Introduction
Primary liver cancer is the fourth most common
malignant tumor and the second leading cause of cancer death in
China (1). Hepatocellular carcinoma
(HCC) is the most common pathological type of primary liver cancer
and has a high incidence rate. The incidence rate of HCC in China
ranks first in the world, and the annual new HCC cases in China
account for ~45% of total new HCC cases worldwide (2). HCC is usually diagnosed at an advanced
stage, and conventional radiotherapy, chemotherapy and molecular
targeting have not demonstrated satisfactory therapeutic effects.
Although liver ultrasonography every 6 months with or without serum
alpha-fetoprotein (AFP) level, computerized tomography, and
magnetic resonance imaging are currently accepted methods for HCC
surveillance, their effectiveness has been controversial due to
sex, physical habits, cost and other limitations (3). Moreover, even after surgical
treatment, the recurrence rate remains high. The majority of HCC
cases in China develop from hepatitis; multiple factors, such as
the interaction of multiple inflammatory cells and the formation of
tumor angiogenesis, result in an immunosuppressive
microenvironment, limiting the clinical efficacy of immune
checkpoint inhibitors. As HCC has an insidious onset and difficulty
in early diagnosis, timely and effective treatment after diagnosis
is essential for inhibiting disease progression. Therefore, there
is an urgent need to find new diagnostic and prognostic indicators
for HCC. In recent years, immunotherapy drugs have been gradually
applied in the clinical treatment of HCC, which has expanded the
treatment possibilities for HCC. Therefore, a comprehensive
understanding of the immune infiltration in HCC is also
particularly important to select the most effective immunotherapy
strategy.
Long non-coding RNAs (lncRNAs) are non-coding RNAs
>200 nucleotides (4). lncRNAs
specifically bind to various proteins and nucleic acids through
their secondary structures. With the development of next-generation
sequencing technology and the large amount of high-throughput
sequencing data on tumors, these advances have made it possible to
predict the function of lncRNAs in tumors using bioinformatics
analysis. Previous studies have demonstrated that some
tissue-specifically lncRNAs cis-regulate the transcription of
adjacent protein-coding genes, and this co-expression pattern has
important implications for other biological processes (5). lncRNAs also play key transcriptional
regulatory roles by influencing gene expression by recruiting
protein complexes or competing for transcription factors. The
function of lncRNAs in diseases is closely related to their
subcellular localization. Wang et al (6) reported that cytoplasmic lnc-IGFL2-AS1
acts as a competing endogenous (ceRNA) to bind microRNA (miRNA or
miR)-4795-3p, promoting the expression of IGFL1, while in the
nucleus, it promotes the formation of the KLF5/TEAD4
transcriptional complex at the enhancer of IGFL2.
lnc-MAPKAPK5-AS1 (hereinafter referred to as
MK5-AS1) is located on chromosome 12:112280206-112282706 with a
length of 2,390 nucleotides. It has been demonstrated that it is
strongly linked to the clinicopathological characteristics and
prognosis of various patients with cancer. Research has revealed
that MK5-AS1 is highly expressed in colorectal cancer and can
promote the proliferation of cells by inhibiting the expression of
p21 (7). Yang et al
(8) also confirmed the adverse
mechanism of MK5-AS1 in colorectal cancer, including the formation
of a MK5-AS1-let-7f-1-3p-SNAI1 ceRNA network and cis-regulation of
its adjacent gene MAPKAPK5 (henceforth called MK5). Zhang et
al (9) reported that MK5-AS1 is
an independent risk factor for lung adenocarcinoma and silencing
its expression has been validated to markedly inhibit the
proliferation of lung adenocarcinoma cells. However, there were few
studies on the association between MK5-AS1 and the tumor
microenvironment (TME) in HCC. Therefore, the role of MK5-AS1 in
HCC and its regulatory relationship with downstream target gene MK5
was elucidated in the present study. Furthermore, the role of
MK5-AS1 in the immune microenvironment was investigated through
multiple public databases by bioinformatics analysis, aiming to
provide new clues for individualized treatment of HCC cases.
Materials and methods
Data collection and processing
The transcriptome data of The Cancer Genome Atlas
Liver Hepatocellular Carcinoma program ‘(TCGA)-LIHC’ and the
corresponding clinical data updated, was downloaded on November 3,
2022 from TCGA (https://portal.gdc.cancer.gov/repository) (10), which contains expression profiles of
mRNAs and lncRNAs of patients with HCC. After removing duplicate
samples, the expression data of 369 patients with HCC and 50
non-tumor liver patients were obtained and converted into TPM
format after preprocessing. The GSE144269 dataset was downloaded
from Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/, accessed on
November 5, 2022) (11) for
validation of gene expression and prognosis. Mutation data of HCC
were collected and visualized in R using ‘maftools’, and the
characteristics of the mutation status were investigated.
Correlation analysis of
lnc-MAPKAPK5-AS1 expression and clinicopathological factors
The expression patterns of MK5-AS1 and MK5 in HCC
tissues and normal tissues from TCGA were compared. GSE144269 was
used as an external validation dataset to verify the expression
status of the two genes. The 369 patients with HCC were divided
into high expression group and low expression group using the
medians of MK5-AS1 and MK5, and the relationships between gene
expression and clinical parameters were analyzed.
Gene Ontology (GO) functional
annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG)
pathway enrichment analysis
The expression profiles were compared between the
MK5-AS1 high and low expression groups; a total of two sets of
differentially expressed genes (DEGs) were obtained from TCGA-LIHC
and GSE144269. lncRNAs and mRNAs in both gene sets were screened
and imported into Metascape (http://metascape.org, accessed on 19 December 2022)
(12) for functional enrichment
analysis. KEGG enrichment analysis was performed using the
‘clusterProfiler’ R package to screen out relevant pathways
(13).
Gene Set Enrichment Analysis
(GSEA)
GSEA (14) was
performed using the ‘clusterProfiler’ R package in the Molecular
Signatures Database (MSigDB)_v7.0_GMTs with the reference dataset
‘c5.all.v7.0.entrez.gmt’. Significantly altered pathways were
validated with 1,000 iterative calculations, and the expression
level of MK5-AS1 was considered as a phenotypic marker. According
to the reference information of GSEA official software, it is
generally considered that the NES absolute value is greater than 1,
NOM P<0.05, false discovery rate (FDR) q-value <0.25 of the
enrichment pathways are significantly enriched between high and low
MK5-AS1 expression groups in HCC (15–18).
Immunohistochemistry (IHC) assay
HCC tissue microarrays used for analysis were
commercial products purchased from Shanghai Xinchao Biological
Technology Co. Ltd. (cat. no. hlivh180su15). All experiments were
performed following the manufacturer's instructions. The
corresponding clinical information was also provided by the
company. The protocol was approved (approval no. SHYJS-CP-1901001
on 11th January 2019, and extended as approval no. SHYJS-BC-2310001
on 20th October, 2023) by the ethics committee of Shanghai Xinchao
Biological Technology Co., Ltd. (Shanghai, China). The tissue
microarray contained tumor samples from 90 patients with HCC who
received surgical treatment from June 2007 to October 2008.
Clinical data included HBsAg, anti-hepatitis C (HCV), alanine
aminotransferase (ALT), AFP, and other liver indicators, as well as
pathological characteristics such as TNM stage and histological
grade. The follow-up period was 3 to 5 years, and data on the
overall survival (OS) and disease-free survival (DFS) were
collected.
Tissue sections were deparaffinized in xylene for
0.5 h twice. The sections were hydrated in ethanol, followed by
high-pressure antigen recovery by heating the tissue with EnVision
FLEX TARGET RETRIEVAL SOLUTION LOW pH (pH, 6.1, 3 min; cat. no.
K8005; Dako; Agilent Technologies, Inc.). Endogenous peroxidase was
blocked by incubating the sections with 3% hydrogen peroxide at
room temperature for 15 min. In addition, the sections were blocked
with 1X Antibody Diluent/Block (cat. no. ARD1001EA; Akoya
Biosciences) at room temperature for 30 min. Subsequently, the
slides were stained with a primary antibody against MAPKAPK5
(1:100; cat. no. HPA015515; Atlas Antibodies) at 4°C overnight. The
slides were then incubated with an appropriate HRP-conjugated
secondary antibody (EnVision FLEX+, Mouse, High pH; no
dilution required; cat. no. SM802; Dako; Agilent Technologies,
Inc.) at 37°C for 30 min. Furthermore, DAB (25°C, 5 min) and
hematoxylin (25°C, 1 min) were used for visual antibody staining.
An optical microscope was used for observation and to capture
images. MAPKAPK5 cytoplasmic staining was scored using four grades
(0: negative, 1: weakly positive, 2: moderately positive, 3:
strongly positive). The percentage of positive cells was
categorized into five grades (0: 0%, 1: 1–5%, 2: 6–25%, 3: 26–50%,
4: 51–100%). The final IHC score was calculated as follows:
intensity grad × positive cell percentage grade. A score of 0–3
indicated low expression, and a score of 4–9 indicated high
expression.
Relationship between lnc-MAPKAPK5-AS1
and the TME
CIBERSORT is an accurate and robust algorithm that
calculates the immune cell composition of tumor tissues by the gene
expression profiles (13). The
normalized mRNA expression matrix of patients with HCC in TCGA was
analyzed using the ‘CIBERSORT’ source R package, which was obtained
from the website https://rdrr.io/github/singha53/amritr/src/R/supportFunc_cibersort.R.
Other required data were obtained from the supplementary data of a
previously published article (13)
(https://www.nature.com/articles/nmeth.3337#MOESM207),
which included the gene expression matrix for 22 types of immune
cells. The number of permutations was 1,000, and the components of
various immune cells were compared in MK5-AS1 high and low
expression groups. The single sample gene set enrichment analysis
(ssGSEA) algorithm was used to assess the infiltration degree of 28
different immune cells. A matrix of immune cell (B cells,
CD4+ T cells, CD8+ T cells, neutrophils,
macrophages and dendritic cells) infiltration levels of TCGA-LIHC
samples were also downloaded using the TIMER database (https://cistrome.shinyapps.io/timer/
(19), accessed on 21 December
2022) and the correlation between the expression status of
MK5-AS1 and immune cell infiltration level in HCC was
detected.
Correlation between expression of
lnc-MAPKAPK5-AS1 and tumor mutational burden (TMB), microsatellite
instability (MSI) and immune checkpoint inhibitors (ICIs)
A number of clinical studies have demonstrated an
association between TMB, MSI and the effect of immunotherapy
(20–22). Therefore, such emerging biomarkers
may be implicated in the regulation of TME, therefore the
relationship between these biomarkers and the expression of MK5-AS1
were explored. Spearman correlation analysis was conducted using
SangerBox (http://vip.sangerbox.com/home.html, accessed on 18
October 2022). The TMB value of patients with HCC and high and low
expression of MK5-AS1 was calculated. A boxplot of TMB value was
generated to visualize the results using the ‘ggbetweenstats’
function of ‘ggstatsplot’ package. Immunotherapy is a strategy for
tumor treatment and includes immunotherapy targeting programmed
death protein-1 (PD-1)/programmed death protein ligand-1 (PD-L1)
and cytotoxic T lymphocyte-associated antigen (CTLA-4). These
immunotherapies have demonstrated great efficacy and application
prospect in the treatment of advanced HCC (23). Immune checkpoint molecules play an
important role in maintaining immune homeostasis and can be
exploited for the immune evasion of tumor cells. The relationship
between MK5-AS1 and eight immune checkpoints were examined to
identify its potential effect in immunotherapy.
Drug susceptibility analysis
The ‘pRRophetic’ R package, developed by Professors
Paul Geeleher, Nancy Cox and R. Stephanie Huang at the University
of Minnesota, was designed to predict phenotypes by gene expression
data (24). It uses data of the CGP
cell line from the Cancer Genome Project to predict clinical
outcomes and drug sensitivity of external cell lines (Cancer Cell
Line Encyclopedia) (25). The
potential chemotherapeutic efficacy of 16 common drugs in HCC was
examined on the basis of gene expression profiles in TCGA and the
differences in the sensitivity of the drugs between MK5-AS1 high
and low expression groups were compared.
Methylation status of the promoter
region of lnc-MAPKAPK5-AS1
Diseasemeth 2.0 (http://bio-bigdata.hrbmu.edu.cn/diseasemeth/, accessed
on 18 November 2022) (26) was used
to examine the methylation pattern of the promoter region of
MK5-AS1 and its relation to different clinical stages and
histological grades. The somatic mutation data of HCC was
downloaded from TCGA and the overall mutation status was analyzed.
The results were visualized using the cBioPortal database
(http://www.cbioportal.org (27), accessed on 1 February 2023).
