Introduction
Gastric cancer (GC) is one of the most common
malignancies, ranking as the fifth most common type of cancer
globally, with ~1,089,103 new diagnoses each year. GC is also the
fourth leading cause of cancer-related deaths and is responsible
for 768,793 fatalities worldwide (1), and chemotherapy and radical surgery
both serve important roles in standard treatment. Despite the
recent availability of additional chemotherapy regimens, the
combination of 5-fluorouracil and cisplatin (CDDP) is still the
most commonly used chemotherapy regimen (2); however, resistance to chemotherapy may
occur, leading to treatment failure and GC recurrence (3). Therefore, it is essential to
investigate the mechanisms of drug resistance in stomach neoplasms
to formulate a treatment strategy.
MicroRNAs (miRNAs/miRs) are an important group of
noncoding RNA molecules that are characterized by short nucleotide
sequences (20–24 nt), and are involved in the post-transcriptional
regulation of gene expression in multicellular organisms by
affecting both the stability and translation of mRNAs (4). Studies have shown that miRNAs are
often involved in regulating tumor development as either tumor
suppressors or promoters (5,6). It
has also been reported that exogenous miRNA mimics regulate mRNA
expression in vivo to achieve effective cancer treatment
(7–9). miR-424-5p has been reported to be
associated with the development of breast, bladder and colorectal
cancer, may be a prognostic biomarker in melanoma, and may serve an
anticancer role by inhibiting the proliferation of cervical cancer
cells (10). However, miR-424-5p
has not been well studied in GC. Our previous analysis of the Gene
Expression Omnibus (GEO) database revealed that miR-424-5p may be a
tumor suppressor and be associated with CDDP resistance (11).
The ubiquitin-proteasome system is an important
method of protein degradation and E3 ubiquitination ligases are
vital to this process. Inhibition of the ubiquitin-proteasome
system may represent a new strategy to overcome chemotherapy
resistance (12). SMAD-specific E3
ubiquitin protein ligase 1 (SMURF1), a member of the NEDD4 family,
is a HECT-type E3 ubiquitin ligase that has an oncogenic role in
various types of cancer, such as colon cancer and pancreatic ductal
adenocarcinoma, through mediating ubiquitination (13).
Our previous study revealed that SMURF1 may be a
target gene of miR-424-5p (11) and
Nie et al (14) reported
that the HECT-type ubiquitin ligase SMURF1 could target the tumor
suppressor ING2 for ubiquitination and proteasome-dependent
degradation, further influencing the effect of ING2 on p53
activity, and suggested that SMURF1 may be involved in CDDP
resistance. High SMURF1 expression is often associated with
increased malignancy and poor prognosis in cancer (11,13). A
growing body of evidence has verified that SMURF1 promotes tumor
proliferation, migration and invasion both in vitro and
in vivo (15). With further
research, its role in drug resistance in tumors is gradually being
investigated (16,17).
In the present study, the effects of miR-424-5p on
SMURF1 expression in GC cells and its molecular role in regulating
chemotherapeutic sensitivity were investigated. The present study
aimed to gain a deeper insight into the molecular mechanisms
underlying CDDP resistance in GC and to provide more information on
targeted therapy for patients with GC.
Materials and methods
Tissue samples
Tumor tissues and adjacent tissues were collected
from 80 patients who underwent radical gastrectomy for GC at
Tianjin Medical University General Hospital (Tianjin, China). The
tissues were stored in liquid nitrogen for RNA extraction and were
fixed in 4% formalin at room temperature for 24 h and embedded in
paraffin for immunohistochemistry (IHC). Median patient age was 67
years (range, 39–85 years). The histopathological diagnosis and
grading were performed by two experienced pathologists, and
clinicopathological characteristics were recorded. Survival data,
such as overall survival (OS) and recurrence-free survival (RFS),
were also collected to perform survival analysis via the
Kaplan-Meier method and values were compared using the log-rank
test (18). Written informed
consent was obtained from all patients or their relatives for
specimen collection. Ethics approval for this project was granted
by the Investigation and Ethics Committee of Tianjin Medical
University General Hospital (approval no. IRB2020-KY-640).
GC cell lines
The human GC cell lines NCI-N87 (cat. no. CL-0169)
and MKN45 (cat. no. CL-0292), and the noncancerous gastric
epithelial cell line GES-1 (cat no. AY09234) were obtained from the
Laboratory of General Surgery, Tianjin Medical University General
Hospital which purchased them from Procell Life Science &
Technology Co., Ltd. (NCI-N87 and MKN45) and Ai-yan Biotechnology
Co. Ltd. (GES-1). The human GC cell lines HGC27 (cat. no. CL-0107)
and AGS (cat. no. CL-0022) were provided by Procell Life Science
& Technology Co., Ltd. Short tandem repeat analysis was
performed on these human cell lines for authentication. The GES-1,
NCI-N87, MKN45 and HGC27 cells were cultured in RPMI-1640 (cat no.
