinized in a xylene and an ethanol gradient at room
temperature. Antigen retrieval was performed with citrate buffer pH
6.0 at 95°C for 10 min, followed by incubating with an endogenous
peroxidase blocker (cat. no. ZLI-9310; ZSGB-Bio) for 10 min at room
temperature. The sections were then washed with PBST (0.1%
Tween-20) for 5 min three times and incubated with 5% goat serum
(cat. no. ZLI-9021; ZSGB-Bio) for 1 h at room temperature. The
sections were incubated with the following primary antibodies at
4°C overnight: chemokine (CXC motif) ligand 8 (CXCL8; 1:500; cat.
no. 27095-1-AP; Proteintech Group, Inc.) and CMTM4 (1:500; cat. no.
HPA014704; Sigma-Aldrich; Merck KGaA). Following washing with PBST
(0.1% Tween-20) three times, the slices were incubated with the
secondary antibody (undiluted; cat. no. SAP-9100; ZSGB-Bio) at room
temperature for 40 min. Diaminobenzidine (Dako; Agilent
Technologies, Inc) substrate was used to observe staining, and the
sections were re-stained with hematoxylin for 5 min at room
temperature (Beyotime Institute of Biotechnology). A bright field
microscope (Leica Microsystems, Inc.) was used to capture images.
The H score is a reliable method that can effectively quantify
protein expression by two independent pathologists (21). The H score was calculated as
follows: (Percentage of weak staining) + (Percentage of moderate
staining) + (Percentage of strong staining) within the target
region, with scores ranging from 0 to 300 (22). The of CMTM4 and CXCL8 had different
expression pattern, therefore the cut-off value is different. H
scores of 0–120 were defined as the CXCL8 low group and H scores of
121–300 as the CMTM4 high group. H scores of 0–150 were defined as
the CMTM4 low group and H scores of 151–300 as the CXCL8 high
group.
Cell culture
The human colon cancer LoVo cell line purchased from
the American Type Culture Collection (cat. no. CCL-229) was
cultured in a hot cell incubator (5% CO2, 37°C) in
RPMI-1640 (Invitrogen; Thermo Fisher Scientific, Inc.) supplemented
with 10% bovine serum (cat. no. 164210; Procell Life Science &
Technology Co., Ltd.) and 100 U/ml streptomycin. When the cells
reached 80–90% confluency, pancreatin digestion was carried out and
the cells were resuspended in fresh culture medium. The consumables
used for cell culture were sterilized by high pressure and the
cells were regularly examined for mycoplasma contamination by
reverse transcription-quantitative PCR (RT-qPCR).
Cell transfections
The lentivirus targeting human CMTM4 and an shRNA
scramble sequence (negative control; NC) were generated and
synthesized by GenePharma Co., Ltd. The short hairpin (sh)RNAs used
in the present study were as follows: shCM4-3,
GAAAUUGCUGCCGUGAUAUTT; shCM4-6, GCAUAUGCAGUGAACACAUTT; and sh NC,
UUCUCCGAACGUGUCACGUTT. Briefly, the shRNAs targeting human CMTM4
were cloned into a 3rd generation lentiviral transfer plasmid,
pLV-eGFP (vector backbone, pLenti-MP2), generating a lenti-shCMTM4
construct for expression knockdown. The virus was added at a volume
ratio of 1:100 and at a titer of 1×108 TU/ml into LoVo
cells, while simultaneously adding polybrene at a volume ratio of
1:500 at 37°C and 5% CO2. Fresh and complete culture
medium was replaced within 24 h from infection, and the infection
efficiency was detected using western blotting and
immunofluorescence after 72 h.
Adenovirus carrying the human CMTM4 gene and the
empty adenovirus were packaged by GenePharma Co., Ltd. LoVo cells
were infected with the adenovirus at a multiplicity of infection of
100 for 72 h. All other steps were as described above.
Irradiation
The LoVo cells were infected with the indicated
lentivirus for 72 h before irradiation. X-ray irradiation was
performed using an X-ray generator (EDGE™ Radiosurgery system;
Varian Medical Systems, Inc.) with gantry 0°, collimator 0°, field
30×30 cm, energy 6 Mv with extra-fine 2.5 mm MLC leaves. Indicated
doses were shown in each experiment.
Western blotting
A total of 2×106 cells with the indicated
treatments were harvested and lysed using RIPA buffer containing 50
mM Tris-HCl pH 7.4, 150 mM NaCl, 50 mM NaF, 1 mM EDTA, 1 mM DTT, 1%
TritonX-100, 0.1% sodium dodecyl sulphate (SDS) and 1X protease
inhibitor cocktail. After a 20-min incubation on ice, the cell
lysates were centrifuged at 13,800 × g for 20 min at 4°C, and the
supernatants were recovered. Protein samples (20 µg) were resolved
by 12% SDS-polyacrylamide gel electrophoresis and blotted onto
polyvinylidene fluoride membrane (cat. no. 88520; Thermo Fisher
Scientific, Inc.), which was blocked in 5% skim milk in TBST
(Tris-buffered saline containing 0.1% Tween-20) at room temperature
for 45 min. The membrane was then probed with rabbit anti-CMTM4
(1:500; cat. no. HPA014704; Sigma-Aldrich; Merck KGaA) and mouse
anti-GAPDH (1:1,000; cat. no. TA-082519; OriGene Technologies,
Inc.) antibodies overnight at 4°C. Following washing with TBST, the
membrane was incubated with the HRP-anti-mouse (1:2,000; cat. no.
ab6789; Abcam), HRP-anti-rabbit (1:2,000; cat. no. ab6721; Abcam)
secondary antibody at room temperature for 45 min. The band strips
were visualized using enhanced chemiluminescence detection system
(cat. no. 34580; Thermo Fisher Scientific, Inc). and the images
were captured using chemiluminescence imaging systems (Azure
Biosystems, Inc.).
Colony formation assay
The cells were digested and counted, and 800 cells
from each group were resuspended in fresh culture medium. After
10–14 days of cell culture, the cells were fixed at room
temperature with precooled methanol for 30 min, the methanol was
discarded, the cells were washed with PBS three times and then
incubated with 1% crystal violet dye solution for 30 min at room
temperature. The cells were then washed with PBS three times and
dried in a fume hood. The bottom of the cell plate was scanned, and
the number of clusters with >50 cells were counted using ImageJ
software (National Institutes of Health; V48.1).
Transwell assay
Migration (cat. no. 3422; Corning, Inc.) and
invasion assays (cat. no. 354480; Corning, Inc.) were performed
using 8-µm pore size plates with a filter insert. The invasion
inserts precoated with diluted Matrigel, were preheated at 37°C for
1 h before the invasion experiments. Briefly, 2×105
cells resuspended in 200 µl medium without serum were inoculated
into the upper chamber of each well; 800 µl medium containing 10%
FBS was then added to the lower chamber. The cells were allowed to
migrate for 36–48 h at 37°C. The rotating pores were fixed in 100%
methanol and dyed in 0.1% crystal violet solution at room
temperature for 30 min. The cells in the upper chamber were then
removed with absorbent cotton. The polycarbonate membrane was then
removed and sealed on a glass slide with resin, and the cells
penetrating the underside of the membrane were counted in four
randomly selected visual fields.
Tumor immune Estimation Resource
(TIMER) database analysis
The TIMER database (https://cistrome.shinyapps.io/timer) is a
comprehensive online analysis software for investigating the gene
expression and immune cell infiltration in different cancer types
(23). The TIMER database contains
166 rectum adenocarcinoma (READ) tumor samples and 10 normal
samples; 457 colon adenocarcinoma (COAD) tumor samples and 41
normal samples. TIMER was used to explore the correlation between
CMTM4 and the levels of immune cell infiltration. The association
of CMTM4 expression with gene marker of different immune cell
including CD4+ T cells, CD8+ T cells, B
cells, macrophages, neutrophils and monocytes were further
investigated (24). P≤0.05 was
considered to indicate a statistically significant difference.
Microarray analysis and Kyoto
Encyclopedia of Genomics and Genomics (KEGG) enrichment
analysis
To use high-throughput methods to analyze gene
expression patterns under different experimental conditions,
microarray analysis was performed. Total RNA was extracted from the
indicated samples using Qiagen RNeasy kit (cat. no. 74104; Qiagen
GmbH). The mRNA library construction and RNA-seq analysis were
constructed and performed by Shenzhen BGI Co., Ltd. using the
Illumina Genome Analyzer platform. The differentially expresses
genes with false discovery rate<0.01, fold change >1.5 were
determined. Principal component analysis was performed using the
‘stats’ package and plotted with the ‘ggplot2’ package in R
(version 3.5) (25). Gene Set
Enrichment Analysis (GSEA) was performed using the GSEA software
(Broad Institute) as previously described (26). DAVID analysis was performed for
transcription factor enrichment as previously described (27,28).
RT-qPCR
Total RNA was extracted from cell lines and FFPE
tissues using TRIzol® reagent (cat. no. 10057821; Thermo
Fisher Scientific, Inc.) and RNeasy FFPE Kit (cat. no. 73504;
Qiagen GmbH), respectively, according to the manufacturer's
instructions. cDNA was synthesized using a GoScript™ reverse
transcription system (cat. no. A5001; Promega Corporation)
according to the manufacturer's instructions. Each assay was tested
in duplicate. The expression of CMTM4 and CXCL8 were assessed by
SYBR GREEN Mixture (ROX reference dye; cat. no. QPK-201; Toyobo
Co., Ltd.). For the RT-qPCR experiments of cell lines and FFPE,
GAPDH served as the internal control. Relative mRNA expression was
calculated using 2−ΔΔCq (29). The thermocycling conditions were as
follows: Pre-denatured at 95°C for 5 min, 40 cycles at 95°C for 10
sec, 60°C for 20 sec and 72°C for 20 sec. The primers used are
listed in Table SI.
Statistical analysis
Data was analyzed using GraphPad Prism 8.3
(Dotmatics) and SPSS 25 (IBM Corp.). Comparison of multiple groups
was performed using one-way ANOVA and Tukey's post hoc test.