Prediction of the relationship between
lnc-MAPKAPK5-AS1 and miRNAs
Based on high-throughput sequencing data from 15
cell lines, lncATLAS (http://lncatlas.crg.eu/, accessed on 5 September 2022)
(28) was used to collect data of
6,768 lncRNAs and to evaluate specific subcellular localization
using the ‘relative concentration index’. This database was used to
predict the localization of MK5-AS1. Target miRNAs that interact
with MK5-AS1 were predicted using the ENCORI platform (http://starbase.sysu.edu.cn/, 5 January 2023)
(29), which contains networks of
interactions among RNAs. The screening conditions were as follows:
i) ‘miRNA-Target: miRNA-lncRNA’; ii) ‘Genome: human’; and iii)
‘Target Gene: lnc-MAPKAPK5-AS1’. The expression of the miRNAs in
HCC and the correlation with MK5-AS1 expression using data from
TCGA were explored.
Weighted correlation network analysis
(WGCNA) screening of key modules and hub genes related to immune
activity
WGCNA is an algorithm widely used to identify
biomarkers through clustering sets of genes with similar expression
patterns (30). It calculates the
associations between distinct modules and specific clinical
features. A total of 8,900 DEGs were identified from HCC tissues
and normal tissues and 5,100 DEGs from the high MK5-AS1 expression
group and low MK5-AS1 expression group. A total of 3,255 DEGs were
obtained by intersection of the two gene sets. The ‘WGCNA’ R
package was applied to construct the co-expression network of 3,255
genes and 369 HCC samples were clustered. β=3 was selected as the
soft threshold power to construct the scale-free network. A
hierarchical clustering tree was constructed using the dynamic
hybrid cutting technology to gather genes with similar expression
patterns. The STRING (http://string.embl.de/) (31) database was used to explore the
interaction network between proteins, which helps to identify the
key regulatory genes.
The cytolytic activity score (CYT) is a robust
transcriptome-based immune signature across multiple cancer types
and defined as the mean of GZMA and PRF1 expression (TPM format).
It has been revealed that a higher CYT is associated with improved
outcomes (32).
The IFNG6 score can reflect the overall immune
activity and predict the therapeutic effect of pembrolizumab in
patients with HCC. This score is calculated from the average
expression of six genes (CXCL9, CXCL10, IDO1, IFNG, HLA-DRA and
STAT1) (33).
Statistical methods
R-4.1.2 and GraphPad Prism-8.00 were used for data
cleaning, statistical analysis and graphing. A Kolmogorov Smirnov
test was performed to determine the distribution and then based on
that, an unpaired t-test was used to explore the difference in
MK5-AS1 expression between the HCC and normal groups in the
TCGA-LIHC dataset. As for the GSE144269 dataset, the tumor and
normal groups were originated from the same patients and then a
paired t-test was used to analyze the differences in MK5-AS1
expression between the aforementioned two groups. The histogram was
visualized using the median gene expression with an interquartile
range. Kruskal-Wallis test with post hoc test (Dunn's or
Steel-Dwass) for multiple testing correction was used for
multi-group comparison. The chi-square test and Fisher's exact test
were employed in the analysis of the relationship between gene
expression and clinicopathological characteristics of patients with
HCC. Survival curves were drawn using Kaplan-Meier method and
comparison among different groups was performed using the log-rank
test. Cox regression models were utilized for univariate and
multivariate analysis. The prognostic ability of the two genes was
assessed by the area under the ROC curve (AUC). Correlations
between genes and immune infiltrating cells were compared using
Spearman correlation analysis. Two-sided P<0.05 were considered
statistically significant in all analyses. Bonferroni correction
was used for pairwise comparisons between multiple groups.
Results
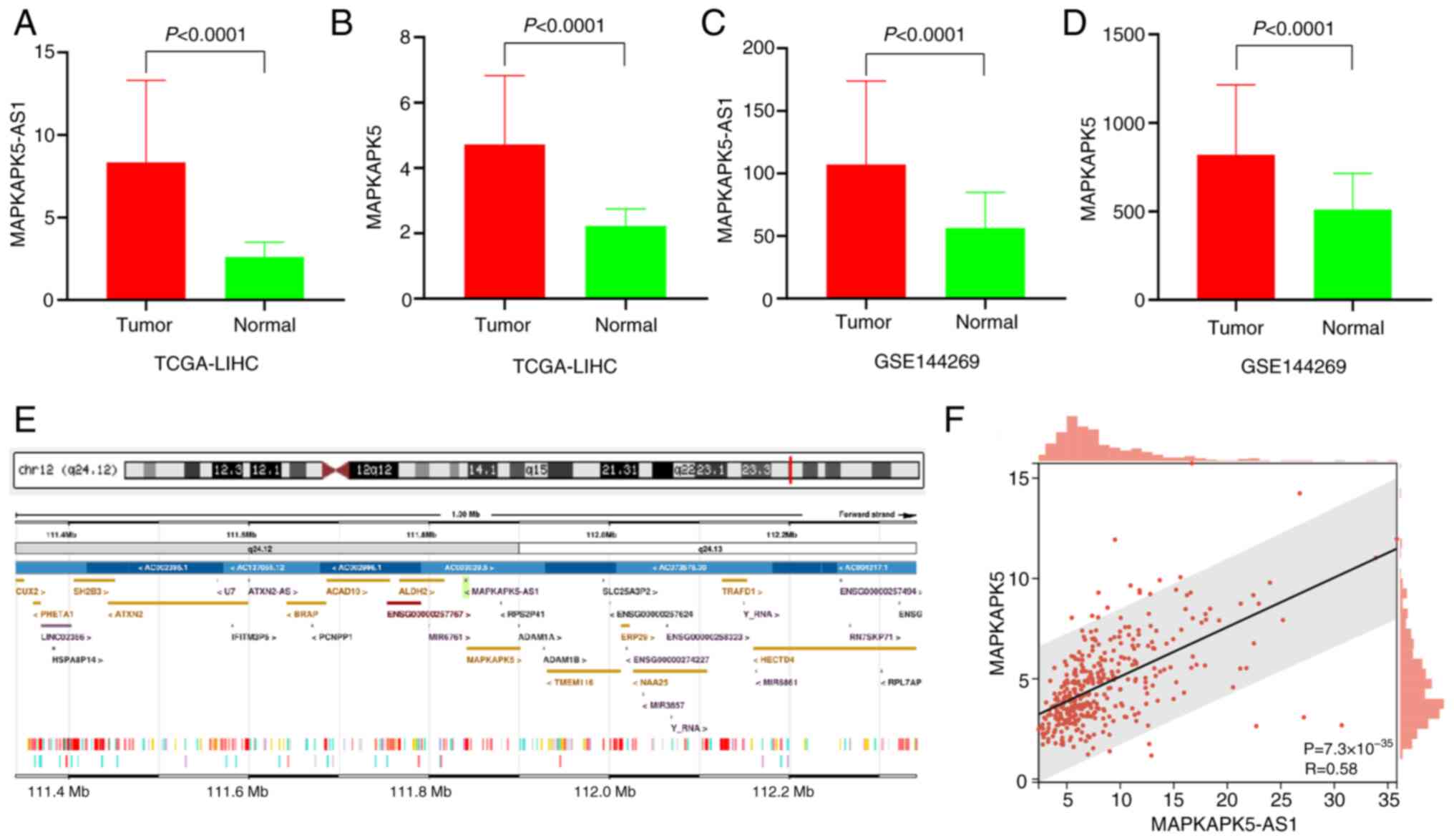
Expression of lnc-MAPKAPK5-AS1 in HCC
tissues
As depicted in Fig. 1A
and B, the expression levels of MK5-AS1 and MK5 showed
significant upregulation in HCC tissues compared with normal liver
tissues in TCGA (both P<0.0001). The median with interquartile
range of MK5-AS1 in HCC and normal liver tissues was 6.965 and
5.247–10.010 vs. 2.536 and 2.003–3.094, respectively; for MK5, they
were 4.268 and 3.199–5.719 vs. 2.180 and 1.870–2.525, respectively.
Similar results were observed in the GSE144269 dataset (both
P<0.0001, Fig. 1C and D).
Antisense lncRNAs are often correlated with the
expression of their sense strand genes, suggesting that they
probably be widely involved in the expression regulation of
protein-coding genes (5). MK5-AS1
is transcribed from the antisense strand of its protein-coding gene
MK5, and the two genes have partially overlapping sequences
(Fig. 1E). Spearman's correlation
analysis revealed a positive correlation between the two genes
(Fig. 1F).
Correlation of gene expression with
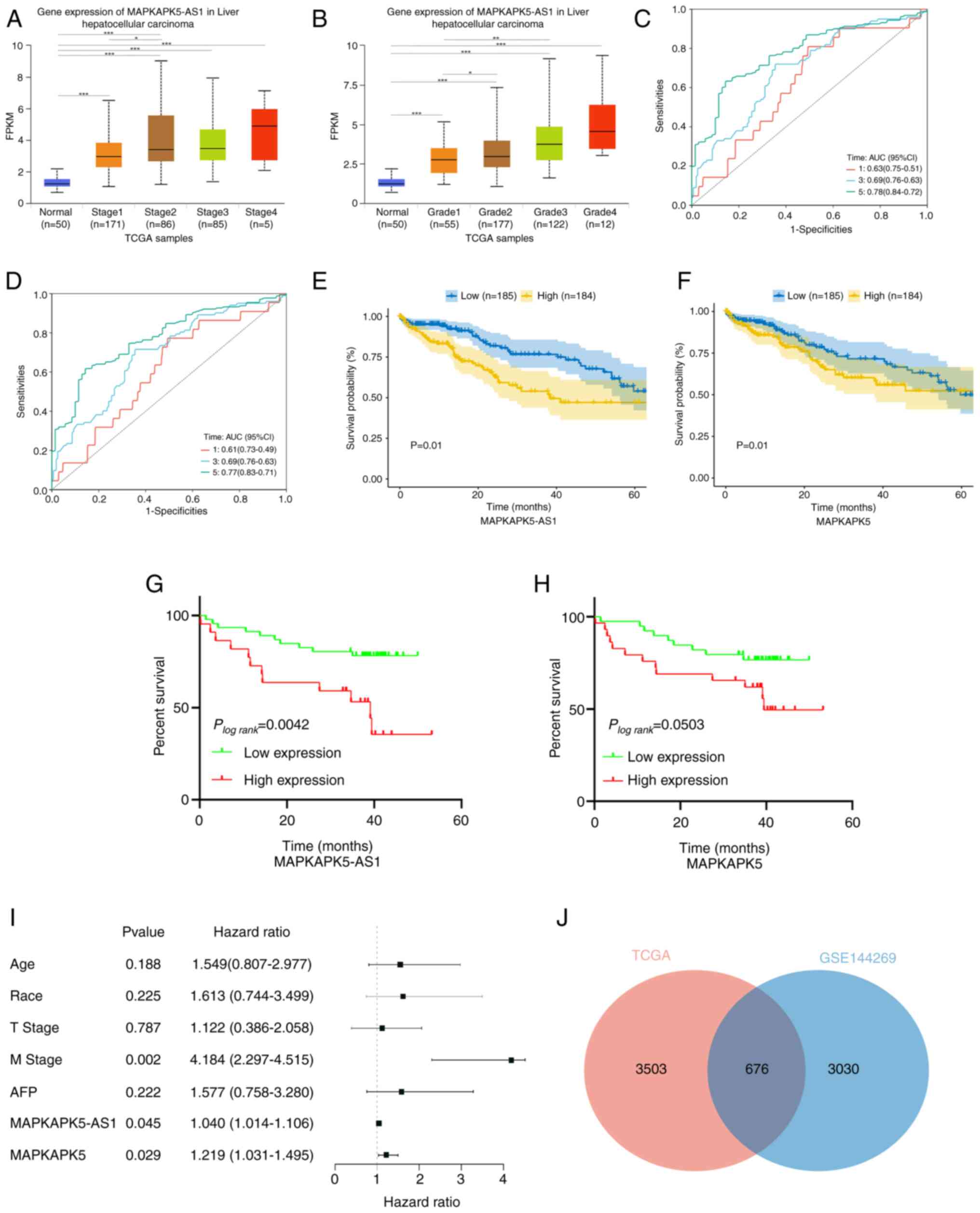
clinicopathological factors in HCC
The relationship between the expression levels of
MK5-AS1 and MK5 and several widely recognized clinicopathological
factors was explored. Analysis of UALCAN (http://ualcan.path.uab.edu/, accessed on 20 October
2022) (34) revealed that a higher
expression level of MK5-AS1 was associated with higher clinical
stage and histological grade of HCC tissues (Fig. 2A and B). The 369 patients with HCC
were divided into the high expression groups and low expression
groups using the median of gene expression. As shown in Table I, higher expression of MK5-AS1 was
significantly associated with advanced clinical stage
(χ2=5.372, P=0.020), T stage (χ2=5.280,
P=0.022), histological grade (χ2=17.825, P<0.01) and
higher AFP (χ2=29.950, P<0.01). Notably, MK5
exhibited similar tendencies: MK5 expression was negatively linked
with the clinical stage (χ2=4.554, P=0.033), T stage
(χ2=3.983, P=0.046), histological grade
(χ2=17.825, P<0.01) and higher AFP
(χ2=23.348, P<0.01) in patients with HCC (Table II). These findings indicated that
high expression of MK5-AS1 and MK5 may be potential risk factors
for HCC.
 | Table I.Relationship between lnc-MAPKAPK5-AS1
expression and clinicopathological parameters of hepatocellular
carcinoma samples in The Cancer Genome Atlas. |
Table I.