11875093), whereas AGS cells were cultured in F12 (cat no.
11765054) supplemented with 10% fetal bovine serum (cat no.
A5670701) and 1% penicillin/streptomycin (cat no. 15140122) (all
from Gibco; Thermo Fisher Scientific, Inc.) at 37°C in a humidified
atmosphere with 5% CO2.
Reverse transcription-quantitative
(RT-q) PCR
Total RNA was extracted using RNA Isolater Total RNA
Extraction Reagent (cat no. R401; Vazyme Biotech Co., Ltd.), and
was converted to cDNA with the HiScript® III 1st Strand
cDNA Synthesis Kit (cat no. R312; Vazyme Biotech Co., Ltd.) or the
miRcute Plus miRNA First-Strand cDNA Kit (cat no. KR211; Tiangen
Biotech Co., Ltd.), according to manufacturers' protocols. qPCR was
performed using ChamQ Universal SYBR RT-qPCR Master Mix (cat no.
Q711; Vazyme Biotech Co., Ltd.) or the miRcute Plus miRNA RT-qPCR
Kit (cat no. FP411; Tiangen Biotech Co., Ltd.) according to the
manufacturer's protocol. For the detection of miRNA, the qPCR
cycling conditions were as follows: Initial denaturation step at
95°C for 15 min, followed by 45 cycles at 94°C for 20 sec
(denaturation) and 60°C for 34 sec (annealing and extension),
concluding with a melting curve analysis. For mRNA detection, the
qPCR cycling conditions included an initial denaturation step at
95°C for 30 sec, followed by 40 cycles at 95°C for 20 sec
(denaturation) and 60°C for 30 sec (annealing and extension), also
concluding with a melting curve analysis. The reverse primers for
miRNA were provided in the qPCR kit. The copy number of miR-424-5p
in patient tissues was calculated via absolute quantification
methods (19), and relative
quantification was performed to evaluate the expression of miRNAs
and mRNAs in cell lines, which was analyzed using the comparative
2−ΔΔCq method (20). The
amplification curve and standard curve of miR-424-5p are shown in
Fig. S1. The primer sequences are
shown in Table SI, and GAPDH and
U6 were used as housekeeping genes for mRNA and miRNA expression,
respectively. All procedures were performed in triplicate.
Vector construction and cell
transfection
The miR-424-5p mimics and inhibitor, and the
negative controls (ncs) were constructed by Shanghai GenePharma
Co., Ltd. HGC27 and AGS cells were transfected in 6-well plates
with the miR-424-5p inhibitor, mimics or ncs (50 pmol/ml) using
Lipofectamine® 2000 (cat no. 11668019; Invitrogen;
Thermo Fisher Scientific, Inc.) once they reached 30–50%
confluence, following the manufacturer's instructions, at 37°C in
CO2 for 24 h. After 24 h, the cells were harvested for
subsequent experiments. The lentivirus used to knock down SMURF1
[short hairpin (sh)SMURF1] and its negative control (sh-nc; cat.
no. 20221126) was constructed using the
pCDH-CMV-MCS-EF1-copGFP-T2A-Puro vector (Shanghai GenePharma Co.,
Ltd.) as a backbone, whereas the SMURF1 overexpression lentivirus
(LV-SMURF1) and its negative control (LV-nc) were constructed using
the pLKO.1-copGFP-2A-PURO vector (Shanghai GenePharma Co., Ltd.).
All lentiviral constructs were produced by Shanghai GenePharma Co.,
Ltd. using the 3rd generation system. The AGS and HGC27 cells were
then infected with lentiviral particles (MOI=10) with the addition
of 5 µg/ml polybrene (cat no. H8761; Beijing Solarbio Science &
Technology Co., Ltd) for 8 h at 37°C in a humidified atmosphere
containing 5% CO2. The culture solution was fully
replaced at 24 h. The stable cell lines were selected with
puromycin (cat. no. HY-B1743; MedChemExpress) after 3 days. RT-qPCR
and western blotting were used to determine the transfection and
infection efficiencies. All of the aforementioned sequences are
presented in Table SII.
Colony formation assay
HGC27 and AGS cells transfected with miR-424-5p
inhibitors, mimics or ncs were seeded into 6-well plates at a
density of 1,000 cells/well, and were cultured at 37°C in a
humidified atmosphere containing 5% CO2, with the
culture media replaced every 3 days. Colony formation was assessed
after culturing the cells for 14 days. After washing the cells
three times with phosphate-buffered saline, the colonies were fixed
with 4% paraformaldehyde for 20 min and stained with 0.1% crystal
violet (cat no. G1063; Beijing Solarbio Science & Technology
Co., Ltd.) staining solution for 5 min at room temperature.