Comparisons of CMTM4 or CXCL8 expression from pre-operative with
post-operative tissues were performed using paired t-test.
Comparison of the RT-qPCR results between the CMTM4 control and
overexpression/shRNA groups was performed using unpaired t-test.
The results are presented as the mean ± standard deviation from at
least three independent experiments. The optimal cut-off value and
Kaplan-Meier curves of CMTM4 and CXCL8 in the survival analysis
were determined with the ‘survival’ (R version 3.7.0) and
‘survminer’ package (R version 4.3.2; http://www.R-project.org). Following cut-off value
determination, patients were stratified into high- and
low-expression groups for each biomarker. Disease-free survival
(DFS) and overall survival (OS) were evaluated using Kaplan-Meier
curves to illustrate differences in survival distributions, with
group comparisons performed via the log-rank test. The univariable
and multivariable Cox proportional hazards regression models were
applied to assess the prognostic significance of CMTM4, adjusting
for relevant clinical covariates where appropriate. For all
statistical tests, including the log-rank comparisons and Cox
regression analyses, a single-sided P<0.05 was considered to
indicate a statistically significant difference.
Results
CMTM4 expression indicates
radiotherapy resistance in rectal cancer
In total, 228 consecutive patients with rectal
cancer (152 men and 76 women) were included in the present study.
The median age of the patients was 57 years (range, 25–80 years),
the median follow-up time was 54.7 months and 73 (32.0%) patients
experienced recurrence or metastasis. Overall, 21.8% of patients
reached pathological complete response [pCR; no observed
adenocarcinoma cells in the surgical resection specimen,
pathological stage after nCRT (yp)T0N0M0 (30)] following nCRT. Downstaging
(ypT0-2N0M0) occurred in 134 patients (58.8%). Additional
post-operative characteristics and distribution of relevant
parameters are listed in Table I.
Patients with pN+, pT3 or nerve invasion were significantly
associated with a poorer overall survival (OS; P=0.001, P=0.042 and
P<0.001, respectively) and DFS (P<0.001, P=0.001 and P=0.008,
respectively) (Table II).
 | Table I.Distribution of relevant parameters
after nCRT. |
Table I.
Distribution of relevant parameters
after nCRT.
| Variables | No. of
patients | % of patients |
|---|
| Age, years |
|
|
|
≤58 | 114 | 50.0 |
|
>58 | 114 | 50.0 |
| Sex |
|
|
|
Male | 152 | 66.7 |
|
Female | 76 | 33.3 |
| TRG |
|
|
| 0 | 50 | 21.9 |
| 1 | 61 | 26.8 |
| 2 | 103 | 45.2 |
| 3 | 14 | 6.1 |
| pCR |
|
|
|
Yes | 50 | 21.9 |
| No | 178 | 78.1 |
| Down staging
(pT-cT) |
|
|
|
Yes | 134 | 58.8 |
| No | 94 | 41.2 |
| Lymph node
sampling |
|
|
| ≥8 | 103 | 45.2 |
|
<8 | 125 | 54.8 |
| Vascular
invasion |
|
|
|
Yes | 6 | 2.6 |
| No | 222 | 97.4 |
| Nerve invasion |
|
|
|
Yes | 4 | 1.8 |
| No | 224 | 98.2 |
| Clinical T
stage |
|
|
| 2 | 28 | 12.3 |
| 3 | 172 | 75.4 |
| 4 | 28 | 12.3 |
| Pathological T
stage |
|
|
|
ypT0 | 50 | 21.9 |
|
ypT1 | 14 | 5.7 |
|
ypT2 | 63 | 28.5 |
|
ypT3 | 101 | 43.9 |
| Pathological N
stage |
|
|
|
ypN0 | 188 | 82.5 |
|
ypN+ | 40 | 17.5 |
| Tumor deposit |
|
|
|
Yes | 15 | 6.6 |
| No | 213 | 93.4 |
| MMR status |
|
|
|
pMMR | 202 | 88.6 |
|
dMMR | 26 | 11.4 |
 | Table II.Univariate analysis to identify
prognosis-related factors. |
Table II.
Univariate analysis to identify
prognosis-related factors.
| Variable | No. of
patients | OS, % | HR (95% CI) | P-value | DFS, % | HR (95% CI) | P-value |
|---|
| Age, years |
|
|
| 0.594 |
|
| 0.809 |
|
≤58 | 114 | 75.4 | 1 |
| 66.7 | 1 |
|
|
>58 | 114 | 79.8 | 0.860
(0.495–1.496) |
| 68.4 | 0.945
(0.599–1.492) |
|
| Sex |
|
|
| 0.694 |
|
| 0.890 |
|
Male | 152 | 77.0 | 1 |
| 67.1 | 1 |
|
|
Female | 77 | 78.9 | 0.888
(0.491–1.605) |
| 68.4 | 0.966
(0.594–1.572) |
|
| pCR |
|
|
| 0.058 |
|
| 0.004 |
|
Yes | 50 | 88.0 | 1 |
| 86.0 | 1 |
|
| No | 178 | 74.7 | 2.282
(0.973–5.350) |
| 62.4 | 3.171
(1.455–6.912) |
|
| Vascular
invasion |
|
|
| 0.730 |
|
| 0.954 |
|
Yes | 6 | 83.3 | 1 |
| 66.7 | 1 |
|
| No | 222 | 77.5 | 0.706
(0.097–5.111) |
| 67.6 | 0.959
(0.235–3.910) |
|
| Nerve invasion |
|
|
| <0.001 |
|
| 0.008 |
|
Yes | 4 | 25.0 | 1 |
| 25.0 | 1 |
|
| No | 224 | 78.6 | 0.119
(0.036–0.390) |
| 68.3 | 0.206
(0.064–0.657) |
|
| Pathological T
stage |
|
|
| 0.042 |
|
| 0.001 |
|
ypT0 | 50 | 88.0 | 1 |
| 86.0 | 1 |
|
|
ypT1 | 14 | 69.2 | 2.773
(0.782–9.783) |
| 76.9 | 1.756
(0.454–6.795) |
|
|
ypT2 | 63 | 84.6 | 1.362
(0.495–3.750) |
| 73.8 | 2.092
(0.867–5.045) |
|
|
ypT3 | 101 | 69.0 | 2.837
(1.183–6.803) |
| 53.0 | 4.177
(1.886–9.248) |
|
| Pathological N
stage |
|
|
| 0.001 |
|
| <0.001 |
|
ypN+ | 40 | 57.5 | 1 |
| 40.1 | 1 |
|
|
ypN0 | 188 | 81.9 | 0.373
(0.212–0.656) |
| 73.4 | 0.321
(0.197–0.525) |
|
| CMTM4
(pre-nCRT) |
|
|
| 0.131 |
|
| 0.025 |
|
Low | 81 | 82.7 | 1 |
| 76.5 | 1 |
|
|
High | 147 | 74.8 | 1.607
(0.868–2.974) |
| 62.6 | 1.820
(1.080–3.068) |
|
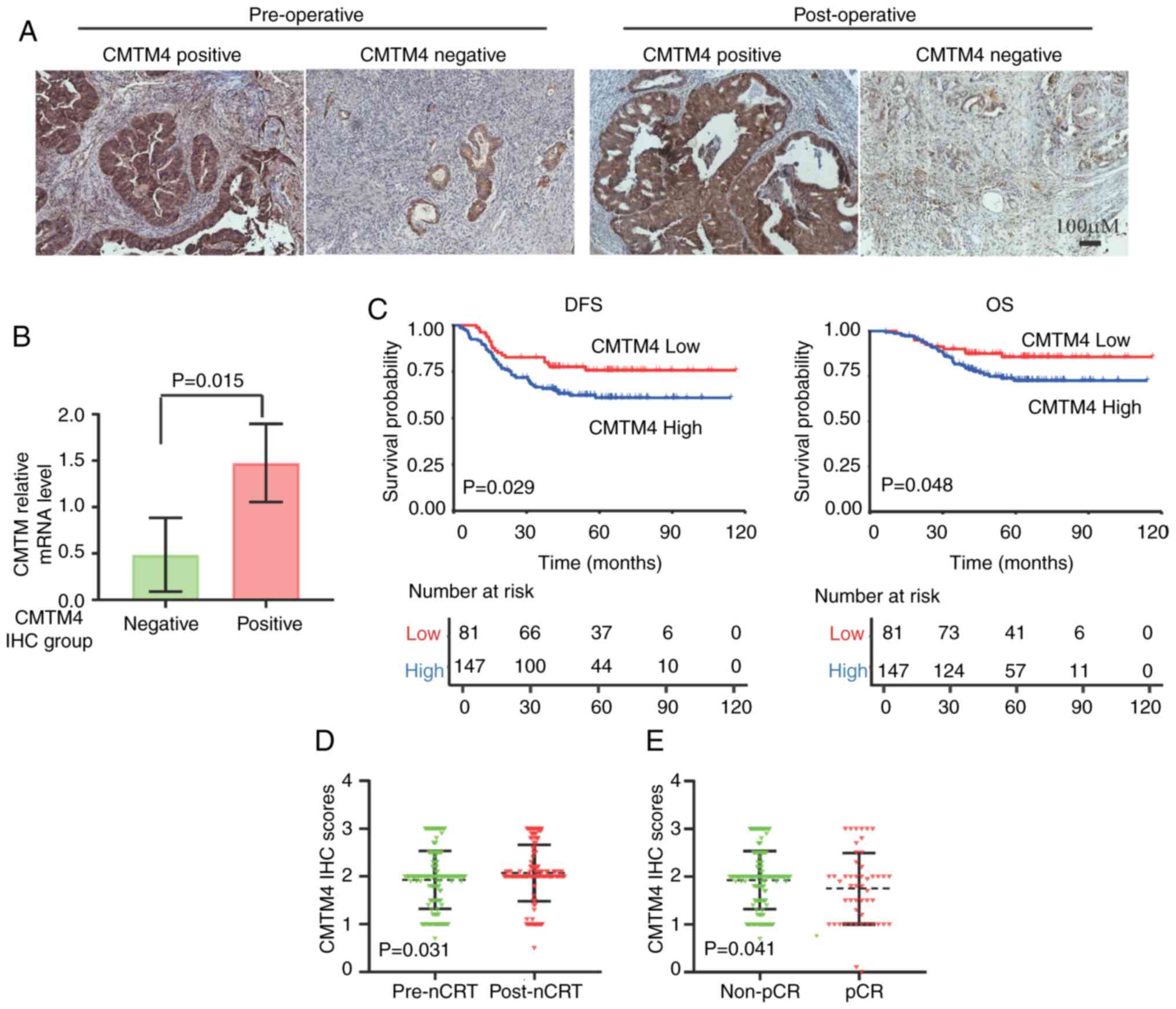
CMTM4 staining was conducted on pre- and
post-operative tissues from patients with LARC. CMTM4 was localized
in the cell membrane and cytoplasm in patients with LARC and
chemoradiotherapy did not change the CMTM4 localization (Fig. 1A). To confirm the IHC observations,
five samples with high and three with low CMTM4 expression were
selected for analysis, and DNA was extracted from these samples.