Relationship between lnc-MAPKAPK5-AS1
expression and clinicopathological parameters of hepatocellular
carcinoma samples in The Cancer Genome Atlas.
|
|
| Expression level of
lnc-MAPKAPK5-AS1 |
|
|
|---|
|
|
|
|
|
|
|---|
|
Characteristics | n | High (n=184) | Low (n=185) | χ2 | P-value |
|---|
| Sex |
|
|
| 0.856 | 0.355 |
|
Male | 249 | 120 (48.2) | 129 (51.8) |
|
|
|
Female | 120 | 64 (53.3) | 56 (46.7) |
|
|
| Age, years |
|
|
| 0.171 | 0.679 |
|
≤60 | 177 | 90 (50.8) | 87 (49.2) |
|
|
|
>60 | 191 | 93 (48.7) | 98 (51.3) |
|
|
| Ethnicity |
|
|
| 0.807 | 0.668 |
|
Asian | 158 | 83 (52.5) | 75 (47.5) |
|
|
|
White | 182 | 87 (47.8) | 95 (52.2) |
|
|
|
Others | 19 | 10 (52.6) | 9 (47.4) |
|
|
| BMI |
|
|
| 1.162 | 0.281 |
|
<24 | 160 | 84 (52.5) | 76 (47.5) |
|
|
|
≥24 | 178 | 83 (46.6) | 95 (53.4) |
|
|
| Historical risk
factors |
|
|
| 0.738 | 0.691 |
| Alcohol
consumption | 117 | 61 (52.1) | 56 (47.9) |
|
|
|
Hepatitis virus | 114 | 53 (46.5) | 61 (53.5) |
|
|
|
Others | 119 | 59 (49.6) | 60 (50.4) |
|
|
| Clinical stage |
|
|
| 5.372 | 0.020 |
| I,
II | 257 | 118 (45.9) | 139 (54.1) |
|
|
| III,
IV | 88 | 53 (60.2) | 35 (39.8) |
|
|
| T |
|
|
| 5.280 | 0.022 |
| T1,
T2 | 275 | 128 (46.5) | 147 (53.5) |
|
|
| T3,
T4 | 91 | 55 (60.4) | 36 (39.6) |
|
|
| N |
|
|
|
| 0.364 |
| N0 | 250 | 122 (48.8) | 128 (51.2) |
|
|
| N1 | 4 | 3 (75.0) | 1 (25.0) |
|
|
| M |
|
|
|
| 0.622 |
| M0 | 265 | 134 (50.6) | 131 (49.4) |
|
|
| M1 | 4 | 3 (75.0) | 1 (25.0) |
|
|
| Histologic
grade |
|
|
| 17.825 | <0.01 |
| G1,
G2 | 232 | 96 (41.4) | 136 (58.6) |
|
|
| G3,
G4 | 132 | 85 (64.4) | 47 (35.6) |
|
|
| AFP |
|
|
| 29.950 | <0.01 |
|
<20 | 147 | 51
(34.7)a | 96
(65.3)a |
|
|
| ≥20 and
<400 | 66 | 31
(46.9)a | 35
(53.1)a |
|
|
|
≥400 | 65 | 49
(75.4)b | 16
(24.6)b |
|
|
| Child pugh
grade |
|
|
| 0.023 | 0.880 |
| A | 217 | 95 (43.8) | 122 (56.2) |
|
|
| B,
C | 22 | 10 (45.5) | 12 (54.5) |
|
|
| Treatment type |
|
|
| 0.024 | 0.876 |
|
Pharmaceutical therapy | 184 | 91 (49.5) | 93 (50.5) |
|
|
|
Radiation therapy | 185 | 93 (50.3) | 92 (49.7) |
|
|
 | Table II.Relationship between MAPKAPK5
expression and clinicopathological parameters of hepatocellular
carcinoma samples in The Cancer Genome Atlas. |
Table II.
Relationship between MAPKAPK5
expression and clinicopathological parameters of hepatocellular
carcinoma samples in The Cancer Genome Atlas.
|
|
| Expression level of
MAPKAPK5 |
|
|
|---|
|
|
|
|
|
|
|---|
|
Characteristics | n | High (n=184) | Low(n=185) | χ2 | P-value |
|---|
| Sex |
|
|
| 0.167 | 0.683 |
|
Male | 249 | 126 (50.6) | 123 (49.4) |
|
|
|
Female | 120 | 58 (48.3) | 62 (51.7) |
|
|
| Age, years |
|
|
| 1.080 | 0.299 |
|
≤60 | 177 | 93 (52.5) | 84 (47.5) |
|
|
|
≥60 | 191 | 90 (47.1) | 101 (52.9) |
|
|
| Ethnicity |
|
|
| 2.532 | 0.282 |
|
Asian | 158 | 79 (50.0) | 79 (50.0) |
|
|
|
White | 182 | 90 (49.5) | 92 (50.5) |
|
|
|
Others | 19 | 13 (68.4) | 6 (31.6) |
|
|
| BMI |
|
|
| 0.019 | 0.890 |
|
≤24 | 160 | 77 (48.1) | 83 (51.9) |
|
|
|
≥24 | 178 | 87 (48.9) | 91 (51.1) |
|
|
| Historical risk
factors |
|
|
| 0.324 | 0.850 |
| Alcohol
consumption | 117 | 56 (47.9) | 61 (52.1) |
|
|
|
Hepatitis virus | 114 | 58 (40.3) | 56 (59.7) |
|
|
|
Others | 119 | 61 (51.3) | 58 (48.7) |
|
|
| Clinical stage |
|
|
| 4.554 | 0.033 |
| I,
II | 257 | 118 (45.9) | 139 (54.1) |
|
|
| III,
IV | 88 | 52 (59.1) | 36 (40.9) |
|
|
| T |
|
|
| 3.983 | 0.046 |
| T1,
T2 | 275 | 130 (47.3) | 145 (52.7) |
|
|
| T3,
T4 | 91 | 54 (59.3) | 37 (40.7) |
|
|
| N |
|
|
| - | 0.622 |
| N0 | 250 | 125 (50.0) | 125 (50.0) |
|
|
| N1 | 4 | 3 (75.0) | 1 (25.0) |
|
|
| M |
|
|
| 0.970 | 0.622 |
| M0 | 265 | 132 (49.8) | 133 (50.2) |
|
|
| M1 | 4 | 1 (25.0) | 3 (75.0) |
|
|
| Histologic
grade |
|
|
| 17.825 | <0.01 |
| G1,
G2 | 232 | 96 (41.4) | 136 (58.6) |
|
|
| G3,
G4 | 132 | 85 (64.4) | 47 (35.6) |
|
|
| AFP |
|
|
| 24.348 | <0.01 |
|
≤20 | 147 | 50
(34.0)a | 97
(66.0)a |
|
|
| ≥20 and
<400 | 66 | 36
(54.5)b | 30
(45.5)b |
|
|
|
≥400 | 65 | 45
(69.2)b | 20
(30.8)b |
|
|
| Child pugh
grade |
|
|
| 1.499 | 0.221 |
| A | 217 | 89 (41.0) | 128 (59.0) |
|
|
| B,
C | 22 | 12 (54.5) | 10 (45.5) |
|
|
| Treatment type |
|
|
| 0.610 | 0.435 |
|
Pharmaceutical therapy | 184 | 88 (47.8) | 96 (52.2) |
|
|
|
Radiation therapy | 185 | 96 (51.9) | 89 (48.1) |
|
|
Association between the expression of
two genes and the diagnosis and prognosis in patients with HCC
Time-dependent ROC curve analysis revealed that
MK5-AS1 had high sensitivity and specificity for the five-year
survival rate of patients with HCC (AUC=0.78, Fig. 2C). The AUC of MK5 for predicting
five-year survival rate was 0.77 (Fig.
2D). To further examine the relationship between gene
expression and survival, Kaplan-Meier survival curves were used to
examine the effect of MK5-AS1 expression on the overall survival
rate of patients with HCC in TCGA and GEO. The median survival time
of patients with HCC with high MK5-AS1 expression in TCGA was only
39 months, while it was ~70 months for patients with low MK5-AS1
expression (P=0.01, Fig. 2E).
Analysis of GEO data also suggested that the high expression group
had improved survival outcomes (P=0.013, Fig. 2F). The results of the survival
analysis of MK5 were consistent with these findings; its high
expression was an adverse factor for the prognosis of patients with
HCC (P=0.013, P=0.05; Fig. 2G and
H).
Cox regression analysis can be used to examine
whether the gene expression level is a risk factor that affects
survival. Univariate Cox regression analysis showed that compared
with patients with low MK5-AS1 and MK5 expression, patients with
high expression of MK5-AS1 and MK5 indicated a substantially higher
risk of mortality. The variables with a statistically significant
effect on survival were further included in multivariate Cox
regression analysis. The results revealed that MK5-AS1 and MK5 may
be independent risk factors for poor survival when M stage was
contained (Table III, Fig. 2I). These results suggested that high
expression of MK5-AS1 in patients with HCC was associated with
tumor progression and adverse prognosis.
 | Table III.Cox regression analysis of
independent risk factors affecting the prognosis of patients with
hepatocellular carcinoma. |
Table III.
Cox regression analysis of
independent risk factors affecting the prognosis of patients with
hepatocellular carcinoma.