Formations with >50 cells were identified as colonies under a
dissecting microscope.
Wound healing assay
Cells were seeded into 6-well plates at a density of
2×105 cells/well and were allowed to attach until they
reached 80–90% confluence at 37°C in a humidified atmosphere with
5% CO2. Subsequently, a straight scratch was made using
a 20-µl pipette tip in the cell monolayer, and cell migration was
monitored and documented every 24 h under an inverted fluorescence
microscope. The migration of the cells was calculated by measuring
the distance covered by the cells at each time point via ImageJ
1.54g (National Institutes of Health). The cells were serum starved
for 12 h prior to the migration assay to negate the effect of cell
proliferation before migration, and then cultured with fresh medium
containing 1% fetal calf serum.
Transwell and invasion assays
Cells (5×104/well) were seeded into the
upper chambers of a 24-well Corning® 6.5 mm
Transwell® with 5.0 µm Pore Polycarbonate Membrane
Insert (cat. no. 3421; Corning, Inc.) to detect cell migration and
invasion. To assess cell invasion, the insert was pre-coated with
Matrigel (cat no. 354248; BD Biosciences) at 37°C for 1 h. In
addition, medium containing 10% fetal bovine serum was added to the
lower chambers. After incubation for 24 h at 37°C in a humidified
atmosphere containing 5% CO2, the migratory and invasive
cells were stained with 0.1% crystal violet at room temperature for
30 min, and images were captured under an inverted fluorescence
microscope. All assays were repeated at least three times.
Western blotting
Total protein was isolated from cells using RIPA
buffer (cat no. R0010; Beijing Solarbio Science & Technology
Co., Ltd.) supplemented with protease inhibitor (cat no. A32955;
Thermo Fisher Scientific, Inc.). After the protein concentration
was measured using the BCA assay, the protein samples were boiled
and ~15 µg protein was separated by SDS-PAGE on 10% gels and
transferred to PVDF membranes. After blocking non-specific binding
sites with 5% non-fat dry milk (cat. no. D8340; Beijing Solarbio
Science & Technology Co., Ltd.) at room temperature for 1 h.
The membranes were probed with antibodies against SMURF1 (1:1,000;
cat. no. 2174; Cell Signaling Technology, Inc.), caspase 3
(1:1,000; cat. no. 9662; Cell Signaling Technology, Inc.), ING2
(1:1,000; cat. no. 11560-1-AP; Proteintech Group, Inc.), p53
(1:5,000; cat. no. 10442-1-AP; Proteintech Group, Inc.) and β-actin
(1:20,000; cat. no. 66009-1-Ig; Proteintech Group, Inc.) overnight
at 4°C. After washing, the membranes were incubated with the HRP
goat anti-rabbit IgG (1:5,000; cat no. SA00001-2; Proteintech Group
Inc.) or HRP-conjugated goat anti-mouse IgG (1:5,000; cat no.
SA00001-1; Proteintech Group Inc.) at room temperature for 1 h. An
ECL substrate (cat. no. E411-04; Vazyme Biotech Co., Ltd.) was used
to visualize the protein bands and the data were analyzed using
ImageJ 1.54g.
IHC
GC and adjacent noncancerous tissues previously
preserved in 4% formalin were embedded, then the sections were
prepared to a thickness of 5 µm. Subsequently, sections were
permeabilized with 0.1% Triton X-100 (cat. no. T8200; Beijing
Solarbio Science & Technology Co., Ltd.) at room temperature
for 30 min, allowing antibodies to enter the cells, and endogenous
peroxidase was blocked with 3% hydrogen peroxide at room
temperature for 10 min. BSA (5%; cat. no. SW3015; Beijing Solarbio
Science & Technology Co., Ltd.) was used to block non-specific
binding sites at room temperature for 1 h. SMURF1 antibody (1:200;
cat no. ab57573; Abcam) was used for IHC staining, and sections
were incubated with this antibody overnight at 4°C. The horseradish
peroxidase-conjugated goat anti-mouse IgG (1:5,000; cat. no.
ZB2305; Beijing Zhongshan Jinqiao Biotechnology Co., Ltd.) was used
to incubate the sections at room temperature for 1 h. Chromogenic
detection was performed using DAB (cat no. DA1010; Beijing Solarbio
Science & Technology Co., Ltd.). Three fields were randomly
selected to determine the percentage of positive cells and the
staining intensity by light microscope. The expression grade of
SMURF1 was calculated by multiplying the positivity score by the
intensity score (21). Cells with
<10% staining were rated as 1, cells with 10–49% staining were
rated as 2, cells with 50–74% staining were rated as 3 and cells
with 75–100% staining were rated as 4. The staining color was
scored as light-yellow particles, 1; brown-yellow particles, 2; and
brown particles, 3. The final score was defined as staining number
score multiplied by staining color score. Patients were
subsequently divided into high expression level (grade >3) and
low expression level (grade ≤3) groups.