The average relative level of CMTM4 in the IHC high group was 1.477
and in the IHC low group was 0.487 (P=0.015; Fig. 1B). The results of the RT-qPCR
experiments indicated that the protein expression detected by IHC
was consistent with the mRNA expression level. Therefore, the
expression detected by IHC was reliable in the downstream analysis.
CMTM4 exhibited lower expression in tumor tissues compared with
adjacent normal tissues in the postoperative samples (P<0.0001;
Fig. S1A). Lower CMTM4 expression
in pre-operative tissues was significantly associated with improved
DFS and OS (P=0.029 and P=0.048, respectively; Fig. 1C). CMTM4 expression in LARC tissues
following TME surgery was not associated with OS or DFS (P=0.18 and
P=0.168, respectively; Fig. S1B).
The multivariate model was employed to evaluate the comprehensive
prognostic value of features obtained from the univariate analysis.
CMTM4 was an independent prognostic factor of DFS [hazard ratio
(HR), 1.759; 95% confidence interval (CI), 1.037–2.984; P=0.036] in
patients with LARC and nerve invasion and pN+ in post-nCRT was an
independent prognostic factor of OS and DFS in patients with LARC
(Table III). The changes in CMTM4
expression were compared between pre-nCRT and post-nCRT and
radiation therapy significantly increased CMTM4 expression
(P=0.031; Fig. 1D). Further
analysis indicated that CMTM4 expression in the pCR group was lower
than that in the non-pCR groups (P=0.041; Fig. 1E). In summary, CMTM4, a potential
new biomarker for patients with LARC before nCRT, was negatively
associated with chemoradiotherapy response and prognosis.
 | Table III.Multivariate analysis to identify
prognosis-related factors. |
Table III.
Multivariate analysis to identify
prognosis-related factors.
|
| OS | DFS |
|---|
|
|
|
|
|---|
| Variable | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Nerve invasion |
| 0.012 |
| 0.041 |
|
Yes | 1 |
| 1 |
|
| No | 0.212
(0.064–0.709) |
| 0.289
(0.088–0.953) |
|
| pCR |
| 0.412 |
| 0.006 |
|
Yes | 1 |
| 1 |
|
| No | 1.451
(0.596–3.534) |
| 2.031
(0.904–4.564) |
|
| Pathological N
stage |
| 0.002 |
| <0.001 |
|
ypN0 | 1 |
| 1 |
|
|
ypN+ | 2.681
(1.458–4.933) |
| 2.709
(1.634–4.492) |
|
| CMTM4
(pre-nCRT) |
|
|
| 0.036 |
|
Low |
|
| 1 |
|
|
High |
|
| 1.759
(1.037–2.984) |
|
CMTM4 knockdown impaired cell
migration and invasion triggered by radiation
To further investigate the role of CMTM4 in
chemoradiotherapy, a CMTM4 knockdown cell line was established
using lentivirus shRNA. The knockdown efficiency was verified by
immunofluorescence, western blotting and RT-qPCR (Fig. 2A-C). Compared with the NC cells,
CMTM4 knockdown significantly increased cell proliferation,
migration and invasion (Fig. 2D-F).
Following radiation exposure (2 and 5 Gy), there was no significant
difference between the NC and CMTM4 knockdown groups in terms of
colony formation ability (Fig. 2D).
However, compared with the NC group, interfering with CMTM4
knockdown significantly decreased the cell migration and invasion
under IR treatment (Fig. 2E and F).
These in vitro experiments were consistent with the findings
in the clinical data; CMTM4 expression induced radiotherapy
resistance in colon cancer cells.
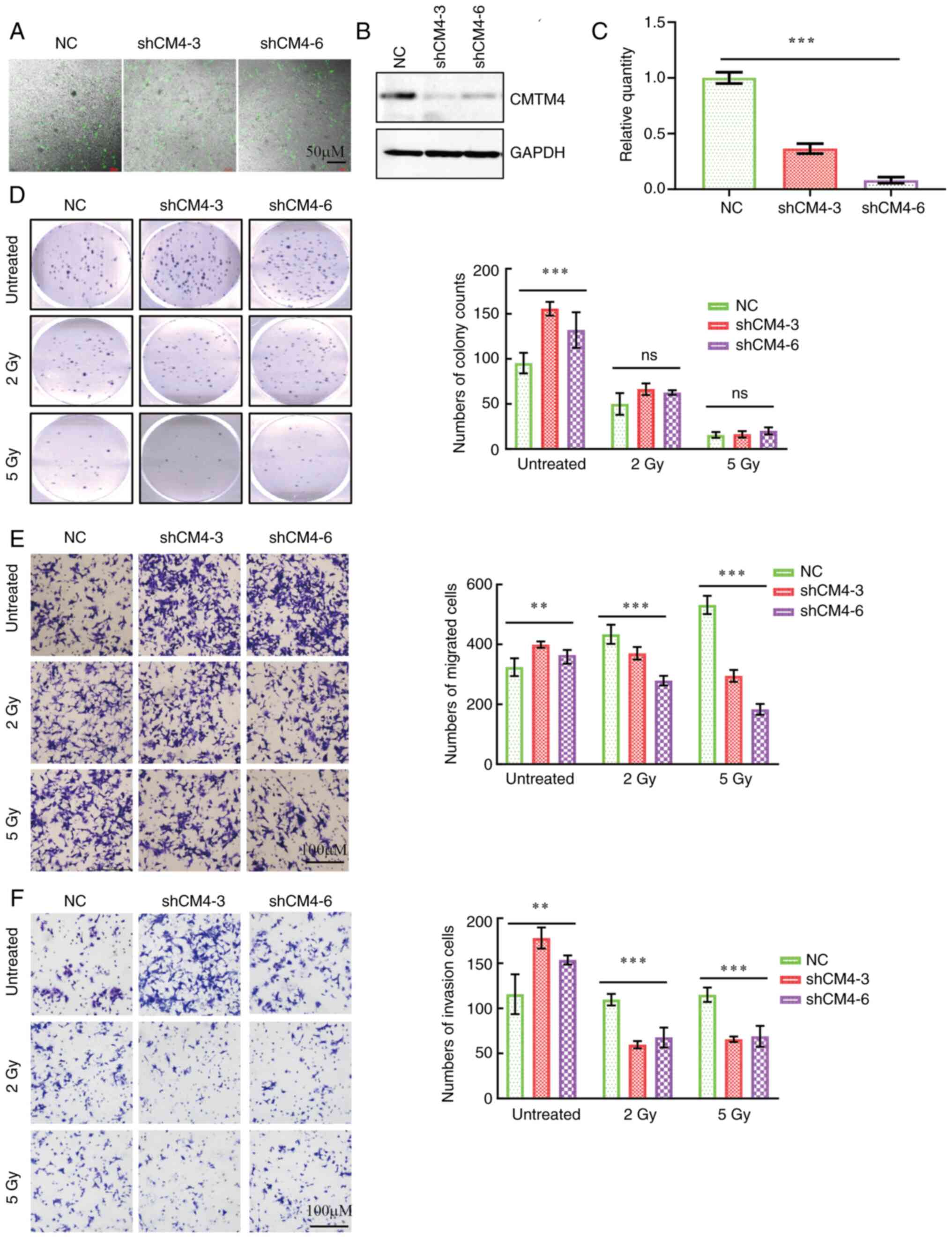 | Figure 2.Radiation impairs cell proliferation,
migration and invasion triggered by CMTM4 knockdown. (A)
Representative immunofluorescence images of CMTM4 knockdown in LoVo
cells. Scale bar, 50 µM. (B) Western blotting and (C) reverse
transcription-quantitative PCR of CMTM4 expression in LoVo cells
transfected with lenti-shCM4-3, lenti-shCM4-6 and NC. (D) IR
impaired the cell proliferation induced by CMTM4 knockdown. Left
panel, representative images of colony formations; right panel,
quantification of cell count in colony formation experiments. IR
inhibited the cell (E) migration and (F) invasion of lenti-shCMTM4
cells compared with the control cells. Left panel, representative
images of cell migration and invasion; right panel, quantification
of cell counts in Transwell experiments. Scale bar, 100 µM. Data
are presented as the mean ± SEM from three independent experiments.