| A, The relationship
between overall survival and clinicopathologic feature in patients
with hepatocellular carcinoma using Univariate Cox regression. |
|---|
|
|---|
| Variable | Hazard ratio | 95% confidence
interval | P-value |
|---|
| Sex | 1.303 | 0.849–1.997 | 0.225 |
| Age | 1.564 | 1.013–2.417 | 0.044 |
| Body mass
index | 0.834 | 0.529–1.315 | 0.435 |
| Race | 2.317 | 1.358–3.954 | 0.002 |
| Clinical stage | 1.484 | 0.896–2.458 | 0.125 |
| T | 1.812 | 1.147–2.860 | 0.011 |
| N | 1.095 | 0.899–27.02 | 0.997 |
| M | 5.296 | 1.631–17.19 | 0.006 |
| Histologic
grade | 1.269 | 0.817–1.973 | 0.288 |
| AFP | 2.063 | 1.194–3.564 | 0.009 |
| Child pugh
grade | 1.577 | 0.711–3.499 | 0.262 |
| Treatment type | 1.289 | 0.843–1.970 | 0.241 |
|
Lnc-MAPKAPK5-AS1 | 1.039 | 1.002–1.078 | 0.039 |
|
MAPKAPK5 | 1.148 | 1.028–1.282 | 0.015 |
|
| B, The
relationship between overall survival and clinicopathologic feature
in patients with hepatocellular carcinoma using Multivariate Cox
regression. |
|
|
Variable | Hazard
ratio | 95% confidence
interval | P-value |
|
| Age | 1.549 | 0.807–2.977 | 0.188 |
| Race | 1.613 | 0.744–3.499 | 0.255 |
| T | 1.122 | 0.386–2.058 | 0.787 |
| M | 4.184 | 2.297–4.515 | 0.002 |
| AFP | 1.577 | 0.758–3.280 | 0.222 |
|
lnc-MAPKAPK5-AS1 | 1.040 | 1.014–1.106 | 0.045 |
|
MAPKAPK5 | 1.219 | 1.031–1.495 | 0.029 |
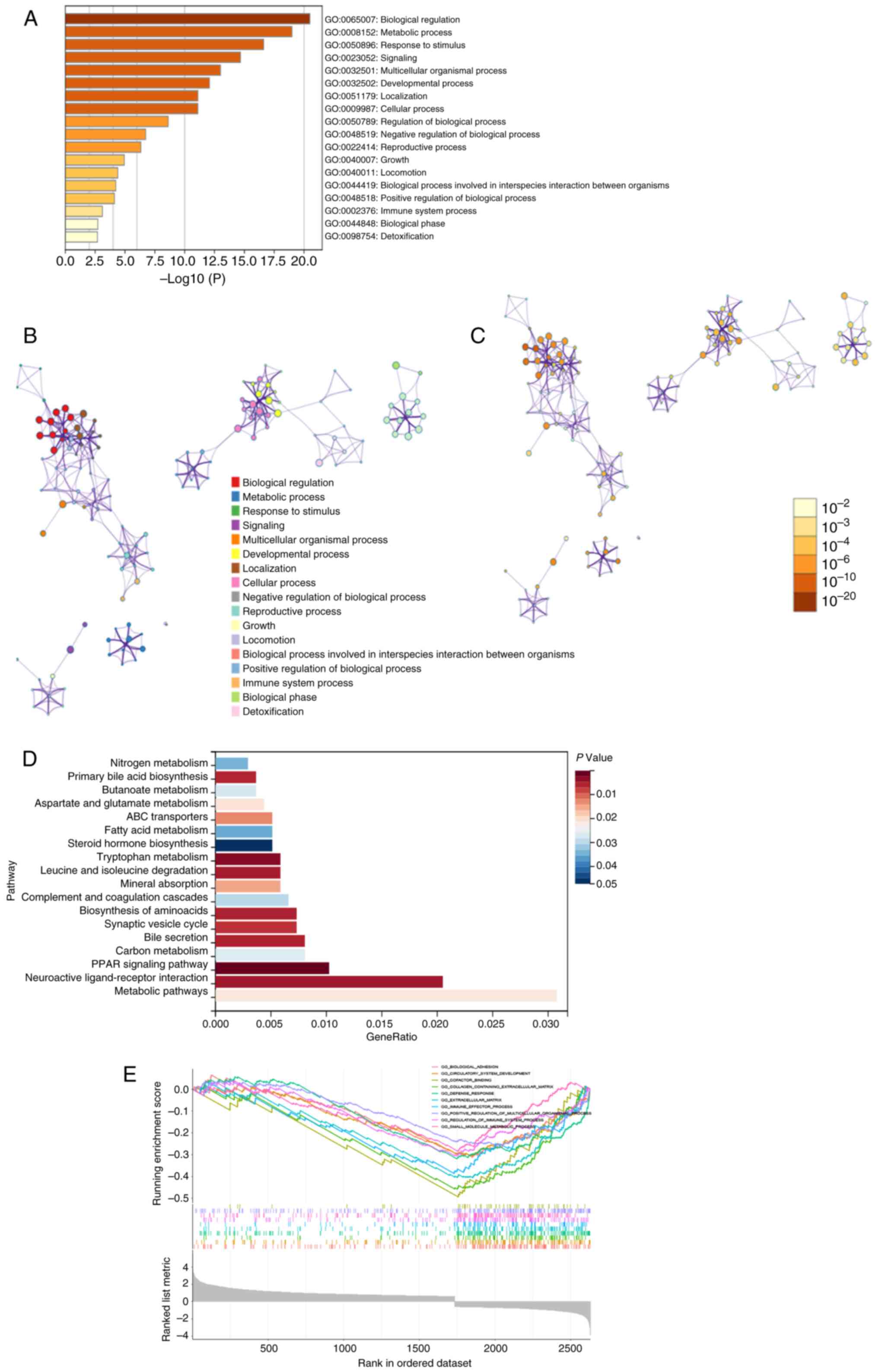
Enrichment analysis of overlapping
DEGs in TCGA and GSE144269
Using ‘DESeq2’ R package, 4,179 and 3,706
differentially expressed lncRNAs and mRNAs (|logFC|≥0.6, FDR
<0.25, P<0.05) were identified in the TCGA-LIHC and GSE144269
datasets, respectively. A Venn diagram was plotted to select the
intersecting genes of the aforementioned two gene sets and 676
genes co-expressed with MK5-AS1 were finally obtained (Fig. 2J). GO analysis demonstrated that
MK5-AS1-related genes were primarily enriched in biological
regulation, metabolic progress, response to stimulus, multicellular
organismal process and immune system process (Fig. 3A). Based on the functional
correlation, a network of enriched terms colored by cluster ID was
constructed in accordance with correlation and similarity, where
nodes that share the same cluster ID were typically close to each
other (Fig. 3B). The relative
number of genes in each pathway is demonstrated in Fig. 3C; a darker color indicates a greater
number of genes, as observed in the pathway ‘biological
regulation’. KEGG analysis results suggested that DEGs may be
involved in various metabolic-related pathways, such as the PRAK
signaling pathway, cell adhesion molecules, carbon metabolism, bile
secretion, synaptic vesicle cycle, biosynthesis of amino-acids,
complement and coagulation cascades and fatty acid metabolism
(Fig. 3D).
GSEA was performed on the basis of normalized
enrichment score and the FDR. Regulation of immune system process,
biological adhesion, collagen containing extracellular matrix,
immune effector process, small molecule metabolic process and
defense response were significantly enriched signaling pathways
(P<0.05, Fig. 3E).
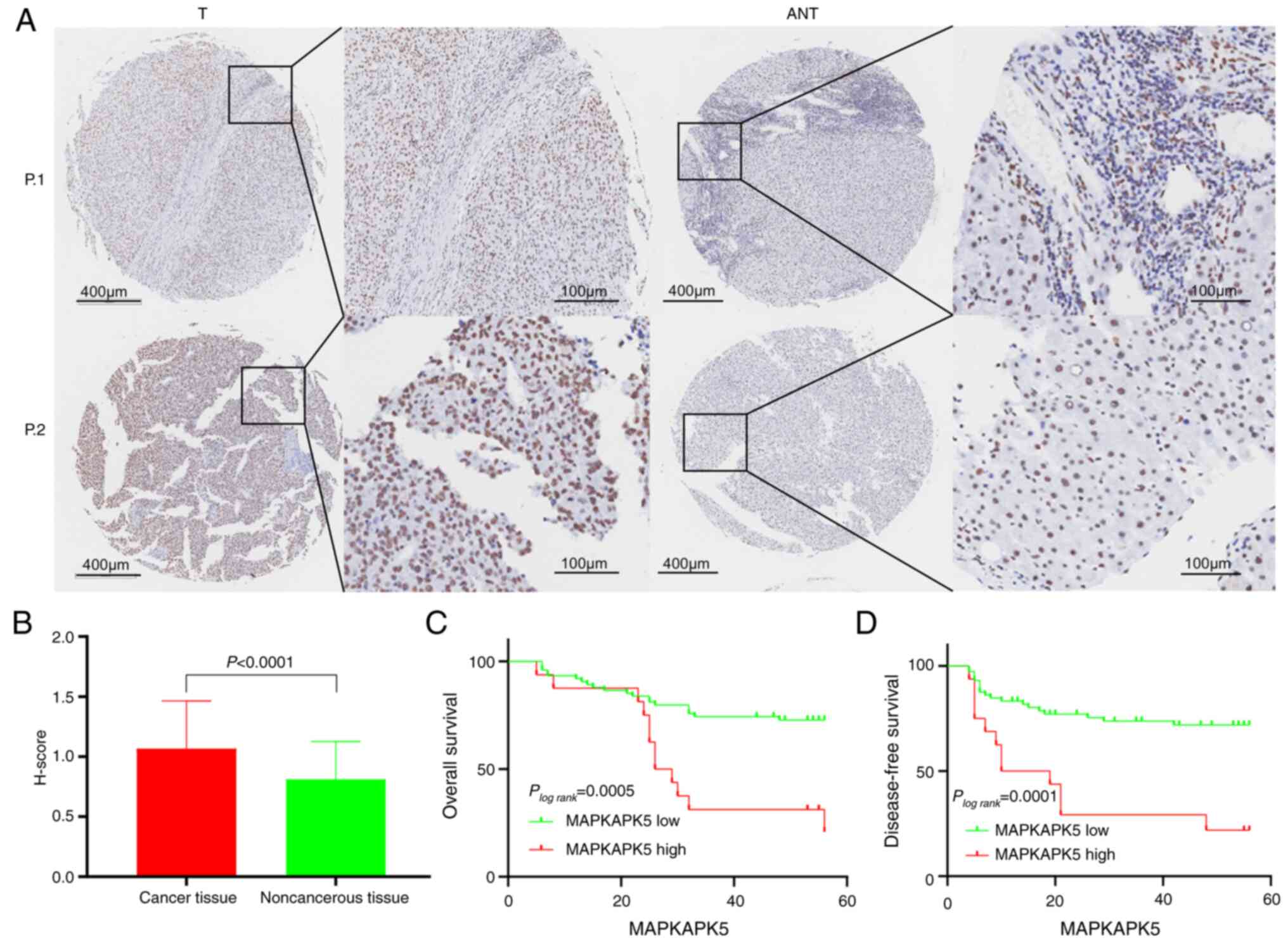
Relationship between MAPKAPK5
expression at protein level and clinicopathological parameters in
HCC tissue chips
To make our results more credible at the
histological level, an external validation of the gene expression
pattern and prognostic significance from 90 HCC tissue chips was
carried out. Immunohistochemical semi-quantitative evaluation of
clinicopathological specimens showed that the expression score of
MK5 in HCC was significantly higher than that in normal tissues
(Fig. 4A and B); the median with
interquartile range of MK5 in HCC and normal tissues were 1.067 and
1.000–1.500 vs. 0.811 and 0.500–1.000, respectively. Based on a
threshold of P<0.05, it was found that the clinical stage
(χ2=7.701, P=0.006), ALT (χ2=5.011, P=0.025),
and PD-L1 expression (χ2=7.003, P=0.008) was strikingly
associated with the expression level of MK5 (Table IV). Nevertheless, the remaining
clinicopathological factors such as sex, age, pathology grade,
tumor size, recurrence, HBsAg, HBcAb, AntiHCV, AFP and CTLA4 were
not statistically significant. As detailed in Fig. 4C and D, patients with HCC with
decreased MK5 expression had longer OS and DFS, which shed light on
the probability that MK5 acted as a risk factor in the development
of HCC.
 | Table IV.Correlation analysis between MAPKAPK5
expression at protein level and clinicopathological feature in
hepatocellular carcinoma. |
Table IV.
Correlation analysis between MAPKAPK5
expression at protein level and clinicopathological feature in
hepatocellular carcinoma.