Dual-luciferase reporter assay
The miR-424-5p binding site in the 3′-UTR of SMURF1
was predicted using starBase v2.0 (http://starbase.sysu.edu.cn/). psiCHECK2-wild type
(WT)-SMURF1 and psiCHECK2-mutant-SMURF1 were synthesized by Hanbio
Biotechnology Co., Ltd. and were cotransfected with miR-424-5p
mimics or nc into 293T cells (cat. no. CL-0005; Procell Life
Science & Technology Co., Ltd) Lipofectamine 2000. After
culture for 48 h, the dual-luciferase system (cat no. E2920;
Promega Corporation) was used to measure the firefly luciferase
(F-Luc) and Renilla luciferase (R-Luc) values, and the
relative fluorescence values of firefly luciferase were calculated
as F-Luc/R-Luc.
CCK-8 assay
The cells were inoculated into 96-well plates (4,000
cells/well) (Corning, Inc.) for 24 h. Then, the cells were treated
with different concentrations (0, 0.005, 0.05, 0.5, 5, 50 and 500
µg/ml) of CDDP (cat. no. P4394; MilliporeSigma) for 48 h at 37°C in
a humidified atmosphere containing 5% CO2. The CCK-8
assay (cat. no. CA1210; Beijing Solarbio Science & Technology
Co., Ltd.) was used to measure cell viability and the half maximal
inhibitory concentration (IC50) was determined using
GraphPad Prism 8 software (Dotmatics).
Flow cytometry
The cells were divided into an experimental group
and a control group, and were seeded into 6-well plates
(1×105 cells/well; Corning Life Sciences) for 24 h. The
experimental group was treated with CDDP (10 µg/ml in AGS cells and
2 µg/ml in HGC27 cells) for 48 h at 37°C and the control group was
treated with DMSO for the same duration. Annexin V-FITC/7-AAD (cat
no. 40311ES60; Shanghai Yeasen Biotechnology Co., Ltd.) was used
for apoptosis analysis; the samples were stained with Annexin
V-FITC at room temperature for 5 min in the dark, after which,
7-AAD was added and the cells were immediately analyzed by flow
cytometry (BD FACSCanto II; BD Biosciences). The data were analyzed
using FlowJo (v10.8.1; FlowJo LLC) and GraphPad Prism 8 software.
The apoptosis rate was represented by the sum of the early
apoptotic rate and the late apoptotic rate.
Statistical analysis
SPSS version 26 (IBM Corp.) and GraphPad Prism 8
were used to analyze the data. The numerical data are presented as
the mean ± standard deviation of at least three experimental
repeats. The normality of the data was tested using the
Kolmogorov-Smirnov test, and the comparisons among groups were
analyzed using either unpaired or paired Student's t-test or
one-way ANOVA followed by Bonferroni's post hoc test.
Clinicopathological data were analyzed using the χ2 test
or Fisher's exact test. Correlations between variables were tested
using linear regression and Spearman rank tests. P<0.05 was
considered to indicate a statistically significant difference.