**P<0.01 and ***P<0.001, determined by one-way ANOVA. ns, not
significant. NC, negative control; CMTM4, CKLF-like MARVEL
transmembrane domain member 4; IR, irradiation. |
CMTM4 participates in multiple
pathways in colon cancer and regulates the expression of
immune-related cytokines
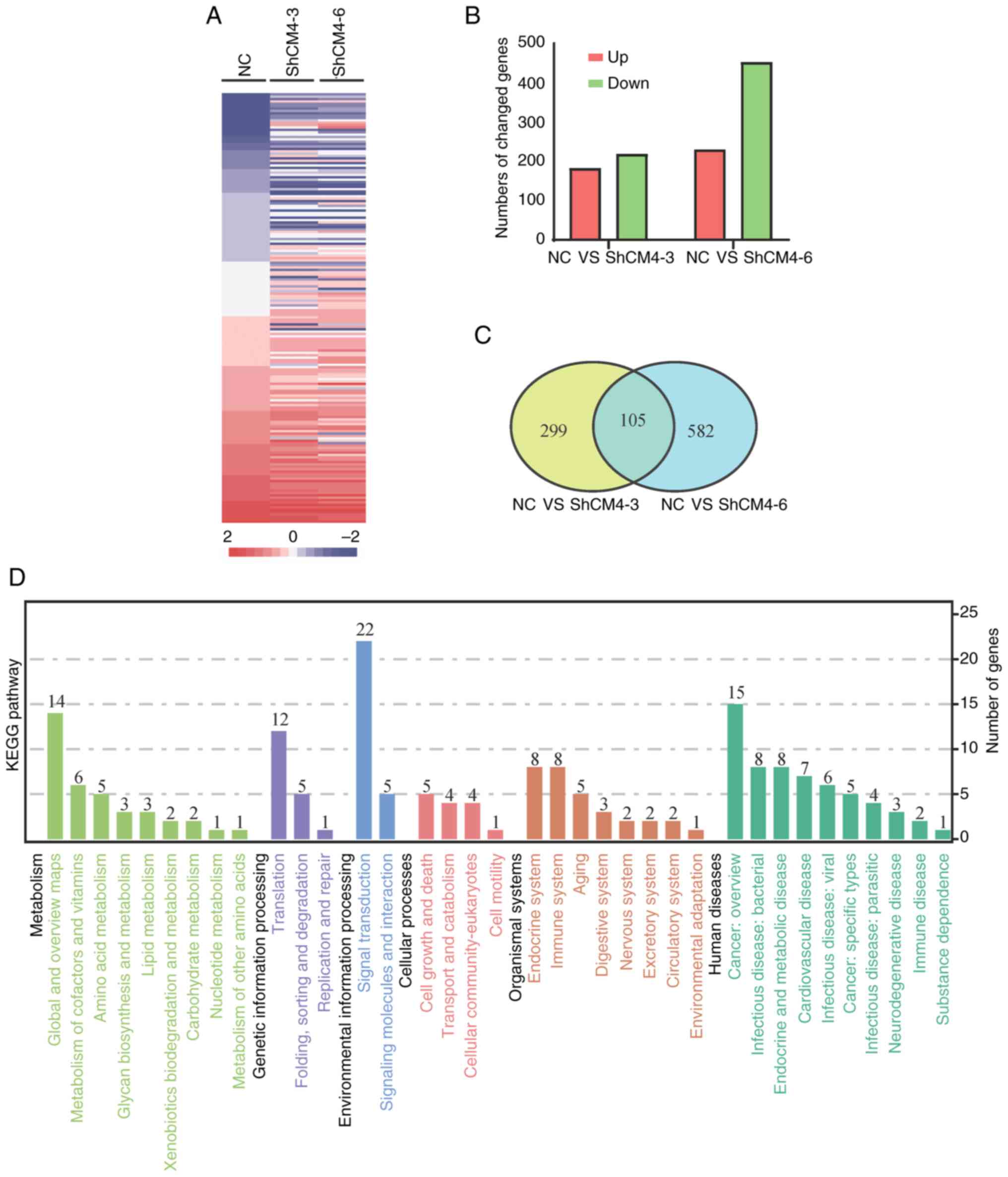
RNA samples were extracted from LoVo-NC,
LoVo-shCM4-3 and LoVo-shCM4-6 cells, and microarray analysis was
performed to examine the effect of CMTM4 on the gene expression
profiles (Fig. 3A). Compared with
the LoVo-NC group, the LoVo-shCM4-3 group had 184 upregulated and
220 downregulated genes, while the LoVo-shCM4-6 group had 232
upregulated and 455 downregulated genes (Fig. 3B). The expression of 105 genes was
mutually altered by CMTM4 knockdown, including 38 upregulated and
67 downregulated genes. CXCL8 was shown to be upregulated by CMTM4
knockdown in the RNA-seq analysis (Fig.
3C and Table SII). Gene
ontology function analysis indicated that CMTM4 knockdown altered
different pathways involved in ‘Metabolism’, ‘Genetic Information
Processing’, ‘Environmental Information Processing’, ‘Cellular
Processes’, ‘Organismal Systems and ‘Human Diseases’ (Fig. 3D). These 105 genes were shown to
participate in the KEGG pathways associated with different
physiological and pathological processes including ‘Metabolism’,
‘Genetic Information Processing’, ‘Cellular Processes’ and ‘Human
Diseases’. The profiles of the representative top 19 KEGG
enrichment pathways (P<0.05) are listed in Table SIII, such as ‘Aminoacyl-tRNA
biosynthesis’ in translation, ‘FoxO signaling pathway’, ‘PI3K-Akt
signaling pathway’ in signal transduction, ‘Mitophagy’ in transport
and catabolism, ‘Apoptosis’ in cell growth and death, ‘NOD-like
receptor signaling pathway’ in immune system.
nCRT typically triggers metabolism, inflammation and
an immune system response (31,32).
Therefore, the TIMER database was used to explore the association
between CMTM4 and immune cell infiltration in READ. In the READ
dataset, a significant although weak correlation was observed
between CMTM4 expression and immune cell infiltration in B cells
(ρ=0.231, P=6.13×10−03), CD8+ T cells
(ρ=0.214, P=1.13×10−02) but not with purity (ρ=−0.105,
P=2.16×10−01), CD4+ T cells (ρ=0.033,
P=7.03×10−01), macrophages (ρ=0.071,
P=4.09×10−01), neutrophils (ρ=0.064,
P=4.57×10−01) and dendritic cells (ρ=0.056,
P=5.15×10−01) (Fig.
S2). The TIMER database was also used to further evaluate the
association between CMTM4 expression and immune marker sets in READ
and colon adenocarcinoma (COAD). The association between CMTM4 and
immune cell markers of B cells, monocytes, tumor-associated
macrophages (TAMs), M1 macrophages, M2 macrophages, neutrophils,
natural killer cells, dendritic cells, general T cells and
CD8+ T cells were examined. Potential CMTM4-related
immune gene markers in READ were selected as follows: Cor>0.15
and P<0.05, which were found only in the READ dataset and not in
COAD. As shown in Table IV, with
or without tumor purity adjustment, negative weak correlations were
observed between CMTM4 and IL-10 in TAMs (ρ=−0.229,
P=2.97×10−03; ρ=−0.21, P=1.30×10−02), cluster
of differentiation 33 (CD33; ρ=−0.186, P=1.66×10−02;
ρ=−0.180, P=3.41×10−02) in neutrophils and transforming
growth factor β1 in T follicular helper (Tfh) cells (ρ=−0.221,
P=4.31×10−03; ρ=−0.275, P=1.04×10−03).
Without tumor purity adjustment, CMTM4 expression was weakly
negatively correlated with CXCL8 (ρ=−0.159,
P=4.10×10−02), IL13 in T helper (Th)2 (ρ=−0.186,
P=1.64×10−02) and granzyme B in exhausted T cells (GZMB;
ρ=−0.158, P=4.15×10−02). Taking tumor purity into
consideration, CD66b in neutrophils (ρ=−0.257,
P=2.27×10−03) and programmed cell death 1 in T cell
exhaustion (ρ=−0.172, P=4.24×10−02) were negatively
correlated with CMTM4 expression in READ. BDCA-4, otherwise known
as neuropilin-1, (ρ=0.169, P=2.95×10−02; ρ=0.271,
P=1.24×10−03) in dendritic cells, STAT1 in Th1 cells
(ρ=0.178, P=2.15×10−02; ρ=0.253, P=2.6×10−03)
and B-cell lymphoma 6 (ρ=0.191, P=1.36×10−02; ρ=0.219,
P=9.46×10−03) were weakly positively correlated with
CMTM4 expression in READ with or without tumor purity adjustment.
Without tumor purity adjustment, CMTM4 expression was only weakly
positively correlated with that of IL-21 (ρ=0.154;
P=4.77×10−02) in Tfh cells. The association between
CMTM4 and other gene markers, as well as B cells, monocytes, TAMs,
M1 macrophages, M2 macrophages, nature killer cells, dendritic
cells and CD8 cells are shown in Table
SIV. V-set and immunoglobulin domain containing 4 and membrane
spanning 4-domains A4A in M2 macrophages, HLA-DPB1, HLA-DQB1,
HLA-DRA and HLA-DPA1 in dendritic cells and CD3D in general T cells
were related to CMTM4 expression in both COAD and READ. Meanwhile,
STAT3 in T helper 17 and STAT5B in regulatory T cells were
positively correlated with CMTM4 expression in both COAD and
READ.
 | Table IV.Correlation of CMTM4 and gene markers
on immune cell infiltration. |
Table IV.