|
|
| Expression level of
MAPKAPK5 |
|
|
|---|
|
|
|
|
|
|
|---|
|
Characteristics | n | High (n=24) | Low (n=66) | χ2 | P-value |
|---|
| Sex |
|
|
| 0.256 | 1.000 |
|
Male | 80 | 22 (27.5) | 58 (72.5) |
|
|
|
Female | 10 | 2 (20.0) | 8 (80.0) |
|
|
| Age, years |
|
|
| 0.297 | 0.586 |
|
<60 | 71 | 18 (25.4) | 53 (74.6) |
|
|
|
>60 | 19 | 6 (31.6) | 13 (58.4) |
|
|
| Pathology
grade |
|
|
| 5.415 | 0.074 |
| I | 4 | 0 | 4 |
|
|
| II | 63 | 14 (22.2) | 49 (77.8) |
|
|
|
III | 23 | 10 (43.5) | 13 (56.5) |
|
|
| Tumor size(cm) |
|
|
| 0.623 | 0.430 |
| ≤5 | 62 | 15 (24.2) | 47 (75.8) |
|
|
|
>5 | 28 | 9 (32.1) | 19 (67.9) |
|
|
| Number of
tumors |
|
|
| 0.603 | 0.475 |
|
Single | 79 | 20 (25.3) | 59 (74.7) |
|
|
|
Multiple | 11 | 4 (36.4) | 7 (63.6) |
|
|
| Liver cirrhosis
nodules |
|
|
| 1.553 | 0.224 |
| ≤1 | 9 | 4 (44.4) | 5 (55.6) |
|
|
|
>1 | 86 | 20 (23.3) | 66 (76.7) |
|
|
| Tumor
encapsulation |
|
|
| 0.104 | 0.747 |
|
Complete | 42 | 12 (28.6) | 30 (71.4) |
|
|
|
Incomplete | 47 | 12 (25.5) | 35 (74.5) |
|
|
| Clinical stage |
|
|
| 7.701 | 0.017 |
| I | 85 | 20 (23.5) | 65(76.5) |
|
|
| II +
III | 5 | 4 (80.0) | 1(20.0) |
|
|
| T |
|
|
| 0.173 | 0.677 |
| T1 | 63 | 16 (25.4) | 47 (74.6) |
|
|
| T2 +
T3 | 27 | 8 (29.6) | 19 (70.4) |
|
|
| Recurrence |
|
|
| 0.001 | 0.974 |
|
Yes | 49 | 13 (26.5) | 36 (73.5) |
|
|
| No | 41 | 11 (26.8) | 30 (73.2) |
|
|
| HBsAg |
|
|
| 1.904 | 0.274 |
|
Positive | 70 | 17 (24.3) | 53 (75.7) |
|
|
|
Negative | 19 | 7 (36.8) | 12 (63.2) |
|
|
| HBcAb |
|
|
| 0.043 | 1.000 |
|
Positive | 80 | 20 (25.0) | 60 (75.0) |
|
|
|
Negative | 7 | 2 (28.6) | 5 (71.4) |
|
|
| Anti-Hepatitis
C |
|
|
| 0.342 | 0.558 |
|
Positive | 1 | 0 | 1 |
|
|
|
Negative | 86 | 22 (25.6) | 64 (74.4) |
|
|
| T-Bil (µmol/l) |
|
|
| 0.232 | 0.752 |
| Medical
reference value | 76 | 21 (31.8) | 55 (68.2) |
|
|
|
Abnormal value | 14 | 3 (21.4) | 11 (78.6) |
|
|
| ALT (U/l) |
|
|
| 5.011 | 0.025 |
| Medical
reference value | 50 | 18 (36.0) | 32 (64.0) |
|
|
|
Abnormal value | 40 | 6 (15.0) | 34 (85.0) |
|
|
| AFP (µg/l) |
|
|
| 0.691 | 0.406 |
|
≤20 | 36 | 8 (22.2) | 28 (77.8) |
|
|
|
>20 | 53 | 16 (30.2) | 37 (69.8) |
|
|
| GGT (U/l) |
|
|
| 0.135 | 0.701 |
|
≤40 | 31 | 9 (29.0) | 22 (71.0) |
|
|
|
>40 | 59 | 15 (25.4) | 44 (74.6) |
|
|
| PD-L1
expression |
|
|
| 7.003 | 0.008 |
|
Low | 67 | 14 (20.9) | 53 (79.1) |
|
|
|
High | 17 | 9 (52.9) | 8 (47.1) |
|
|
| CTLA4
expression |
|
|
| 0.174 | 0.677 |
|
Low | 15 | 5 (33.3) | 10 (66.7) |
|
|
|
High | 68 | 19 (27.9) | 49 (72.1) |
|
|
Analysis of tumor-infiltrating immune
cells
To clarify the relationship between MK5-AS1 and
tumor infiltrating immune cells, the association between MK5-AS1
expression and the infiltration levels of six immune cells in HCC
was investigated using the immune cell infiltration data downloaded
from the TIMER online database. All correlation analysis conducted
with Spearman's test exhibited statistically significant positive
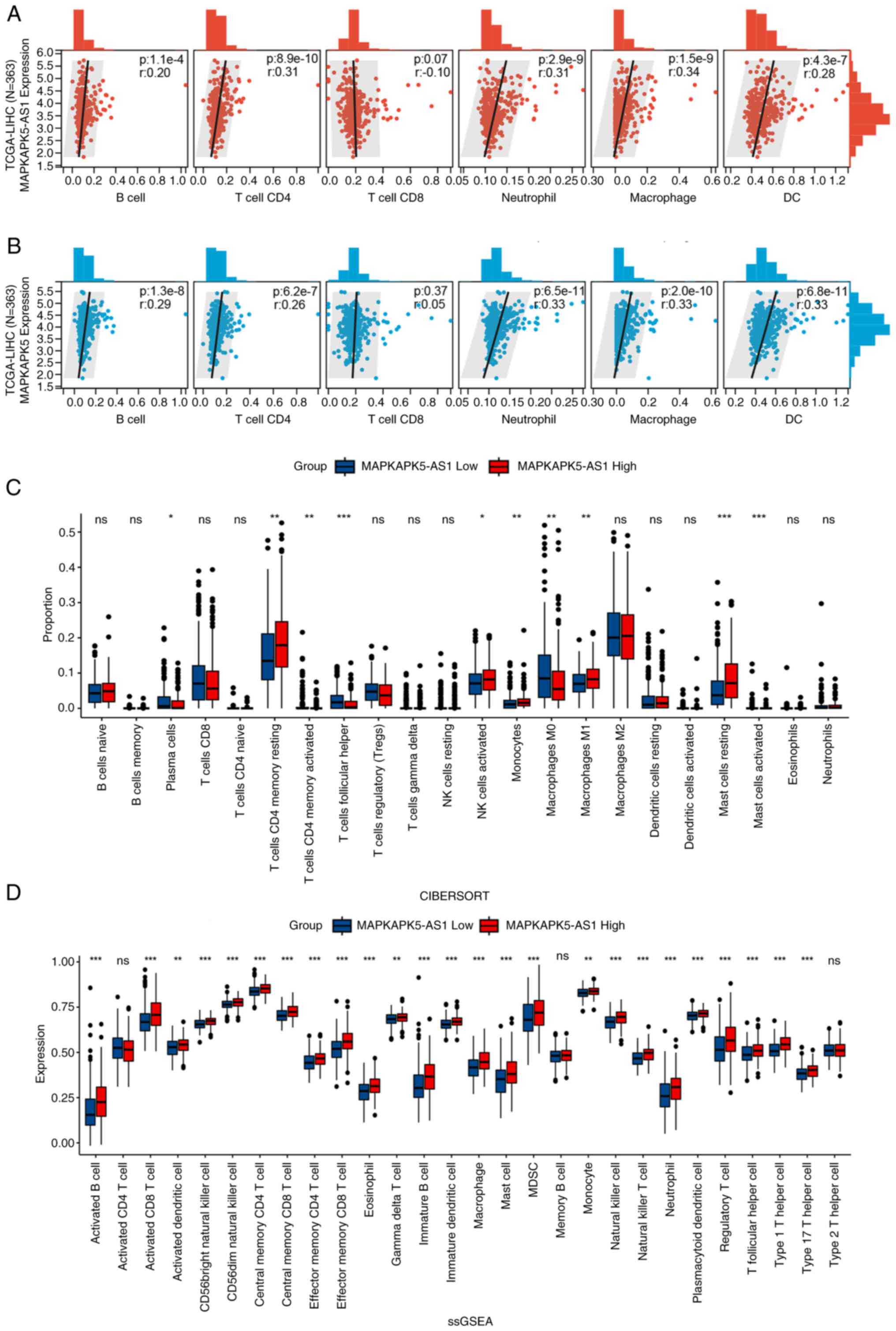
correlations, including B cells (R=0.20, P=1.1×10−4),
CD4+ T (R=0.31, P=8.9×10−10), CD8+
T (R=−0.10, P=0.07), neutrophils (R=0.31, P=2.9×10−9),
macrophages (R=0.34, P=1.5×10−9) and dendritic cells
(R=0.28, P=4.3×10−7) (Fig.
5A). MK5 was also found to be positively associated with six
types of cells of the immune system (Fig. 5B). Furthermore, as suggested in
Table V, MK5-AS1 expression is
positively linked with multiple immune cell biomarkers in HCC. The
aforementioned results demonstrated its effectiveness in regulating
TME of HCC.
 | Table V.The relationship between
lnc-MAPKAPK5-AS1 and biomarkers of immune system cells in
hepatocellular carcinoma. |
Table V.
The relationship between
lnc-MAPKAPK5-AS1 and biomarkers of immune system cells in
hepatocellular carcinoma.
| Immune cell | Biomarker | R-value | P-value |
|---|
| B cell | CD19 | 0.16 | 0.0022 |
|
| MS4A1 | 0.072 | 0.17 |
| CD4+ T
cell | CD4 | 0.2 | 0.067 |
| CD8+ T
cell | CD8A | 0.11 | 0.013 |
|
| CD8B | 0.12 | 0.02 |
| Neutrophil | ITGAM | 0.16 | 0.0016 |
|
| CD177 | 0.078 | 0.14 |
|
| CCR7 | 0.06 | 0.25 |
| Dendritic cell | HLA-DRA | 0.16 |
6.6×10−4 |
|
| HLA-DRA1 | 0.16 | 0.054 |
|
| HLA-DPB1 | 0.077 | 0.14 |
|
| HLA-DQB1 | 0.13 | 0.015 |
|
| BDCA1 | 0.13 | 0.0049 |
|
| ITGAX | 0.18 |
6×10−4 |
|
| NRP1 | 0.17 | 0.0013 |
| M1 macrophage | CD80 | 0.21 |
4.7×10−6 |
|
| CD86 | 0.25 |
3.4×10−8 |
|
| IL-1 | 0.091 | 0.047 |
| M2 macrophage | CD163 | 0.097 | 0.034 |
|
| CD206 | −0.092 | 0.044 |
|
| CD301 | 0.062 | 0.18 |
The immune cell infiltration ratio of TCGA-LIHC was
evaluated based on CIBERSORT algorithm. Patients with HCC were
divided according to the median of MK5-AS1 expression and the
proportions of immune infiltrating cell subtypes in high and low
expression groups were calculated (Fig.
5C). Using the R package ‘GSVA’, ssGSEA was used to calculate
the abundance of immune cells based on the corresponding data set.
For the majority of the 28 types of immune cells, including
myeloid-derived suppressor cells, gamma delta T cells, effector
memory CD4+ T cells, mast cells, memory B cells and
natural killer T cells were more significantly enriched in MK5-AS1
high expression group (Fig. 5D).
The aforementioned findings indicated that MK5-AS1 may regulate the
progression of HCC by affecting cellular infiltration of the immune
system.
lnc-MAPKAPK5-AS1 expression is
positively related to TMB and MSI in HCC
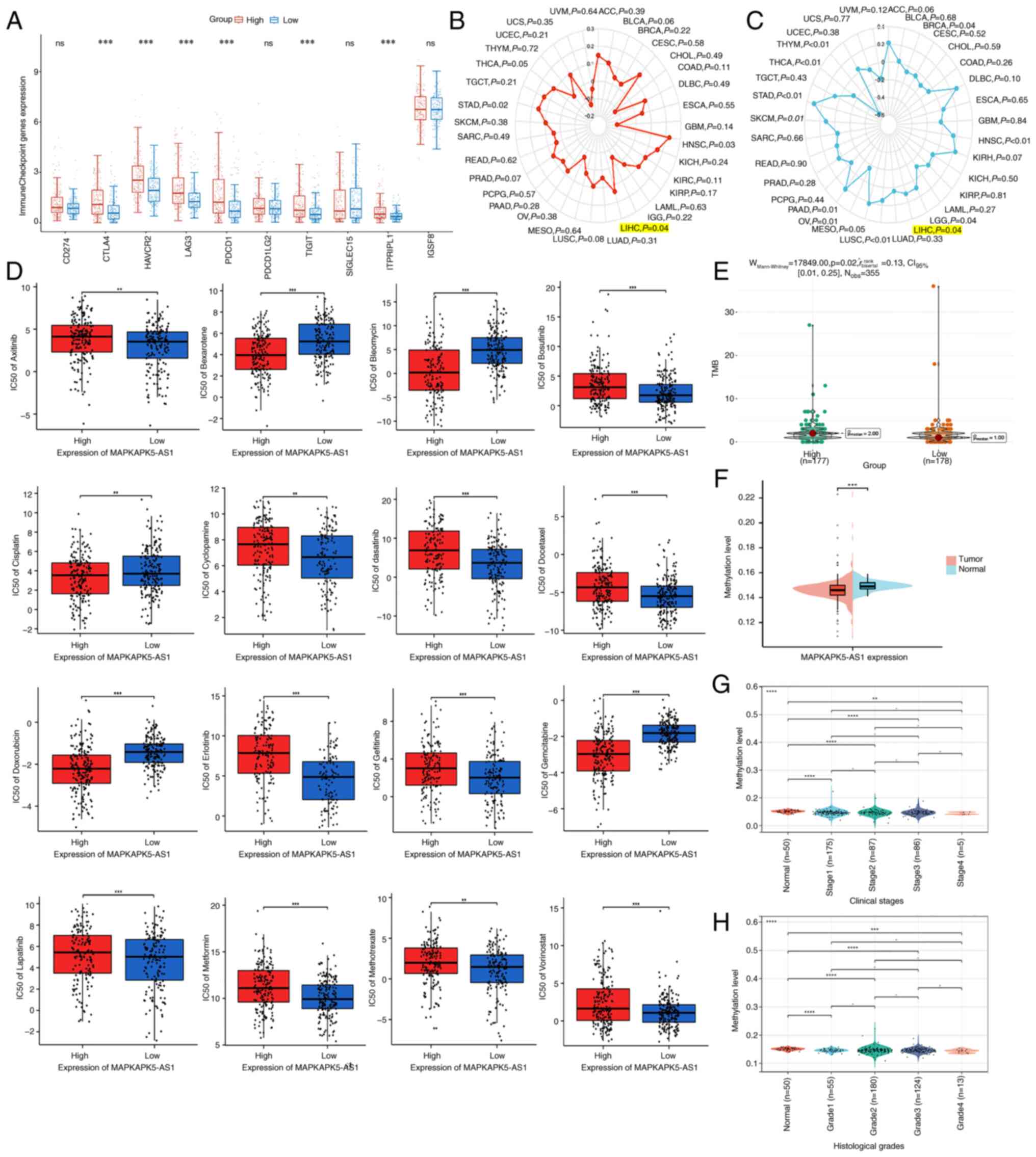
Inhibiting immune checkpoint signaling pathways are
key strategies for the treatment of a range of cancers. The
association of MK5-AS1 with ten common immune checkpoints in human
cancers, including CD274, CTLA4, HAVCR2, LAG3, PDCD1, PDCD1LG2,
TIGIT, SIGLEC15, ITPRIPL1 and IGSF8, was explored. The expression
levels of six immune checkpoints were significantly increased in
the MK5-AS1 high expression group (Fig.