Results
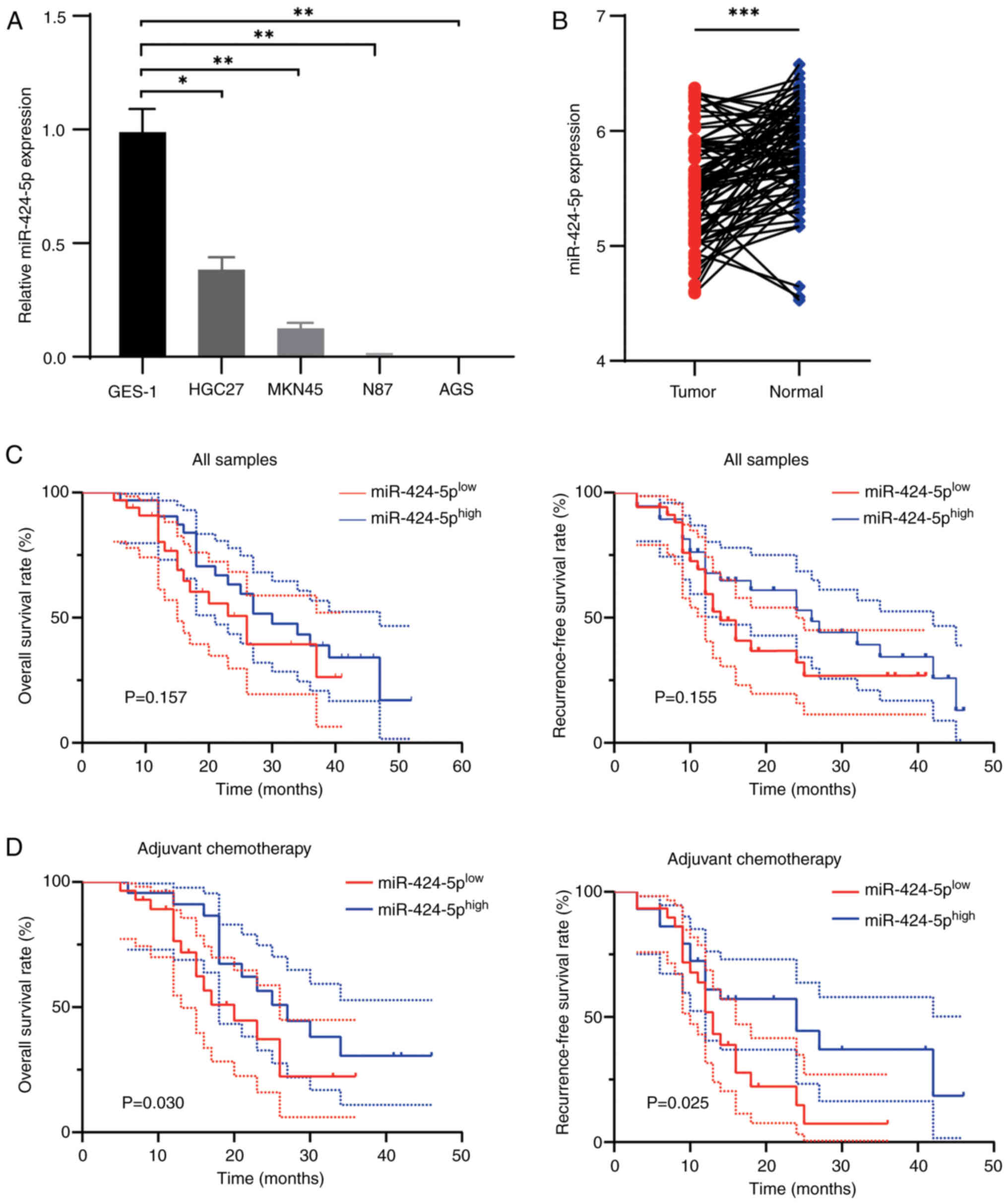
miR-424-5p is downregulated in human
GC tissues and cell lines
The expression of miR-424-5p in noncancerous gastric
epithelial cells (GES-1) and GC cell lines (HGC27, MKN45, NCI-N87
and AGS) was assessed using RT-qPCR. As shown in Fig. 1A, the expression of miR-424-5p in GC
cells was lower than that in GES-1 cells. In addition, 80 pairs of
GC tissues and adjacent noncancerous tissues were collected and the
absolute expression of miR-424-5p was assessed by miRNA RT-qPCR.
Compared with those in paired adjacent tissues, the expression
levels of miR-424-5p in human GC tissues were decreased (Fig. 1B). The samples were subsequently
divided into high- and low-expression groups on the basis of their
miR-424-5p expression levels, according to the median value. As
shown in Table I, owing to one
missing data point, 40 patients were included in the
high-miR-424-5p group and 39 patients were included in the
low-miR-424-5p group. Low miR-424-5p expression was associated with
increased pathological N stage. The survival analysis of all
samples revealed a weak association with poor prognostic outcome,
but the results were not significant (Fig. 1C). Subgroup analysis revealed that
lower expression of miR-424-5p was associated with a poor OS and
RFS in patients treated with adjuvant chemotherapy (Fig. 1D).
 | Table I.Association between expression levels
of miR-424-5p and SMURF1 and the clinicopathological
characteristics of patients. |
Table I.
Association between expression levels
of miR-424-5p and SMURF1 and the clinicopathological
characteristics of patients.