Correlation of CMTM4 and gene markers
on immune cell infiltration.
|
|
| COAD | READ |
|---|
|
|
|
|
|
|---|
|
|
| None | Purity | None | Purity |
|---|
|
|
|
|
|
|
|
|---|
| Cell type | Marker | Cor | P-value | Cor | P-value | Cor | P-value | Cor | P-value |
|---|
| TAM | IL10 | −0.092 |
5.02×10−02 | −0.084 |
8.99×10−2 | −0.229 |
2.97×10−03 | −0.21 |
1.30×10−02 |
| Neutrophils | CD66b | 0.016 |
7.39×10−01 | −0.027 |
5.88×10−01 | −0.134 |
8.62×10−02 | −0.257 |
2.27×10−03 |
|
| (CEACAM8) |
|
|
|
|
|
|
|
|
|
| CXCL8 | −0.098 |
3.63×10−02 | −0.09 |
6.97×10−02 | −0.159 |
4.10×10−02 | −0.117 |
1.71×10−01 |
|
| CD33 | −0.136 |
3.46×10−03 | −0.132 |
7.55×10−03 | −0.186 |
1.66×10−02 | −0.18 |
3.41×10−02 |
| Dendritic
cells | BDCA-4 (NRP1) | 0.015 |
7.54×10−01 | 0.032 |
5.19×10−01 | 0.169 |
2.95×10−02 | 0.271 |
1.24×10−03 |
| Th1 | STAT1 | 0.002 |
9.70×10−01 | −0.107 |
2.16×10−02 | 0.178 |
2.15×10−02 | 0.253 |
2.63×10−03 |
| Th2 | IL13 | −0.076 |
1.06×10−01 | −0.058 |
2.47×10−01 | −0.186 |
1.64×10−02 | −0.142 |
9.52×10−02 |
| Tfh | BCL6 | 0.069 |
1.40×10−01 | 0.085 |
8.90×10−02 | 0.191 |
1.36×10−02 | 0.219 |
9.46×10−03 |
|
| IL21 | −0.069 |
1.39×10−01 | −0.068 |
1.74×10−01 | 0.154 |
4.77×10−02 | 0.134 |
1.17×10−01 |
| Treg | TGFβ (TGFB1) | −0.131 |
5.08×10−03 | −0.122 |
1.39×10−02 | −0.221 |
4.31×10−03 | −0.275 |
1.04×10−03 |
| Exhausted T
cells | PD-1 (PDCD1) | −0.12 |
1.01×10−02 | −0.109 |
2.79×10−02 | −0.130 |
9.39×10−02 | −0.172 |
4.24×10−02 |
|
| GZMB | −0.086 |
6.73×10−02 | 0.094 |
5.92×10−02 | −0.158 |
4.15×10−02 | −0.131 |
1.23×10−01 |
CXCL8 is negatively correlated with
CMTM4 and indicates poor outcomes in patients with LARC
To verify the RNA-seq and TIMER database results,
the CMTM4 overexpression and CMTM4 shRNA knockdown cell lines were
constructed. CMTM4 knockdown by shRNA significantly altered CD66b,
CXCL8, STAT1, PD-1 and GZMB levels (Fig. 4A). However, CMTM4 overexpression did
not change the mRNA level of STAT1 and GZMB (Fig. 4B). There was a negative association
between CMTM4 and CD66b in the TIMER database (Table IV), but the mRNA level of CD66b was
positively associated with CMTM4 in the RT-qPCR experiments
(Fig. 4A and B). Therefore, CD66b
has been excluded from the further analysis and CXCL8 was
specifically chosen as CMTM4-regulated gene of interest. We
hypothesized that CMTM4 negatively regulates CXCL8.
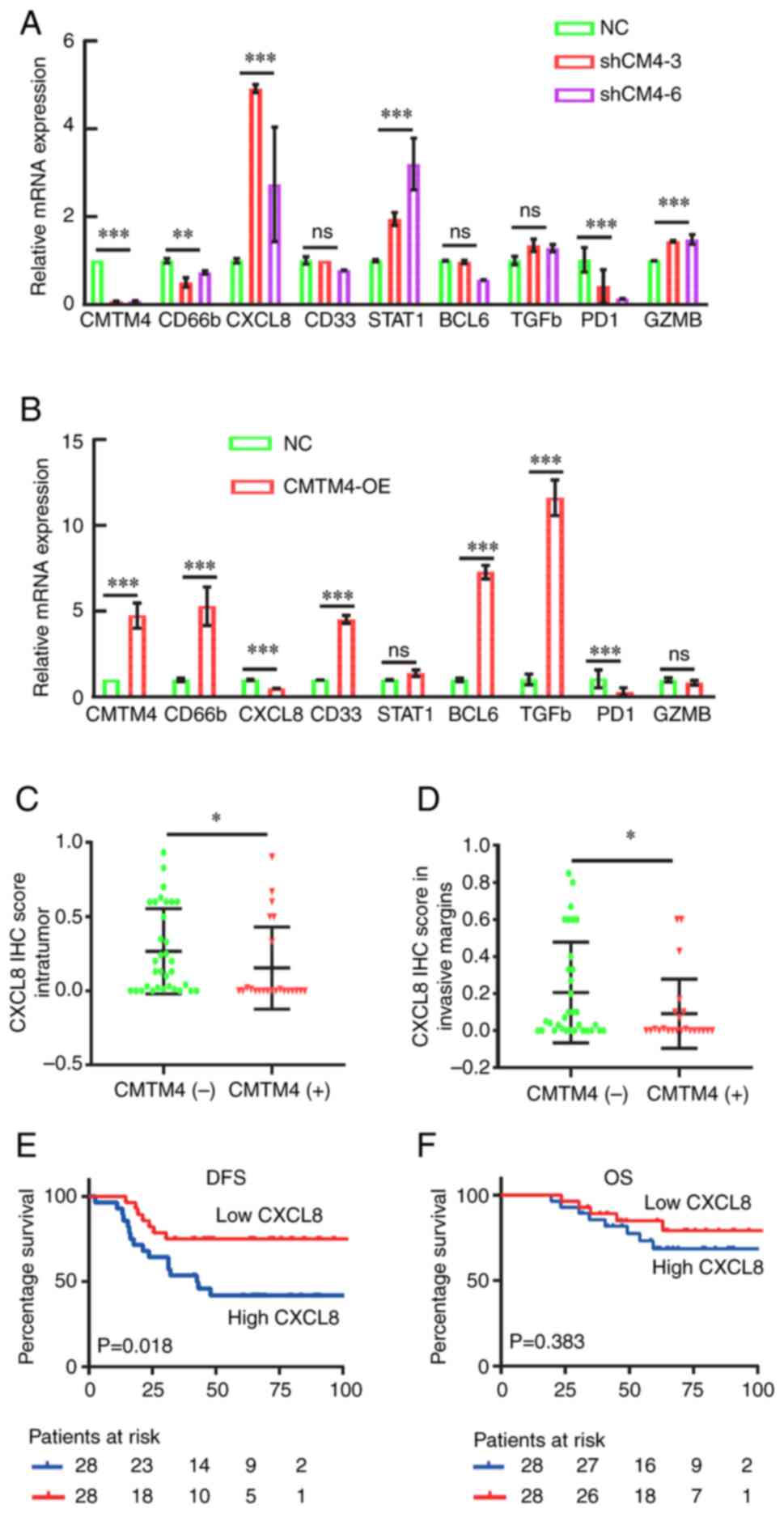 | Figure 4.CXCL8 was negatively associated with
CMTM4 and indicated an inferior outcome in patients with LARC. (A)
RT-qPCR of RNA-seq-identified CMTM4 targets in the shCM4-3 and
shCM4-6 cells, compared with NC cells. The level of GAPDH
transcripts was used for normalization. (B) RT-qPCR analysis of
RNA-seq-identified CMTM4 targets in the CMTM4 overexpression cells,
compared with the NC cells. The level of GAPDH transcripts was used
for normalization. (C) CXCL8 expression in the intratumor region
was negatively correlated with CMTM4 expression in patients with
LARC (n=56; P=0.012). (D) CXCL8 expression in the invasive margin
region was negatively associated with CMTM4 expression in patients
with LARC (n=56; P=0.049). (E) K-M analysis indicated that CXCL8
expression in tumor margins was correlated with a shorter DFS (95%
CI, 1.189–6.166; P=0.018). (F) K-M analysis indicated that CXCL8
expression in tumor margins was not associated with OS in patients
with LARC (P=0.383). *P<0.05, **P<0.01 and ***P<0.001. NC,
negative control. CXCL8, chemokine (CXC motif) ligand 8; CMTM4,
CKLF-like MARVEL transmembrane domain member 4; RT-qPCR, reverse
transcription-quantitative PCR; NC, negative control; K-M,
Kaplan-Meier; CI, confidence interval; DFS, disease-free survival;
OS, overall survival; OE, overexpression; sh, short hairpin. |
Next, CXCL8 expression in the surgical tissues from
patients with LARC (n=56) were detected using IHC. The depth of
immune cell infiltration may exhibit different functions in
antitumor activity. The location of CXCL8 expression was classified
into intratumor and tumor invasive margins (Fig. S3). Both the intratumor CXCL8
expression or the tumor invasive margins in the CMTM4 negative
group were higher than those in the CMTM4 positive group (P=0.012
and P=0.049, respectively; Fig. 4C and
D). This indicated that the location of CXCL8 did not affect
the negative association between CXCL8 and CMTM4. Additionally, 5
post-nCRT tissues with high CMTM4 expression and 3 tissues with low
CMTM4 expression were also collected for RT-qPCR analysis. The
average relative expression of CXCL8 in the CMTM4 high group was
significantly lower than the expression in the CMTM4 low group
(0.075 vs. 0.646; P=0.031; Fig.
S4A). The negative association between CMTM4 and CXCL8 mRNA
expression was also verified. Furthermore, Kaplan-Meier (K-M)
analysis results showed that CXCL8 expression in tumor margins was
linked to a shorter DFS (95% CI, 1.189–6.166; P=0.018; Fig. 4E) but did not significantly affect
the OS (P=0.383; Fig. 4F). There
were no significant associations between CXCL8 expression in the
intratumor region and survival time (DFS, P=0.546; OS, P=0.973;
Fig. S4B and C). In addition,
univariate analysis showed that clinicopathological characteristics
such as nerve invasion, pathological tumor (pT) stage and node (N)
stage indicated a poorer DFS and OS (Table V). Multivariate analysis showed that
CXCL8 expression in the tumor invasive margin regions and
pathological N stage were both independent DFS predictors in
patients with LARC (95% CI, 1.053–10.514, P=0.041; 95% CI
1.586–10.750, P=0.004; Table VI).
Therefore, CMTM4 was negatively associated with the
neutrophil-related cytokine, CXCL8, in LARC tissues and CXCL8
expression in surgical tissues may be an independent prognostic
factor in patients with LARC treated with nCRT.
 | Table V.Univariate analysis to identify
prognosis-related factors in patients with LARC. |
Table V.