6A).
As emerging markers of immunotherapy, the predictive
value of TMB and MSI in certain cancers has been validated in
clinical trials. The effects of TMB and MSI status on the
expression level of MK5-AS1 were assessed using the Sangerbox
database. Radar charts showed that the expression of MK5-AS1 was
positively correlated with TMB and MSI in HCC (R=0.138, P=0.04;
R=0.17, P=0.04, respectively) (Fig. 6B
and C). The expression of MK5-AS1 revealed a significant effect
on the TMB of patients with HCC in the analysis of data from TCGA
(Fig. 6E). Collectively, these
findings suggested that the group with higher TMB may have a
shorter survival time owing to the overexpression of MK5-AS1 in
these patients with HCC.
Drug sensitivity analysis of
lnc-MAPKAPK5-AS1
To further explore the clinical significance and
drug sensitivity of MK5-AS1, based on ‘pRRophetic’ R package, the
potential relationships between MK5-AS1 expression and drug
sensitivity of targeted therapeutic drugs that commonly used in
patients with HCC were further probed. Obviously, patients with low
expression of MK5-AS1 were more sensitive to Axitinib, Bosutinib,
Cyclopamine, dasatinib, Docetaxel, Embelin, Gefitinib, Lapatinib,
Metformin, Methotrexate and Vorinostat. By contrast, the
half-maximal inhibitory concentration (IC50) calculated
utilizing ‘pRRophetic’ R package of Bexarotene, Bleomycin,
Cisplatin, Doxorubicin and Gemcitabine was lower in MK5-AS1 high
expression group (Fig. 6D). These
findings suggested that MK5-AS1 might act as an effective biomarker
for the efficacy of targeted treatment of patients with HCC.
Mechanisms of lnc-MAPKAPK5-AS1
upregulation in HCC
To seek out the possible reasons for the
upregulation of MK5-AS1 in HCC, the methylation level of promoter
region near MK5-AS1 was analyzed based on sample type, clinical
stage and histological grade and the mutation status of two genes
in HCC was probed.
Firstly, the methylation data of the promoter region
near MK5-AS1 was obtained using the Diseasemeth 2.0 database. The
results revealed that methylation level in HCC tissues was
significantly lower than that in normal tissues (Fig. 6F), and there were significant
differences in the methylation expression level of MK5-AS1 in
different clinical stages and histological grades of HCC (Fig. 6G and H), which provided evidence
that the upregulation of MK5-AS1 in HCC might partly due to
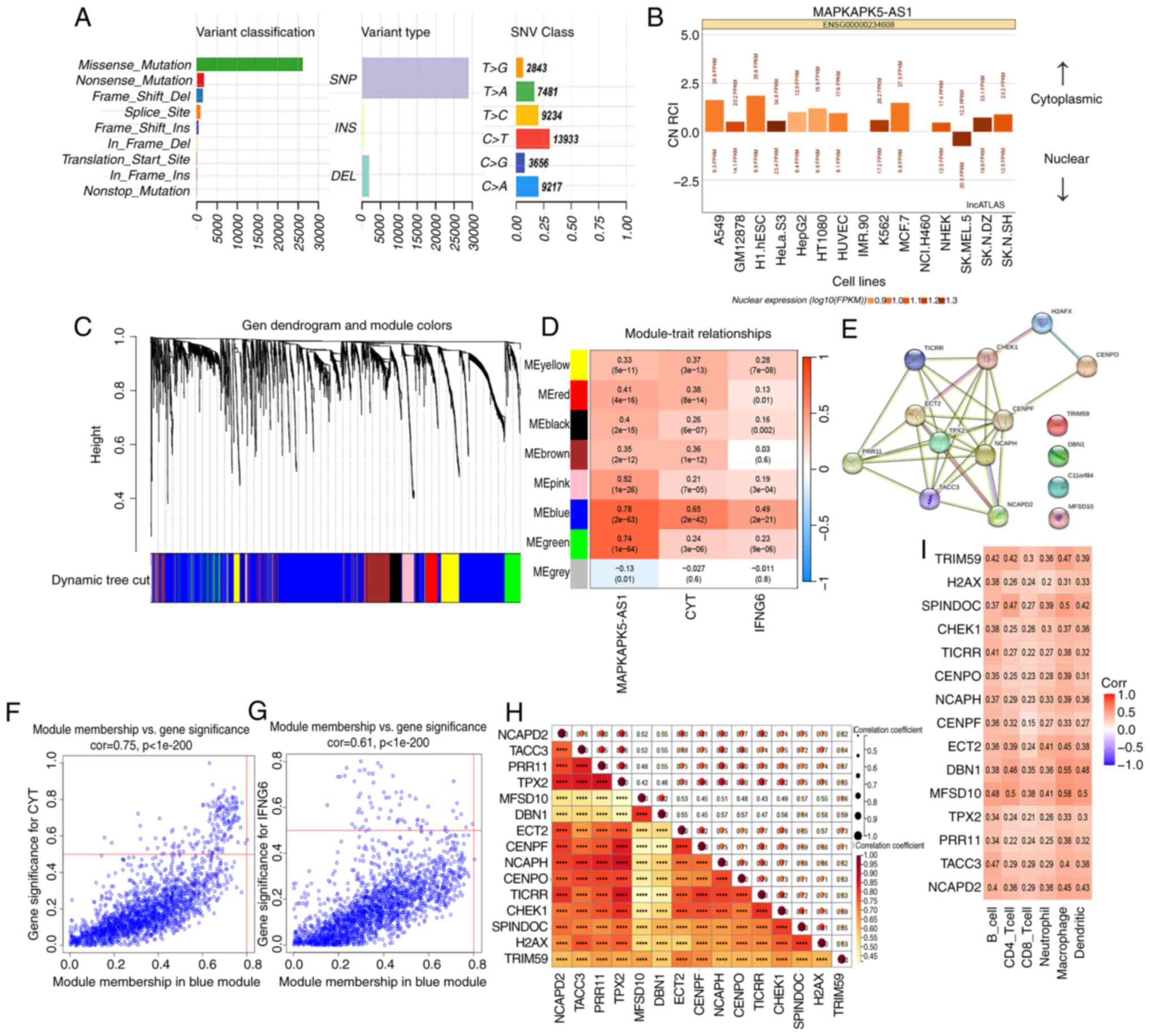
hypomethylation of its promoter. In addition, the overall result of
the ‘MAF’ file was plotted and it was found that missense mutation
accounted for the predominant part when the mutation types were
classified according to different categories (Fig. 7A). Moreover, single nucleotide
polymorphism appeared more frequently than insertions or deletions,
with C>T being the most common mutation in single nucleotide
variants. Then, the mutation status of MK5-AS1 and MK5 was explored
using cBioPortal database, and the results showed that the
incidence of MK5-AS1 mutation was 6% (21/360) in HCC, and only two
of the 360 patients had missense mutation in MK5, demonstrating
that this mutation is fairly unlikely to be the major reason of
MK5-AS1 upregulation (Fig. S1A and
B).
Prediction of miRNAs that probably
interact with lnc-MAPKAPK5-AS1
Evidence suggests that lncRNAs in the cytoplasm can
affect the stability and translation regulation of mRNA mainly
through the ceRNA regulatory mechanism by adsorbing miRNAs. Using
lncATLAS database, the expression pattern of MK5-AS1 in different
cell lines was uncovered, while most of the data exhibited that
MK5-AS1 is primarily located in the cytoplasm (Fig. 7B). Therefore, it was hypothesized
that MK5-AS1 might promote the progression of HCC through the
sponge adsorption of miRNAs. ENCORI database unveiled that four
miRNAs (hsa-miR-452-5p, hsa-miR-556-3p, hsa-miR-4676-3p and
hsa-miR-892c-3p) could directly bind to the gene body of MK5-AS1
and MK5. It has been already noted that hsa-miR-452-5p was
overexpressed in HCC tissues compared with normal tissues, which
lead to a poor prognosis of HCC through modulating the RNA levels
of downstream target genes (35).
Unfortunately, based on data from the LIRI-JP dataset of the ICGC
database (https://dcc.icgc.org/, accessed on 10
January 2021) (36), there was no
discernible variation in the expression pattern of these miRNAs
between HCC tissues and normal tissues (Fig. S2A-D). Furthermore, there was no
concomitant negative association between these miRNAs and the
expression level of MK5-AS1 or MK5 owing to the limitation of
sample size and data source (Fig.
S2E-L); thus, further molecular experiments are required to
confirm the specific mechanism.
Co-expression network construction and
identification of immune-related key genes of HCC
A total of eight modules were generated in the
hierarchical clustering tree (Fig.
7C). The correlations between all feature genes of these
modules and CYT and IFNG6 scores reflecting immune activity are
shown in Fig. 7D, in which the blue
module showed the strongest association with the aforementioned two
scores (R=0.75, P<0.001; R=0.61, P<0.001, Fig. 7F and G). Using MS>0.8 and
GS>0.3, 15 genes (NCAPD2, TACC3, PRR11, TPX2, MFSD10, DBN1,
ECT2, CENPF, NCAPH, CENPO, TICRR, CHEK1, SPINDOC, H2AX and TRIM59)
were screened from the blue module as candidate hub genes. To
further explore the biological function of these genes, a
protein-protein interaction (PPI) network was constructed using the
STRING database (Fig. 7E).
Spearman's correlation analysis revealed strong correlation between
genes and six types of cells of the immune system (Fig. 7H and I).
Discussion
Patients with HCC exhibited a poor survival and lack
effective prognostic biomarkers over a long period of time. The
identification of genes with potential value for the diagnosis and
treatment of HCC will be critical to improving patient treatment.
Studies have revealed that lncRNAs participate in gene regulation
by acting as a miRNA sponge or RBP binding and play critical roles
in tumor angiogenesis, invasion and metastasis (37–39).
Antisense lncRNAs exhibit special structures and represent a class
of lncRNAs that are complementary to other transcript sequences.
lnc-MCM3AP-AS1 (40) and
lnc-AFAP1-AS1 (41) may be novel
molecular tumor markers. Previous research has demonstrated a role
for MK5-AS1 in tumors. Cheng et al (42) demonstrated that MK5-AS1 may be a
hypoxia-related lncRNA in HCC and involved in tumorigenesis and
progression. Wang et al (43) constructed an immune-lncRNAs
signature containing MK5-AS1 in anaplastic gliomas. Several studies
have revealed that MK5-AS1 may be acting as a ceRNA potential in
various tumors (44–46).
Previous studies (7,8,46) on
the role of MK5-AS1 in HCC were mostly focused on cell line
experiments through reverse transcription-quantitative PCR and cell
phenotype experiments and rarely integrated multiple large cancer
databases such as TCGA, GEO and ICGC for overall analysis at the
human tissue level. In the present study, a comprehensive
bioinformatics analysis was performed using the aforementioned
databases, including clinical correlation analysis, enrichment
analysis, methylation analysis, immune infiltration analysis,
association analysis with TMB and MSI, drug sensitivity analysis
and gene mutation analysis. Core immune-related genes co-expressed
with MK5-AS1 were screened by WGCNA analysis, and a PPI network was
constructed to understand the reaction mechanism of gene expression
regulation and biological signal transmission. Analysis of the
external cohort of 90 patients with liver cancer verified that
upregulated MK5-AS1 in liver cancer tissues was associated with
poor prognosis of patients with HCC and revealed the potential
positive regulatory relationship between MK5-AS1 and MK5. These
findings provide substantial evidence of the functional role of
MK5-AS1 in HCC.