|
| miR-424-5p
expression | SMURF1
expression |
|---|
|
|
|
|
|---|
| Characteristic | Low | High group | P-value | Low | High group | P-value |
|---|
| Male sex, n
(%) | 26 (68.4) | 31 (75.6) | 0.476b | 32 (66.7) | 25 (80.6) | 0.176b |
| Aged ≥60 years, n
(%) | 33 (86.8) | 31 (75.6) | 0.203b | 41 (85.4) | 23 (74.2) | 0.214b |
| Location |
|
| 0.635b |
|
| 0.058b |
|
Upper | 5 (13.2) | 8 (19.5) |
| 7 (14.6) | 6 (19.4) |
|
|
Body | 9 (23.7) | 7 (17.1) |
| 6 (12.5) | 10 (32.3) |
|
|
Lower | 24 (63.2) | 26 (63.4) |
| 35 (72.9) | 15 (48.4) |
|
| Sized >3 cm, n
(%) | 20 (52.6) | 20 (48.8) | 0.732b | 21 (43.8) | 19 (61.3) | 0.128b |
| T grade |
|
| 0.767a |
|
| 0.312a |
| I | 3 (7.9) | 6 (14.6) |
| 7 (14.6) | 2 (6.5) |
|
| II | 5 (13.2) | 6 (14.6) |
| 8 (16.7) | 3 (9.7) |
|
|
III | 6 (15.8) | 7 (17.1) |
| 9 (18.8) | 4 (12.9) |
|
| IV | 24 (63.2) | 22 (53.7) |
| 24 (50.0) | 22 (71.0) |
|
| N grade |
|
| 0.009b |
|
| 0.336b |
| 0 | 6 (15.8) | 18 (43.9) |
| 18 (37.5) | 6 (19.4) |
|
| 1 | 5 (13.2) | 9 (22.0) |
| 8 (16.7) | 6 (19.4) |
|
| 2 | 9 (23.7) | 6 (14.6) |
| 9 (18.8) | 6 (19.4) |
|
| 3 | 18 (47.4) | 8 (19.5) |
| 13 (27.1) | 13 (41.9) |
|
| M grade |
|
| 0.140a |
|
| 0.075a |
| 0 | 34 (89.5) | 40 (97.6) |
| 47 (97.9) | 27 (87.1) |
|
| 1 | 4 (10.5) | 1 (2.4) |
| 1 (2.1) | 4 (12.9) |
|
| TNM stage |
|
| 0.058a |
|
| 0.020a |
| I | 4 (10.5) | 9 (22.0) |
| 10 (20.8) | 3 (9.7) |
|
| II | 5 (13.2) | 12 (29.3) |
| 14 (29.2) | 3 (9.7) |
|
|
III | 25 (65.8) | 19 (46.3) |
| 23 (47.9) | 21 (67.7) |
|
| IV | 4 (10.5) | 1 (2.4) |
| 1 (2.1) | 4 (12.9) |
|
| Histological
type |
|
| 0.115a |
|
| 0.141a |
|
Poorly | 21 (55.3) | 30 (73.2) |
| 27 (56.3) | 24 (77.4) |
|
|
Moderately | 12 (31.6) | 10 (24.4) |
| 17 (35.4) | 5 (16.1) |
|
|
Well | 5 (13.2) | 1 (2.4) |
| 4 (8.3) | 2 (6.5) |
|
miR-424-5p inhibits chemoresistance in
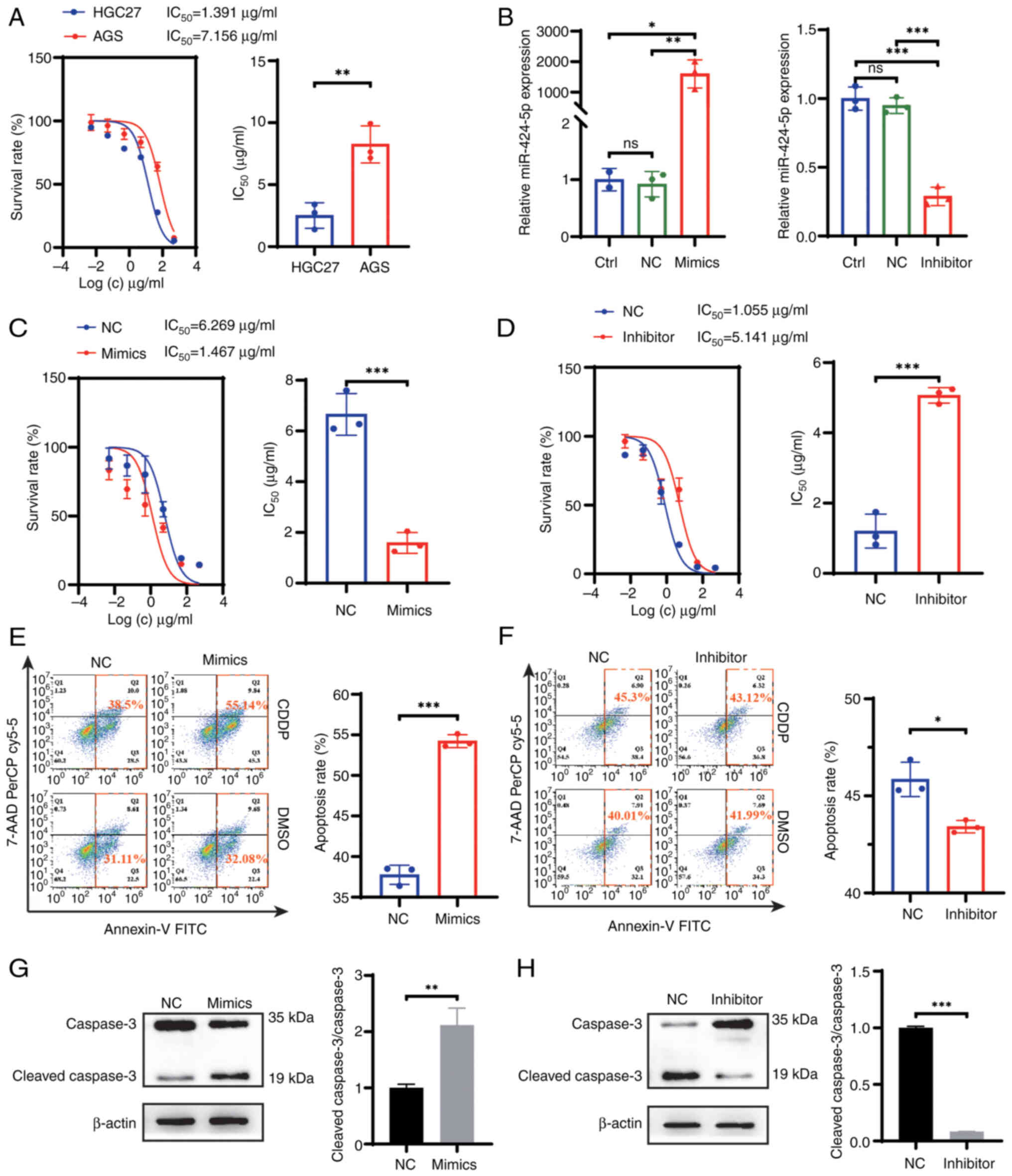
GC cells
To investigate the effect of miR-424-5p on
chemotherapy resistance in GC, the present study used AGS and HGC27
cells to verify the biological function of miR-424-5p. The results
of miR-424-5p expression and the IC50 values of CDDP in
AGS and HCG27 cells are shown in Fig.