Univariate analysis to identify
prognosis-related factors in patients with LARC.
|
| DFS, HR (95%
CI) | OS, HR (95%
CI) |
|---|
|
|
|
|
|---|
| Variable | P-value | HR | Lower | Upper | P-value | HR | Lower | Upper |
|---|
| Sex | 0.833 | 0.905 | 0.356 | 2.297 | 0.741 | 0.802 | 0.217 | 2.965 |
| Age | 0.915 | 0.956 | 0.422 | 12.168 | 0.705 | 0.801 | 0.254 | 2.530 |
| Vascular
invasion | 0.730 | 1.425 | 0.192 | 10.594 | 0.329 | 2.774 | 0.357 | 21.565 |
| Nerve invasion | 0.013 | 17.827 | 1.854 | 171.401 | 0.021 | 13.295 | 1.486 | 118.958 |
| Pathological T
stage | 0.016 | 2.116 | 1.152 | 3.886 | 0.125 | 6.653 | 0.593 | 74.690 |
| Pathological N
stage | 0.000 | 5.530 | 2.280 | 13.410 | 0.026 | 3.930 | 1.175 | 13.143 |
| CXCL8 margins | 0.023 | 2.797 | 1.150 | 6.803 | 0.253 | 2.013 | 0.606 | 6.691 |
| CXCL8
intratumor | 0.269 | 1.604 | 0.694 | 3.708 | 0.988 | 0.991 | 0.319 | 3.074 |
 | Table VI.Multivariate analysis to identify
prognosis-related factors in patients with LARC. |
Table VI.
Multivariate analysis to identify
prognosis-related factors in patients with LARC.
|
| DFS, HR (95%
CI) | OS, HR (95%
CI) |
|---|
|
|
|
|
|---|
| Variable | P-value | HR | Lower | Upper | P-value | HR | Lower | Upper |
|---|
| Nerve invasion | 0.274 | 3.677 | 0.357 | 37.910 | 0.283 | 3.811 | 0.332 | 43.725 |
| Pathological T
stage | 0.059 | 1.834 | 0.976 | 3.446 | 0.950 | 476.286 | 0.000 |
9.16×1086 |
| Pathological N
stage | 0.004 | 4.129 | 1.586 | 10.750 | 0.199 | 2.483 | 0.620 | 9.944 |
| CXCL8 margins | 0.041 | 3.328 | 1.053 | 10.514 | 0.299 | 2.075 | 0.524 | 8.219 |
| CXCL8
intratumor | 0.564 | 0.722 | 0.238 | 2.187 | 0.541 | 0.651 | 0.164 | 2.576 |
Discussion
The CMTM family members have been shown to play
important roles in different types of cancer, including CRC, clear
cell renal carcinoma, HNSCC and HCC (9–11,13,14,33,34).
However, CMTM4 has been much less studied compared with the other
CMTM family members (8,18). Xue et al (9) observed lower CMTM4 expression in
colorectal adenocarcinoma compared with normal tissues. The low
CMTM4 expression was associated with a significantly shorter OS
time based on The Human Protein Atlas database. A similar pattern
was found in lung adenocarcinoma (35). CMTM4 inhibited clear cell renal cell
carcinoma 768-O cell-derived tumor growth in tumor xenograft model
experiments (10). However, high
expression of CMTM4 was found to be associated with poor prognosis
in HNSCC, and interfering with CMTM4 expression inhibited EMT and
expression of the cancer stem cell markers, CD44, aldehyde
dehydrogenase 1, B-cell-specific Moloney murine leukemia virus
integration region 1 and SRY-box 2, through the AKT pathway
(11). High CMTM4 expression was
also shown to be associated with poor DFS and OS in HCC by the TCGA
database (12). However, CMTM4
showed no significant prognostic association with survival in
patients with HCC (n=90) from Guilin Medical University (Guilin,
China) (14). Therefore, the
function of CMTM4 in solid tumors remains unclear, particularly the
expression pattern and role of CMTM4 in rectal cancer.
The treatment of LARC has always been challenging to
uncover, although new treatments such as nCRT or TNT have been
established for decades. The side effects, reduced quality of life
and local recurrence still cause problems in the clinic. The
evaluation of clinical CR using MRI, endoscopy and digital rectal
examination is suboptimal compared with the pCR following TME. The
proposal of the Watch and Waite strategy also accelerated the
demand to explore biomarkers to predict nCRT response in LARC
before TME (36).
In the present study, the application of CMTM4 in
predicting nCRT response was first explored in 228 patients with
LARC, focusing on the patients with LARC (T3-4/N+M0) who received
nCRT following TME. Patients with LARC and high CMTM4 expression in
the biopsy tissue had a lower rate of reaching pCR status after TME
(P=0.041). K-M analysis also showed that high CMTM4 expression in
the biopsy tissues was associated with a lower DFS and OS. In the
postoperative tissues, the expression of CMTM4 was higher in tumor
tissues compared with adjacent normal tissues. No association
between CMTM4 expression and survival was observed in postoperative
tissues from patients with LARC (T3/T4). Considering the analysis
of the published data from the TCGA database, the results indicated
that the function of CMTM4 is varied in different tumor development
stages or treatment stages of rectal cancer. The role of CMTM4 in
treatment response has been reported in gastric cancer and HCC
(12,14,34). A
high percentage of CMTM4+ epithelial cells indicated a
shorter prognosis in gastric cancer, while in mesenchymal regions,
a high percentage of CMTM4+ cells was associated with an
improved OS (34). Furthermore,
CMTM4/6 exhibited higher expression in the partial response group
with PDL1 therapy compared with the stable disease and progressive
disease groups. Above all, radiation or immune checkpoints
inhibitors therapy possibly changed the expression of CMTM4 in LARC
or gastric cancer. These studies suggested the potential
application of CMTM4 in nCRT or PDL1 therapy to achieve treatment
response in gastrointestinal cancer.
According to the World Health Organization
classification of tumors, venous invasion and perineural invasion
in colorectal cancers is 4–40% and 20%, respectively (37–41).
The low frequency of venous invasion and perineural invasion
observed in the present study was lower than previously reported.
There are two reasons for the lower frequency in the data of the
present study. First, the higher the T stage, the higher the
frequency of vascular and nerve invasion. A previous study enrolled
1,142 patients with CRC undergoing resection, and vascular invasion
was present in 40.5% of patients with T4 rectal cancer (37). Nerve invasion in another study was
present in 5.7% of patients with pT1-T2 and 24.0% of patients with
pT3-T4 (37,38). Additionally, venous invasion and
perineural invasion has not been observed in the post-operative
tissues from pCR patients (41). In
the present study, 55.7% of the patients were assessed as pCR, pT1
or pT2 after surgery; therefore, the observed frequency of venous
invasion and perineural invasion was lower. Second, the present
study was a single center retrospective study with a limited sample
number. The percentage of vascular and nerve invasion in patients
with LARC receiving nCRT treatment should be further proved in a
multicenter large enrollment study. Additionally, the surgical team
at Peking University Cancer Hospital (Beijing, China) published a
study in 2018, and the lymphovascular invasion was reported as
5.4%, which is similar to the results of the present study
(39).
It was confirmed in the present study that CMTM4
acted as a tumor suppressor gene in a colon cancer cell line
without IR treatment, consistent with a previous study (9). Following radiation treatment, the
migration and invasion ability of colon cancer cells was markedly
decreased by knocking down CMTM4 expression, which agreed with the
rectal cancer clinical findings. Radiation therapy has been shown
to reduce the tumor size (36),
while it also triggers an immune response and activates
CD8+ T cell infiltration (42). A positive correlation has also been
found between CD8+ infiltration and CMTM4 expression in
the stroma region of HNSCC (11).
In the context of liver cancer, CMTM4 has been shown to be the
primary positive regulator of PDL1 through a post-translational
mechanism (12). CMTM4/6 has been
reported to stabilize PDL1 in both tumor and dendritic cells by
reducing its ubiquitination. CMTM6 interacts with CD58 and PDL1.
Additionally, CMTM6 depletion activates the tumor specific
perforin- and TNFα producing CD8+ T cell activity
through PDL1 (16,17,33).
In the present study, to examine the downstream genes of CMTM4,
RNA-seq and TIMER database analyses were performed. Colon cancer
cells were used in the present study as the knockdown of CMTM4
expression was more notable in LoVo cells than the rectal cell
lines, such as SW480 (data not shown). The expression levels of 105
genes were changed after knocking down CMTM4 expression in the LoVo
cells. CXCL8 was the only gene negatively associated with CMTM4
expression in both the RNA-seq and TIMER analyses. The negative
association was confirmed by RT-qPCR and IHC staining. The
expression levels of CXCL8 and CMTM4 were detected in tissues from
patients with LARC to confirm the results obtained from the colon
cancer cell line.
CXCL8, a proinflammatory chemokine, is expressed in
epithelial and macrophage cells and recruits neutrophils to
inflammatory sites (43). CXCL8 is
induced by immune infiltration cells, stromal cells and tumor cells
in the tumor microenvironment (44,45).
Multiple G protein-mediated signaling pathway cascades are
activated by CXCL8-CXCR1/2 binding, including the PI3K-Akt,
ERK-MAPK, P38-MAPK, FAK-Src and JAK-STAT pathways (46–52).
CXCL8 participates in tumor cell motility, angiogenesis and
survival through these signaling pathways. The expression level of
CXCL8 serves as a prognostic marker in numerous types of cancer.
For instance, the stroma level of CXCL8 was shown to be negatively
associated with the 5-year survival rate in right-side colon cancer
(53), which was consistent with
the results of the present study. High expression of CXCL8 was also
shown to indicate poor prognosis in triple-negative and estrogen
receptor-negative breast cancer (54,55).
CXCL8 expression is also positively correlated with lymph node
metastasis and TNM stage in patients with non-small cell lung
cancer (56). CXCL8 secreted by
tumors through paracrine signaling recruits neutrophils and
macrophages into the tumor microenvironment and inhibits the
antitumor immune activity (57).
Thus, CXCL8 is associated with chemotherapy and immunotherapy
resistance in breast, gastric, prostate and pancreatic cancer
(58–62). In the present study, the prognostic
role of CXCL8 was reported in patients with LARC subjected to nCRT.