Through data mining of TCGA and GEO, MK5-AS1 was
found to be notably increased in HCC tissues and linked to poor
outcome, suggesting that MK5-AS1 may be an independent prognostic
factor in HCC. Unfavorable pathological grade, clinical stage, T
stage and higher AFP was associated with the increased expression
of MK5-AS1 and MK5 in HCC. A positive regulatory relationship
between MK5-AS1 and its antisense transcript MK5 was also observed,
which may be involved in the progression of HCC; however, the
specific mechanism remains to be elucidated. The protein expression
of MK5 in HCC was significantly higher than that in adjacent
non-cancerous tissues, which was in line with previous results.
Correlation analysis revealed that advanced clinical stage,
abnormal ALT value and upregulated PD-L1 expression was more
frequent in the high MK5 group. ALT is a sensitive predictor of
early hepatocyte injury and elevated ALT is associated with
increased mortality of HCC (47).
Patients in the TCGA and GSE144269 datasets were grouped by MK5-AS1
expression. DEG analysis and a Venn diagram were carried out to
obtain intersection genes. In enrichment analysis, the FDR
threshold of 0.25 is set based on a trade-off between statistics,
the error rate of the actual study and the reliability of the
results. This threshold not only guarantees a certain discovery
rate, but also controls the error rate, which is a common balance
point in genomics research. GO and KEGG analysis revealed that the
DEGs may regulate the progression of HCC through pathways such as
biological regulation, metabolic progress, response to stimulus,
multicellular organismal process, immune system process, PRAK
signaling pathway and cell adhesion molecules.
Cell adhesion molecules serve as the molecular
foundation for a variety of critical physiological and pathological
processes, including immunological response, inflammatory response,
coagulation and tumor metastasis. ICAM-1/CD54 is expressed at low
levels on resting vascular endothelial cells, increasing cell
adhesion across HCC cells and endothelial cells by binding to
particular receptors on their surface (48). Synaptic vesicles are involved in
cellular component exchange, signal transduction and pathological
progress, and some have even been linked to the TME (49). Fat is an important energy source and
molecular signal. Dysmetabolism of fatty acids in the TME not only
affects the susceptibility of patients with cancer to radiotherapy
or chemotherapy, but also interferes with their immunotherapy by
affecting the immune response of T cells (50). Regulation of immune system process,
biological adhesion, collagen containing extracellular matrix,
immune effector process and other pathways were enriched in GSEA
analysis. These results suggest that MK5-AS1 is associated with
multiple immune-related metabolic signal channels.
HCC is a tumor driven by chronic inflammation
(51), and the immunosuppressive
microenvironment of HCC is an important factor in disease
progression. Current clinical studies have explored the complex
interplay between NASH, HCC and the immune response, and numerous
therapeutic approaches have focused on targeting immune cells. In
particular, changes in B cells, T cells and dendritic cells in the
adaptive immune system, impaired cytotoxicity of natural killer
cells and the accumulation of neutrophils (52). To explore the specific mechanism of
MK5-AS1 in immune regulation, immune infiltration data from TIMER
was downloaded and it was found that MK5-AS1 was positively
correlated with multiple types of immune system cells and
corresponding biomarkers, suggesting that MK5-AS1 may negatively
affect the prognosis of HCC owing to its regulatory role in the
TME. The results of the present study indicated that MK5-AS1 was
not directly involved in the functional regulation of
CD8+ T cells during certain stages or conditions of the
immune response due to disparities in the specificity and
regulatory mechanisms of gene expression. However, this does not
mean that MK5-AS1 has no effect on CD8+ T cells, because
there may be indirect or conditional interactions; these
possibilities should be further explored in future studies. Using
the CIBERSORT and ssGSEA algorithm, plasma cells, T cells
CD4+ memory resting, macrophages M1 and several immune
cells were found to be present at high levels in the MK5-AS1 high
expression group. Previous research reported that patients with a
higher proportion of plasma cells in HCC have a shorter survival
time (53). Zong et al
(54) demonstrated that M1
macrophages mediate inducible PD-L1 expression in HCC cells and
perform a tumor-promoting role, which also lends credence to our
conclusion.
As emerging biomarkers of cancer immunotherapy, TMB
and MSI are closely related to clinical prognosis. TMB has been
extensively utilized to forecast the effectiveness of immunotherapy
in non-small cell lung cancer and melanoma, but few studies have
focused on its role in HCC. In the present study, it was revealed
that MK5-AS1 expression was positively related to TMB and MSI.
Moreover, survival outcome was found in high TMB group verified by
data in TCGA, which had higher expression of MK5-AS1. Immune
checkpoints are pivotal effector molecules in the immune
microenvironment. Currently, there are no well-established
biomarkers for immunotherapy for tumors of the digestive system,
especially for HCC. Under physiological conditions, PD-1 binds to
PD-L1 to release inhibitory signals while CTLA-4 is present in
regulatory T lymphocytes; they impede autoimmune responses and
participate in the immune evasion process of HCC through various
pathways (55,56). PD-L1 and TMB are not related in
major tumor types such as HCC, head and neck squamous cell
carcinoma, renal cell carcinoma and small cell lung cancer
(57,58). Detection of both factors can provide
guidance for the clinical treatment and application of ICIs
(59). In non-small cell lung
cancer, patients with both high PD-L1 expression and TMB exhibit
the best curative effect from ICIs, with a clinical benefit rate of
50%, while patients with low PD-L1 expression and TMB have a
clinical benefit rate of only 18.2% (60). In the present study, it was found
that the PD-L1 and TMB levels of HCC in the high MK5-AS1 expression
group were considerably increased compared with the low MK5-AS1
expression group, which is consistent with previous studies that
revealed that HCC cases with a high TMB have a shorter OS (61,62).
The infiltration of various types of immune system cells was also
different between the two groups, indicating that TMB may also
determine the efficacy of ICIs by affecting the TME of HCC. As a
result, it would be a new challenge and strategy to screen the
advantage groups or combination therapy for immunotherapy in the
future.
To explore the mechanism of MK5-AS1 overexpression
in HCC tissues, the mutation and methylation status of MK5-AS1 was
examined. DNA methylation is an important form of epigenetic
modification. Aberrant methylation affects the conformation of DNA,
making it difficult for transcription factors to bind and inhibit
gene transcription. Database prediction indicated that the
methylation level of the MK5-AS1 promoter in HCC was lower than
that in normal tissues and correlated with advanced clinical stage
and histological grade, indicating that MK5-AS1 is upregulated in
HCC tissues possibly from hypomethylation. Tao et al
(63) previously confirmed that
MK5-AS1 expression in HBV-related HCC was elevated in M2
macrophages. As a result of N6-methyladenosine modification, the
expression of MK5-AS1 in HCC cells was also increased after the
transfer exosomes, which promotes cell proliferation.
WGCNA was applied to identify modules closely
associated with immune scores in the MK5-AS1 high and low
expression groups. Using MM and GS, 15 hub genes were screened out
of the blue module, including TPX2, CENPO, CENPF and ECT2. Wang
et al (64) demonstrated
that TPX2 regulates CXCR5 through the NF-κB signaling pathway to
improve the anti-tumor function of human CD8+ T cells
and has a synergistic effect with anti-PD-1 therapy. CENPO and
CENPF are key genes related to antitumor immunity in HCC (65,66).
Xu et al (67) reported that
ECT2 promotes the polarization of M2 macrophages, which may be
related to enhanced aerobic glycolysis.
In conclusion, the results of the present study
revealed that the expression of MK5-AS1 was upregulated in HCC
tissues and MK5-AS1 was co-expressed with its protein-coding gene
MK5. Increased expression is linked to a poor prognosis as well as
higher levels of immune infiltration and immune-related genes,
indicating that MK5-AS1 may serve as a prognostic biomarker and
therapeutic target for HCC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science
Foundation of China (grant no. 81803325), the Natural Science
Foundation of Guangdong (grant nos. 2021A1515011175 and
2024A1515011646), the Guangzhou Science and Technology Project
(grant no. 202102080126), the Key Project of Medicine Discipline of
Guangzhou (grant no. 2025-2027-12), the Medical Science and
Technology Foundation of Guangdong (grant no. A2024733) and the
Basic Research Project of Key Laboratory of Guangzhou (grant no.
202102100001).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
DWu and XH conceptualized the study, conducted
investigation and wrote the original draft. Material preparation,
data collection and analysis were performed by DWa, PQ, LZ, XH, BL
and JC. DWu, DWa and PQ reviewed edited the manuscript. DWu and XH
confirm the authenticity of all the raw data. All authors read and
approved the final version of the manuscript.
Ethics approval and consent to
participate
The present study was conducted in accordance with
the Declaration of Helsinki, and was approved (approval no.
SHYJS-CP-1901001 in 11th January 2019; and extended as approval no.
SHYJS-BC-2310001 on 20th October, 2023) by the ethics committee of
Shanghai Outdo Biotech Co., Ltd. (Shanghai, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Xie DY, Ren ZG, Zhou J, Fan J and Gao Q:
2019 Chinese clinical guidelines for the management of
hepatocellular carcinoma: Updates and insights. Hepatobiliary Surg
Nutr. 9:452–463. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Xu XF, Xing H, Han J, Li ZL, Lau WY, Zhou
YH, Gu WM, Wang H, Chen TH, Zeng YY, et al: Risk factors, patterns,
and outcomes of late recurrence after liver resection for
hepatocellular carcinoma: A multicenter study from China. JAMA
Surg. 154:209–217. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ahn JC, Lee YT, Agopian VG, Zhu Y, You S,
Tseng HR and Yang JD: Hepatocellular carcinoma surveillance:
Current practice and future directions. Hepatoma Res.
8:102022.PubMed/NCBI
|
|
4
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Luo S, Lu JY, Liu L, Yin Y, Chen C, Han X,
Wu B, Xu R, Liu W, Yan P, et al: Divergent lncRNAs regulate gene
expression and lineage differentiation in pluripotent cells. Cell
Stem Cell. 18:637–652. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang H, Shi Y, Chen CH, Wen Y, Zhou Z,
Yang C, Sun J, Du G, Wu J, Mao X, et al: KLF5-induced lncRNA
IGFL2-AS1 promotes basal-like breast cancer cell growth and
survival by upregulating the expression of IGFL1. Cancer Lett.
515:49–62. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ji H, Hui B, Wang J, Zhu Y, Tang L, Peng
P, Wang T, Wang L, Xu S, Li J and Wang K: Long noncoding RNA
MAPKAPK5-AS1s promotes colorectal cancer proliferation by partly
silencing p21 expression. Cancer Sci. 110:72–85. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang T, Chen WC, Shi PC, Liu MR, Jiang T,
Song H, Wang JQ, Fan RZ, Pei DS and Song J: Long noncoding RNA
MAPKAPK5-AS1 promotes colorectal cancer progression by
cis-regulating the nearby gene MK5 and acting as a let-7f-1-3p
sponge. J Exp Clin Cancer Res. 39:1392020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang H, Wang Y and Lu J: Identification
of lung-adenocarcinoma-related long non-coding RNAs by random
walking on a competing endogenous RNA network. Ann Transl Med.
7:3392019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gao GF, Parker JS, Reynolds SM, Silva TC,
Wang LB, Zhou W, Akbani R, Bailey M, Balu S, Berman BP, et al:
Before and after: Comparison of legacy and harmonized TCGA genomic
data commons' data. Cell Syst. 9:24–34.e10. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets-update. Nucleic Acids Res. 41((Database issue)):
D991–D995. 2013.PubMed/NCBI
|
|
12
|
Zhou Y, Zhou B, Pache L, Chang M,
Khodabakhshi AH, Tanaseichuk O, Benner C and Chanda SK: Metascape
provides a biologist-oriented resource for the analysis of
systems-level datasets. Nat Commun. 10:15232019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Newman AM, Liu CL, Green MR, Gentles AJ,
Feng W, Xu Y, Hoang CD, Diehn M and Alizadeh AA: Robust enumeration
of cell subsets from tissue expression profiles. Nat Methods.
12:453–457. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li H, Liu C, Huang S, Wang X, Cao M, Gu T,
Ou X, Pan S, Lin Z, Wang X, et al: Multi-omics analyses demonstrate
the modulating role of gut microbiota on the associations of
unbalanced dietary intake with gastrointestinal symptoms in
children with autism spectrum disorder. Gut Microbes.