2A. The IC50 value in AGS cells was greater than
that in HGC27 cells, which indicated that AGS cells may be more
resistant to CDDP than HGC27 cells. According to the miR-424-5p
expression status in GC cells shown in Fig. 1A, miR-424-5p exhibited the highest
expression in HGC27 cells, whereas its expression was the lowest in
AGS cells. Therefore, the HGC27 cells were selected for miR-424-5p
silencing, while AGS cells were chosen for miR-424-5p
overexpression in subsequent experiments. The miR-424-5p mimics and
inhibitor were constructed and transfected into AGS and HGC27
cells, respectively. RT-qPCR was performed to verify the
transfection efficiency in the cell lines (Fig. 2B). As expected, miR-424-5p was
significantly upregulated in AGS cells and significantly
downregulated in HGC27 cells post-transfection compared with that
in the nc groups. First, the colony formation, wound healing,
Transwell and invasion assays revealed that there was no
significant difference in cell proliferation, invasion and
migration after altering the expression of miR-424-5p (Fig. S2). A CCK-8 assay, flow cytometry
and western blotting were used to evaluate the effects of
miR-424-5p on CDDP resistance in GC cells. The CCK-8 assay results
revealed that the sensitivity of AGS cells transfected with the
miR-424-5p mimics to CDDP was increased compared with that of the
cells transfected with miR-nc, whereas HGC27 cells transfected with
the miR-424-5p inhibitor presented the opposite results (Fig. 2C and D). Following CDDP treatment,
flow cytometry revealed that the apoptosis rate of the miR-424-5p
mimics-transfected cells was greater than that of the
miR-nc-transfected cells (Fig. 2E).
By contrast, a lower percentage of apoptotic cells was detected in
the miR-424-5p inhibitor group (Fig.
2F). In addition, to evaluate apoptosis, the expression levels
of caspase 3 and cleaved caspase 3 were assessed. In contrast to
the nc groups, the cleaved caspase 3/caspase 3 rate was elevated in
the miR-424-5p mimics group and was reduced in the miR-424-5p
inhibitor group, which indicated that the miR-424-5p could promote
apoptosis following CDDP treatment (Fig. 2G and H).