CXCL8 expression was negatively associated with CMTM4 expression in
postoperative tissues and CXCL8 expression indicated a shorter DFS
time, but not OS time. The role of CXCL8 in the prediction of nCRT
response in pre-operative tissues was not investigated in the
present study. This is because the role of CXCL8 in radiotherapy or
chemoradiotherapy is unclear, particularly in rectal cancer. In
head and neck cancer, lower salivary CXCL8 levels indicate improved
radiotherapy outcomes (63).
Additionally, higher CXCL8 expression in head and neck squamous
carcinoma tissues is associated with a lower 5-year local
recurrence-free survival (64). In
the present study, CMTM4, which indicated an unfavorable nCRT
response and DFS, was not associated with DFS and OS in
postoperative tissues. Therefore, CXCL8 combined CMTM4 may predict
outcomes in patients with LARC on two independent time points:
Biopsy tissues from pre-nCRT and surgery tissues.
In conclusion, CMTM4 may be a new nCRT response
prediction biomarker in patients with LARC. CMTM4 may serve as a
target to sensitize chemoradiation therapy in patients with LARC.
In the present study, high CXCL8 expression indicated poorer
outcomes in patients with LARC who received TME followed by
nCRT.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
The authors would like to thank Miss Jiayi Ma
(Beijing National Day School, Beijing 100039, P.R. China) for
experimental assistance including cell culture, migration and
invasion.
Funding
This work was supported by PKU-Baidu Fund (grant no. A002292),
the Capital's Funds for Health Improvement and Research (grant no.
2022-2-1024), Peking University Medicine Seed Fund for
Interdisciplinary Research (grant no. BUM2020MX009), the Capital's
Funds for Health Improvement and Research (grant no. 2018-2-1022),
the Beijing Municipal Science and Technology Commission Capital
Characteristic Clinical Application Research (grant no.
Z141107002514077), the National Natural Science Foundation of China
(grant no. 81872309) and the National Natural Science Foundation of
China (grant no. 61501039).
Availability of data and materials
The RNA-sequencing data generated in the present
study may be found in the Sequence Read Archive under accession no.
PRJNA1176295 or at the following URL: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1176295.
All other data generated in the present study may be requested from
the corresponding author.
Authors' contributions
ZWL and SYL contributed to the study conception and
design. LJY, ZTM, NNL and SYL wrote the first draft of the
manuscript. LJY, ZTM, NNL and YHB were responsible for collecting
the patient's information and the IHC investigation and assessment.
ZTM, LM, QF, DBQ, YW and PW were responsible for material
preparation, data collection and analysis. LJY, YHB, ZWL and SYL
provided comments on previous versions of the manuscript. ZWL and
SYL confirm the authenticity of all the raw data. All authors read
and approved the final version of the manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Peking University Cancer Hospital & Institute
(Beijing, China; approval no. 2021KT93).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
LARC
|
locally advanced rectal cancer
|
|
nCRT
|
neoadjuvant chemoradiotherapy
|
|
pCR
|
pathological complete response
|
|
CRC
|
colorectal cancer
|
|
TME
|
total mesorectal excision
|
|
TNT
|
total neoadjuvant therapy
|
|
CR
|
complete response
|
|
CMTM
|
CKLF-like MARVEL transmembrane domain
member
|
|
RT-qPCR
|
reverse transcription-quantitative
qPCR
|
|
OS
|
overall survival
|
|
DFS
|
disease-free survival
|
|
READ
|
rectum adenocarcinoma
|
|
COAD
|
colon adenocarcinoma
|
|
TAMs
|
tumor-associated macrophages
|
|
Th
|
T helper
|
|
Tfh
|
Follicular helper T cell
|
|
ICI
|
immune checkpoints inhibitors
|
|
MRI
|
magnetic resonance imaging
|
|
CXCL8
|
chemokine (CXC motif) ligand 8
|
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Xia C, Dong X, Li H, Cao M, Sun D, He S,
Yang F, Yan X, Zhang S, Li N and Chen W: Cancer statistics in China
and United States, 2022: Profiles, trends, and determinants. Chin
Med J (Engl). 135:584–590. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dekker E, Tanis PJ, Vleugels JLA, Kasi PM
and Wallace MB: Colorectal cancer. Lancet. 394:1467–1480. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang Y, Shen L, Wan J, Zhang H, Wu R, Wang
J, Wang Y, Xu Y, Cai S, Zhang Z and Xia F: Neoadjuvant
chemoradiotherapy combined with immunotherapy for locally advanced
rectal cancer: A new era for anal preservation. Front Immunol.
13:10670362022. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mei WJ, Wang XZ, Li YF, Sun YM, Yang CK,
Lin JZ, Wu ZG, Zhang R, Wang W, Li Y, et al: Neoadjuvant
chemotherapy with CAPOX versus chemoradiation for locally advanced
rectal cancer with uninvolved mesorectal fascia (CONVERT): Initial
results of a phase III trial. Ann Surg. 277:557–564. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Peeters KCMJ, Marijnen CAM, Nagtegaal ID,
Kranenbarg EK, Putter H, Wiggers T, Rutten H, Pahlman L, Glimelius
B, Leer JW, et al: The TME trial after a median follow-up of 6
years: Increased local control but no survival benefit in
irradiated patients with resectable rectal carcinoma. Ann Surg.
246:693–701. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kapiteijn E, Marijnen CA, Nagtegaal ID,
Putter H, Steup WH, Wiggers T, Rutten HJ, Pahlman L, Glimelius B,
van Krieken JH, et al: Preoperative radiotherapy combined with
total mesorectal excision for resectable rectal cancer. N Engl J
Med. 345:638–646. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wu J, Li L, Wu S and Xu B: CMTM family
proteins 1–8: Roles in cancer biological processes and potential
clinical value. Cancer Biol Med. 17:528–542. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xue H, Li T, Wang P, Mo X, Zhang H, Ding
S, Ma D, Lv W, Zhang J and Han W: CMTM4 inhibits cell proliferation
and migration via AKT, ERK1/2, and STAT3 pathway in colorectal
cancer. Acta Biochim Biophys Sin (Shanghai). 51:915–924. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li T, Cheng Y, Wang P, Wang W, Hu F, Mo X,
Lv H, Xu T and Han W: CMTM4 is frequently downregulated and
functions as a tumour suppressor in clear cell renal cell
carcinoma. J Exp Clin Cancer Res. 34:1222015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li H, Liu YT, Chen L, Zhou JJ, Chen DR, Li
SJ and Sun ZJ: CMTM4 regulates epithelial-mesenchymal transition
and PD-L1 expression in head and neck squamous cell carcinoma. Mol
Carcinog. 60:556–566. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chui NN, Cheu JW, Yuen VW, Chiu DK, Goh
CC, Lee D, Zhang MS, Ng IO and Wong CC: Inhibition of CMTM4
sensitizes cholangiocarcinoma and hepatocellular carcinoma to T
cell-mediated antitumor immunity through PD-L1. Hepatol Commun.
6:178–193. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhou HQ, Li JH, Liu LW, Lou JM and Ren ZG:
Increased CMTM4 mRNA expression predicts a poor prognosis in
patients with hepatocellular carcinoma. Hepatobiliary Pancreat Dis
Int. 19:596–601. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tan S, Guo X, Bei C, Zhang H, Li D, Zhu X
and Tan H: Prognostic significance and immune characteristics of
CMTM4 in hepatocellular carcinoma. BMC Cancer. 22:9052022.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bei C, Zhang Y, Wei R, Zhu X, Wang Z, Zeng
W, Chen Q and Tan S: Clinical significance of CMTM4 expression in
hepatocellular carcinoma. Onco Targets Ther. 10:5439–5443. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mezzadra R, Sun C, Jae LT, Gomez-Eerland
R, de Vries E, Wu W, Logtenberg MEW, Slagter M, Rozeman EA, Hofland
I, et al: Identification of CMTM6 and CMTM4 as PD-L1 protein
regulators. Nature. 549:106–110. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Burr ML, Sparbier CE, Chan YC, Williamson
JC, Woods K, Beavis PA, Lam EYN, Henderson MA, Bell CC, Stolzenburg
S, et al: CMTM6 maintains the expression of PD-L1 and regulates
anti-tumour immunity. Nature. 549:101–105. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang T, Yu H, Dai X and Zhang X: CMTM6
and CMTM4 as two novel regulators of PD-L1 modulate the tumor
microenvironment. Front Immunol. 13:9714282022. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang L, Li ZY, Li ZW, Li YH, Sun YS, Ji
JF, Gu J and Cai Y: Efficacy and safety of neoadjuvant
intensity-modulated radiotherapy with concurrent capecitabine for
locally advanced rectal cancer. Dis Colon Rectum. 58:186–192. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cuccurullo V and Mansi L: AJCC cancer
staging handbook: From the AJCC Cancer Staging Manual (7th
edition). Eur J Nucl Med Mol Imaging. 38:4082011. View Article : Google Scholar
|
|
21
|
Wen Z, Luo D, Wang S, Rong R, Evers BM,
Jia L, Fang Y, Daoud EV, Yang S, Gu Z, et al: Deep learning-based
h-score quantification of immunohistochemistry-stained images. Mod
Pathol. 37:1003982024. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Matos LL, Trufelli DC, de Matos MG and da
Silva Pinhal MA: Immunohistochemistry as an important tool in
biomarkers detection and clinical practice. Biomark Insights.