15:22813502023. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Song J, Ren K, Zhang D, Lv X, Sun L, Deng
Y and Zhu H: A novel signature combing cuproptosis- and
ferroptosis-related genes in sepsis-induced cardiomyopathy. Front
Genet. 14:11707372023. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rukhsan A, Supty AT, Hussain M and Lee Y:
STK3 higher expression association with clinical characteristics in
intrinsic subtypes of breast cancer invasive ductal carcinoma
patients. Breast Cancer Res Treat. 206:119–129. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Reimand J, Isserlin R, Voisin V, Kucera M,
Tannus-Lopes C, Rostamianfar A, Wadi L, Meyer M, Wong J, Xu C, et
al: Pathway enrichment analysis and visualization of omics data
using g:Profiler, GSEA, cytoscape and EnrichmentMap. Nat Protoc.
14:482–517. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li T, Fan J, Wang B, Traugh N, Chen Q, Liu
JS, Li B and Liu XS: TIMER: A web server for comprehensive analysis
of tumor-infiltrating immune cells. Cancer Res. 77:e108–e110. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Samstein RM, Lee CH, Shoushtari AN,
Hellmann MD, Shen R, Janjigian YY, Barron DA, Zehir A, Jordan EJ,
Omuro A, et al: Tumor mutational load predicts survival after
immunotherapy across multiple cancer types. Nat Genet. 51:202–206.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Klein O, Kee D, Markman B, Carlino MS,
Underhill C, Palmer J, Power D, Cebon J and Behren A: Evaluation of
TMB as a predictive biomarker in patients with solid cancers
treated with anti-PD-1/CTLA-4 combination immunotherapy. Cancer
Cell. 39:592–593. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Goodman AM, Kato S, Bazhenova L, Patel SP,
Frampton GM, Miller V, Stephens PJ, Daniels GA and Kurzrock R:
Tumor mutational burden as an independent predictor of response to
immunotherapy in diverse cancers. Mol Cancer Ther. 16:2598–2608.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Feng D, Hui X, Shi-Chun L, Yan-Hua B, Li
C, Xiao-Hui L and Jie-Yu Y: Initial experience of anti-PD1 therapy
with nivolumab in advanced hepatocellular carcinoma. Oncotarget.
8:96649–96655. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Geeleher P, Cox NJ and Huang RS: Clinical
drug response can be predicted using baseline gene expression
levels and in vitro drug sensitivity in cell lines. Genome Biol.
15:R472014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Maeser D, Gruener RF and Huang RS:
oncoPredict: An R package for predicting in vivo or cancer patient
drug response and biomarkers from cell line screening data. Brief
Bioinform. 22:bbab2602021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xiong Y, Wei Y, Gu Y, Zhang S, Lyu J,
Zhang B, Chen C, Zhu J, Wang Y, Liu H and Zhang Y: DiseaseMeth
version 2.0: A major expansion and update of the human disease
methylation database. Nucleic Acids Res. 45((D1)): D888–D895. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mas-Ponte D, Carlevaro-Fita J, Palumbo E,
Hermoso Pulido T, Guigo R and Johnson R: LncATLAS database for
subcellular localization of long noncoding RNAs. RNA. 23:1080–1087.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42((Database issue)): D92–D97. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Langfelder P and Horvath S: WGCNA: An R
package for weighted correlation network analysis. BMC
Bioinformatics. 9:5592008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Szklarczyk D, Morris JH, Cook H, Kuhn M,
Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al:
The STRING database in 2017: Quality-controlled protein-protein
association networks, made broadly accessible. Nucleic Acids Res.
45:D362–D368. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wakiyama H, Masuda T, Motomura Y, Hu Q,
Tobo T, Eguchi H, Sakamoto K, Hirakawa M, Honda H and Mimori K:
Cytolytic activity (CYT) score is a prognostic biomarker reflecting
host immune status in hepatocellular Carcinoma (HCC). Anticancer
Res. 38:6631–6638. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Seiwert TY, Burtness B, Mehra R, Weiss J,
Berger R, Eder JP, Heath K, McClanahan T, Lunceford J, Gause C, et
al: Safety and clinical activity of pembrolizumab for treatment of
recurrent or metastatic squamous cell carcinoma of the head and
neck (KEYNOTE-012): An open-label, multicentre, phase 1b trial.
Lancet Oncol. 17:956–965. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chandrashekar DS, Karthikeyan SK, Korla
PK, Patel H, Shovon AR, Athar M, Netto GJ, Qin ZS, Kumar S, Manne
U, et al: UALCAN: An update to the integrated cancer data analysis
platform. Neoplasia. 25:18–27. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zheng Z, Liu J, Yang Z, Wu L, Xie H, Jiang
C, Lin B, Chen T, Xing C, Liu Z, et al: MicroRNA-452 promotes
stem-like cells of hepatocellular carcinoma by inhibiting Sox7
involving Wnt/β-catenin signaling pathway. Oncotarget.
7:28000–28012. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Bertucci F, Chaffanet M and Birnbaum D: An
ICGC major achievement in breast cancer: A comprehensive catalog of
mutations and mutational signatures. Chin Clin Oncol. 6:42017.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li Z, Lin Y, Cheng B, Zhang Q and Cai Y:
Identification and analysis of potential key genes associated with
hepatocellular carcinoma based on integrated bioinformatics
methods. Front Genet. 12:5712312021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Cui XY, Zhan JK and Liu YS: Roles and
functions of antisense lncRNA in vascular aging. Ageing Res Rev.
72:1014802021. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang J, Su Z, Lu S, Fu W, Liu Z, Jiang X
and Tai S: LncRNA HOXA-AS2 and its molecular mechanisms in human
cancer. Clin Chim Acta. 485:229–233. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang Y, Yang L, Chen T, Liu X, Guo Y, Zhu
Q, Tong X, Yang W, Xu Q, Huang D and Tu K: A novel lncRNA
MCM3AP-AS1 promotes the growth of hepatocellular carcinoma by
targeting miR-194-5p/FOXA1 axis. Mol Cancer. 18:282019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang JY, Weng MZ, Song FB, Xu YG, Liu Q,
Wu JY, Qin J, Jin T and Xu JM: Long noncoding RNA AFAP1-AS1
indicates a poor prognosis of hepatocellular carcinoma and promotes
cell proliferation and invasion via upregulation of the RhoA/Rac2
signaling. Int J Oncol. 48:1590–1598. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Cheng M, Zhang J, Cao PB and Zhou GQ:
Prognostic and predictive value of the hypoxia-associated long
non-coding RNA signature in hepatocellular carcinoma. Yi Chuan.
44:153–167. 2022.PubMed/NCBI
|
|
43
|
Wang W, Zhao Z, Yang F, Wang H, Wu F,
Liang T, Yan X, Li J, Lan Q, Wang J and Zhao J: An immune-related
lncRNA signature for patients with anaplastic gliomas. J
Neurooncol. 136:263–271. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhang J, Fan D, Jian Z, Chen GG and Lai
PB: Cancer specific long noncoding RNAs show differential
expression patterns and competing endogenous RNA potential in
hepatocellular carcinoma. PLoS One. 10:e01410422015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yang J, Xu QC, Wang ZY, Lu X, Pan LK, Wu J
and Wang C: Integrated analysis of an lncRNA-associated ceRNA
network reveals potential biomarkers for hepatocellular carcinoma.
J Computat Biol. 28:330–344. 2021. View Article : Google Scholar
|
|
46
|
Peng Z, Ouyang X, Wang Y and Fan Q:
MAPKAPK5-AS1 drives the progression of hepatocellular carcinoma via
regulating miR-429/ZEB1 axis. BMC Mol Cell Biol. 23:212022.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wedemeyer H, Hofmann WP, Lueth S, Malinski
P, Thimme R, Tacke F and Wiegand J: (ALT screening for chronic
liver diseases: Scrutinizing the evidence). Z Gastroenterol.
48:46–55. 2010.(In German). View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Han P, Lei Y, Liu J, Liu J, Huang H, Tian
D and Yan W: Cell adhesion molecule BVES functions as a suppressor
of tumor cells extrusion in hepatocellular carcinoma metastasis.
Cell Commun Signal. 20:1492022. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Xu Y, Feng K, Zhao H, Di L, Wang L and
Wang R: Tumor-derived extracellular vesicles as messengers of
natural products in cancer treatment. Theranostics. 12:1683–1714.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Luu M, Riester Z, Baldrich A, Reichardt N,
Yuille S, Busetti A, Klein M, Wempe A, Leister H, Raifer H, et al:
Microbial short-chain fatty acids modulate CD8(+) T cell responses
and improve adoptive immunotherapy for cancer. Nat Commun.
12:40772021. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Ringelhan M, Pfister D, O'Connor T,
Pikarsky E and Heikenwalder M: The immunology of hepatocellular
carcinoma. Nat Immunol. 19:222–232. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Gregory SN, Perati SR and Brown ZJ:
Alteration in immune function in patients with fatty liver disease.
Hepatoma Res. 8:312022. View Article : Google Scholar
|
|
53
|
Zhang S, Liu Z, Wu D, Chen L and Xie L:
Single-Cell RNA-Seq analysis reveals microenvironmental
infiltration of plasma cells and hepatocytic prognostic markers in
HCC with Cirrhosis. Front Oncol. 10:5963182020. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Zong Z, Zou J, Mao R, Ma C, Li N, Wang J,
Wang X, Zhou H, Zhang L and Shi Y: M1 macrophages induce PD-L1
expression in hepatocellular carcinoma cells through IL-1β
signaling. Front Immunol. 10:16432019. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Yao S and Chen L: PD-1 as an immune
modulatory receptor. Cancer J. 20:262–264. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Chen X, Du Y, Hu Q and Huang Z:
Tumor-derived CD4+CD25+regulatory T cells inhibit dendritic cells
function by CTLA-4. Pathol Res Pract. 213:245–249. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Rizvi H, Sanchez-Vega F, La K, Chatila W,
Jonsson P, Halpenny D, Plodkowski A, Long N, Sauter JL, Rekhtman N,
et al: Molecular determinants of response to anti-programmed cell
death (PD)-1 and anti-programmed death-ligand 1 (PD-L1) blockade in
patients with non-small-cell lung cancer profiled with targeted
next-generation sequencing. J Clin Oncol. 36:633–641. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Yarchoan M, Albacker LA, Hopkins AC,
Montesion M, Murugesan K, Vithayathil TT, Zaidi N, Azad NS, Laheru
DA, Frampton GM and Jaffee EM: PD-L1 expression and tumor
mutational burden are independent biomarkers in most cancers. JCI
Insight. 4:e1269082019. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Yarchoan M, Hopkins A and Jaffee EM: Tumor
mutational burden and response rate to PD-1 inhibition. N Engl J
Med. 377:2500–2501. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Jiang J, Jin Z, Zhang Y, Peng L, Zhang Y,
Zhu Z, Wang Y, Tong D, Yang Y, Wang J, et al: Robust prediction of
immune checkpoint inhibition therapy for non-small cell lung
cancer. Front Immunol. 12:6468742021. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Cai H, Zhang Y, Zhang H, Cui C, Li C and
Lu S: Prognostic role of tumor mutation burden in hepatocellular
carcinoma after radical hepatectomy. J Surg Oncol. 121:1007–1014.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Huo J, Wu L and Zang Y: A prognostic model
of 15 immune-related gene pairs associated with tumor mutation
burden for hepatocellular carcinoma. Front Mol Biosci.
7:5813542020. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Tao L, Li D, Mu S, Tian G and Yan G:
LncRNA MAPKAPK5_AS1 facilitates cell proliferation in hepatitis B
virus -related hepatocellular carcinoma. Lab Invest. 102:494–504.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Wang X, Wang J, Shen H, Luo Z and Lu X:
Downregulation of TPX2 impairs the antitumor activity of CD8+ T
cells in hepatocellular carcinoma. Cell Death Dis. 13:2232022.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
He K, Xie M, Li J, He Y and Yin Y: CENPO
is associated with immune cell infiltration and is a potential
diagnostic and prognostic marker for hepatocellular carcinoma. Int
J Gen Med. 15:7493–7510. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Si T, Huang Z, Jiang Y, Walker-Jacobs A,
Gill S, Hegarty R, Hamza M, Khorsandi SE, Jassem W, Heaton N and Ma
Y: Expression levels of three key genes CCNB1, CDΧ20, and CENPF in
HCC are associated with antitumor Immunity. Front Oncol.
11:7388412021. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Xu D, Wang Y, Wu J, Zhang Z, Chen J, Xie
M, Tang R, Chen C, Chen L, Lin S, et al: ECT2 overexpression
promotes the polarization of tumor-associated macrophages in
hepatocellular carcinoma via the ECT2/PLK1/PTEN pathway. Cell Death
Dis. 12:1622021. View Article : Google Scholar : PubMed/NCBI
|