5:9–20. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li T, Fan J, Wang B, Traugh N, Chen Q, Liu
JS, Li B and Liu XS: TIMER: A web server for comprehensive analysis
of tumor-infiltrating immune cells. Cancer Res. 77:e108–e110. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Danaher P, Warren S, Dennis L, D'Amico L,
White A, Disis ML, Geller MA, Odunsi K, Beechem J and Fling SP:
Gene expression markers of tumor infiltrating leukocytes. J
Immunother Cancer. 5:182017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ginestet C: ggplot2: Elegant graphics for
data analysis. Journal of the Royal Statistical Society Series A:
Stat Soc. 174:245–246. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Huang DW, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Huang DW, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Martin ST, Heneghan HM and Winter DC:
Systematic review and meta-analysis of outcomes following
pathological complete response to neoadjuvant chemoradiotherapy for
rectal cancer. Br J Surg. 99:918–928. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wen J, Fang S, Hu Y, Xi M, Weng Z, Pan C,
Luo K, Ling Y, Lai R, Xie X, et al: Impacts of neoadjuvant
chemoradiotherapy on the immune landscape of esophageal squamous
cell carcinoma. EBioMedicine. 86:1043712022. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shi M, Chen Y and Ji D: The implications
from the interplay of neoadjuvant chemoradiotherapy and the immune
microenvironment in rectal cancer. Future Oncol. 18:3229–3244.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Miao B, Hu Z, Mezzadra R, Hoeijmakers L,
Fauster A, Du S, Yang Z, Sator-Schmitt M, Engel H, Li X, et al:
CMTM6 shapes antitumor T cell response through modulating protein
expression of CD58 and PD-L1. Cancer Cell. 41:1817–1828.e1819.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang Z, Peng Z, Liu Q, Guo Z, Menatola M,
Su J, Li T, Ge Q, Wang P, Shen L and Jin R: Co-expression with
membrane CMTM6/4 on tumor epithelium enhances the prediction value
of PD-L1 on Anti-PD-1/L1 therapeutic efficacy in gastric
adenocarcinoma. Cancers (Basel). 13:51752021. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhu X, Zhang S, Tan S, Li D, Chen X, Kong
J, Fu Y, Wang C and Wen L: Expression of CMTM4 shows clinical
significance in lung cancer. Transl Cancer Res. 9:6214–6220. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu S, Jiang T, Xiao L, Yang S, Liu Q, Gao
Y, Chen G and Xiao W: Total neoadjuvant therapy (TNT) versus
standard neoadjuvant chemoradiotherapy for locally advanced rectal
cancer: A systematic review and meta-analysis. Oncologist.
26:e1555–e1566. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Bhangu A, Fitzgerald JE, Slesser A,
Northover JM, Faiz O and Tekkis P: Prognostic significance of
extramural vascular invasion in T4 rectal cancer. Colorectal Dis.
15:e665–e671. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chablani P, Nguyen P, Pan X, Robinson A,
Walston S, Wu C, Frankel WL, Chen W, Bekaii-Saab T, Chakravarti A,
et al: Perineural invasion predicts for distant metastasis in
locally advanced rectal cancer treated with neoadjuvant
chemoradiation and surgery. Am J Clin Oncol. 40:561–568. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wu AW, Cai Y, Li YH, Wang L, Li ZW, Sun YS
and Ji JF: Pattern and management of recurrence of mid-low rectal
cancer after neoadjuvant intensity-modulated radiotherapy:
Single-center results of 687 cases. Clin Colorectal Cancer.
17:e307–e313. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liu S, Yang S, Yu H, Luo H, Chen G, Gao Y,
Sun R and Xiao W: A nomogram for predicting 10-year cancer specific
survival in patients with pathological T3N0M0 rectal cancer. Front
Med (Lausanne). 9:9776522022. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Shin JK, Huh JW, Lee WY, Yun SH, Kim HC,
Cho YB and Park YA: Clinical prediction model of pathological
response following neoadjuvant chemoradiotherapy for rectal cancer.
Sci Rep. 12:71452022. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Sinha UK, Kast WM and Lin DC: Single-cell
genomics identifies immune response to neoadjuvant
chemoradiotherapy. EBioMed. 86:1043892022. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Holmes WE, Lee J, Kuang WJ, Rice GC and
Wood WI: Structure and functional expression of a human
interleukin-8 receptor. Science. 253:1278–1280. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Waugh DJ and Wilson C: The interleukin-8
pathway in cancer. Clin Cancer Res. 14:6735–6741. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Alfaro C, Sanmamed MF, Rodríguez-Ruiz ME,
Teijeira Á, Oñate C, González Á, Ponz M, Schalper KA, Pérez-Gracia
JL and Melero I: Interleukin-8 in cancer pathogenesis, treatment
and follow-up. Cancer Treat Rev. 60:24–31. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Meng ZW, Zhang L, Cai XR, Wang X, She FF
and Chen YL: IL-8 is a novel prometastatic chemokine in
intrahepatic cholangiocarcinoma that induces CXCR2-PI3K/AKT
signaling upon CD97 activation. Sci Rep. 13:187112023. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Chen F, Aye L, Yu L, Liu L, Liu Y, Lin Y,
Gao D, Gao Q and Zhang S: SSH1 promotes progression of intrahepatic
cholangiocarcinoma via p38 MAPK-CXCL8 axis. Carcinogenesis.
44:232–241. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhang SQ, Wang LL, Li YT, Wang G, Li L,
Sun SZ, Yao LJ and Shen L: MicroRNA-126 attenuates the effect of
chemokine CXCL8 on proliferation, migration, apoptosis, and
MAPK-dependent signaling activity of vascular endothelial cells
cultured in a medium with high glucose concentration. Bull Exp Biol
Med. 171:202–207. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Petreaca ML, Yao M, Liu Y, Defea K and
Martins-Green M: Transactivation of vascular endothelial growth
factor receptor-2 by interleukin-8 (IL-8/CXCL8) is required for
IL-8/CXCL8-induced endothelial permeability. Mol Biol Cell.
18:5014–5023. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Ahmed S, Mohamed HT, El-Husseiny N, El
Mahdy MM, Safwat G, Diab AA, El-Sherif AA, El-Shinawi M and Mohamed
MM: IL-8 secreted by tumor associated macrophages contribute to
lapatinib resistance in HER2-positive locally advanced breast
cancer via activation of Src/STAT3/ERK1/2-mediated EGFR signaling.
Biochim Biophys Acta Mol Cell Res. 1868:1189952021. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Fu XT, Dai Z, Song K, Zhang ZJ, Zhou ZJ,
Zhou SL, Zhao YM, Xiao YS, Sun QM, Ding ZB and Fan J:
Macrophage-secreted IL-8 induces epithelial-mesenchymal transition
in hepatocellular carcinoma cells by activating the
JAK2/STAT3/Snail pathway. Int J Oncol. 46:587–596. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Hu X, Yuan L and Ma T: Mechanisms of
JAK-STAT signaling pathway mediated by CXCL8 gene silencing on
epithelial-mesenchymal transition of human cutaneous melanoma
cells. Oncol Lett. 20:1973–1981. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Pennel KA, Quinn JA, Nixon C, Inthagard J,
van Wyk HC, Chang D, Rebus S; GPOL Group, ; Hay J, Maka NN, et al:
CXCL8 expression is associated with advanced stage, right sided
ness, and distinct histological features of colorectal cancer. J
Pathol Clin Res. 8:509–520. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Shen Y, Zhang B, Wei X, Guan X and Zhang
W: CXCL8 is a prognostic biomarker and correlated with TNBC brain
metastasis and immune infiltration. Int Immunopharmacol.
103:1084542022. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Fang QI, Wang X, Luo G, Yu M, Zhang X and
Xu N: Increased CXCL8 expression is negatively correlated with the
overall survival of patients with er-negative breast cancer.
Anticancer Res. 37:4845–4852. 2017.PubMed/NCBI
|
|
56
|
Gu L, Yao Y and Chen Z: An
inter-correlation among chemokine (C-X-C motif) ligand (CXCL) 1,
CXCL2 and CXCL8, and their diversified potential as biomarkers for
tumor features and survival profiles in non-small cell lung cancer
patients. Transl Cancer Res. 10:748–758. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
David JM, Dominguez C, Hamilton DH and
Palena C: The IL-8/IL-8R axis: A double agent in tumor immune
resistance. Vaccines (Basel). 4:222016. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zhang M, Huang L, Ding G, Huang H, Cao G,
Sun X, Lou N, Wei Q, Shen T, Xu X, et al: Interferon gamma inhibits
CXCL8-CXCR2 axis mediated tumor-associated macrophages tumor
trafficking and enhances anti-PD1 efficacy in pancreatic cancer. J
Immunother Cancer. 8:e0003082020. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Yi M, Peng C, Xia B and Gan L: CXCL8
facilitates the survival and paclitaxel-resistance of
triple-negative breast cancers. Clin Breast Cancer. 22:e191–e198.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Wang RX, Ji P, Gong Y, Shao ZM and Chen S:
Value of CXCL8-CXCR1/2 axis in neoadjuvant chemotherapy for
triple-negative breast cancer patients: A retrospective pilot
study. Breast Cancer Res Treat. 181:561–570. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Wilson C, Purcell C, Seaton A, Oladipo O,
Maxwell PJ, O'Sullivan JM, Wilson RH, Johnston PG and Waugh DJ:
Chemotherapy-induced CXC-chemokine/CXC-chemokine receptor signaling
in metastatic prostate cancer cells confers resistance to
oxaliplatin through potentiation of nuclear factor-kappaB
transcription and evasion of apoptosis. J Pharmacol Exp Ther.
327:746–759. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Lin C, He H, Liu H, Li R, Chen Y, Qi Y,
Jiang Q, Chen L, Zhang P, Zhang H, et al: Tumour-associated
macrophages-derived CXCL8 determines immune evasion through
autonomous PD-L1 expression in gastric cancer. Gut. 68:1764–1773.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Principe S, Zapater-Latorre E, Arribas L,
Garcia-Miragall E and Bagan J: Salivary IL-8 as a putative
predictive biomarker of radiotherapy response in head and neck
cancer patients. Clin Oral Investig. 26:437–448. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
León X, García J, Farré N, Majercakova K,
Avilés-Jurado FX, Quer M and Camacho M: Predictive capacity of IL-8
expression in head and neck squamous carcinoma patients treated
with radiotherapy or chemoradiotherapy. Acta Otorrinolaringol Esp
(Engl Ed). 72:337–343. 2021. View Article : Google Scholar : PubMed/NCBI
|