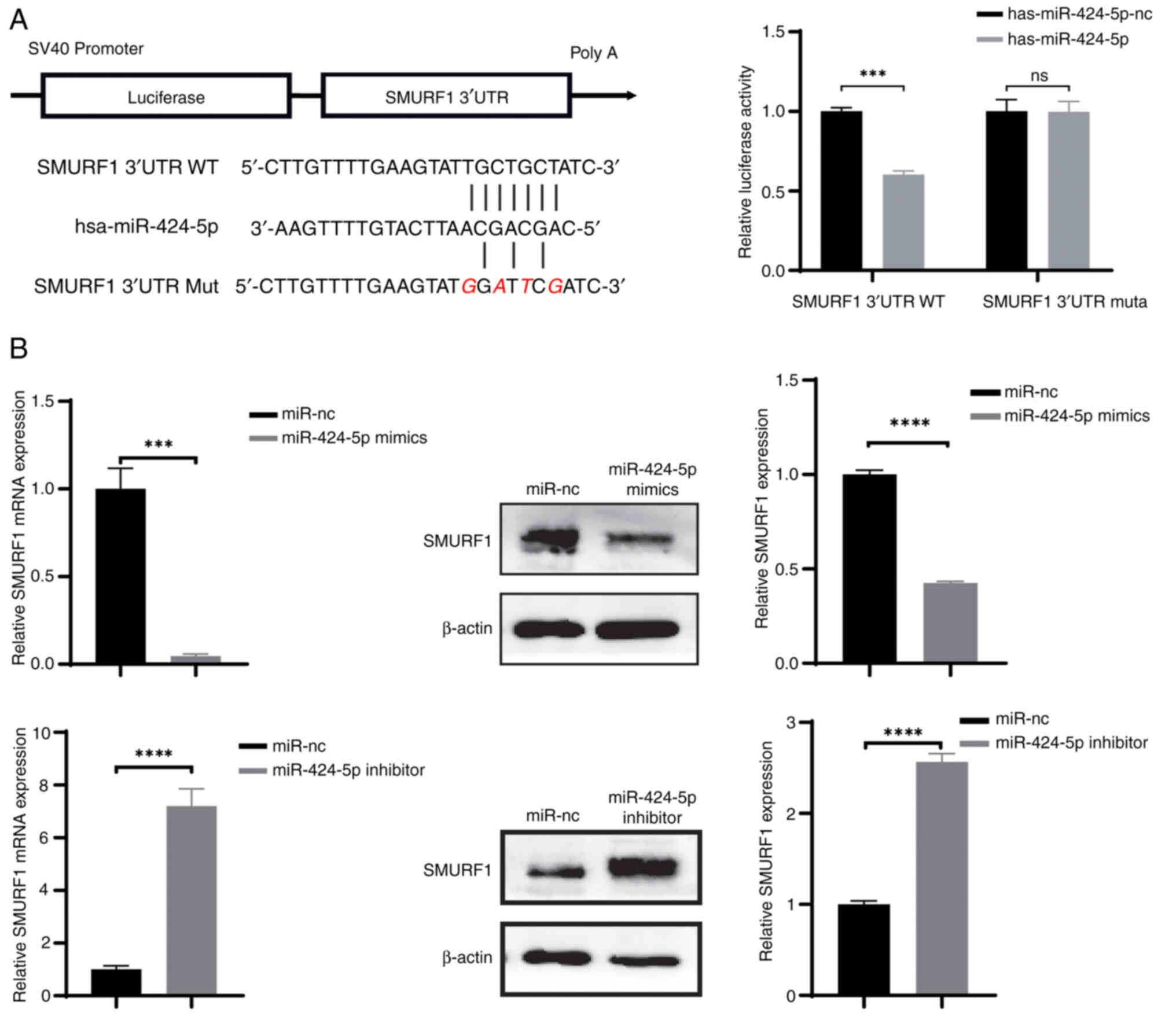
SMURF1 is a target gene of
miR-424-5p
In our previous study, SMURF1 was found to be
downstream of miR-424-5p (11). The
miR-424-5p binding site in the 3′-UTR of SMURF1 was predicted using
starBase v2.0. A dual-luciferase reporter assay revealed that the
luciferase activity in the SMURF1 3′UTR WT group was significantly
lower than that in the nc group (P<0.05; Fig. 3A). Thus, these results confirmed
that miR-424-5p regulated luciferase expression through this
binding site. To clarify the mechanism by which miR-424-5p
regulates SMURF1 in GC, RT-qPCR and western blotting were used to
assess the expression levels of SMURF1 in different treatment
groups. The qPCR results revealed that the mRNA expression levels
of SMURF1 in AGS cells transfected with miR-424-5p mimics were
significantly lower than those in the miR-nc group (Fig. 3B). By contrast, the mRNA expression
levels of SMURF1 in HGC27 cells transfected with the miR-424-5p
inhibitor were significantly increased. Consistently, the western
blotting results were the same as the RT-qPCR results. These
results suggested that miR-424-5p may suppress the expression of
SMURF1.
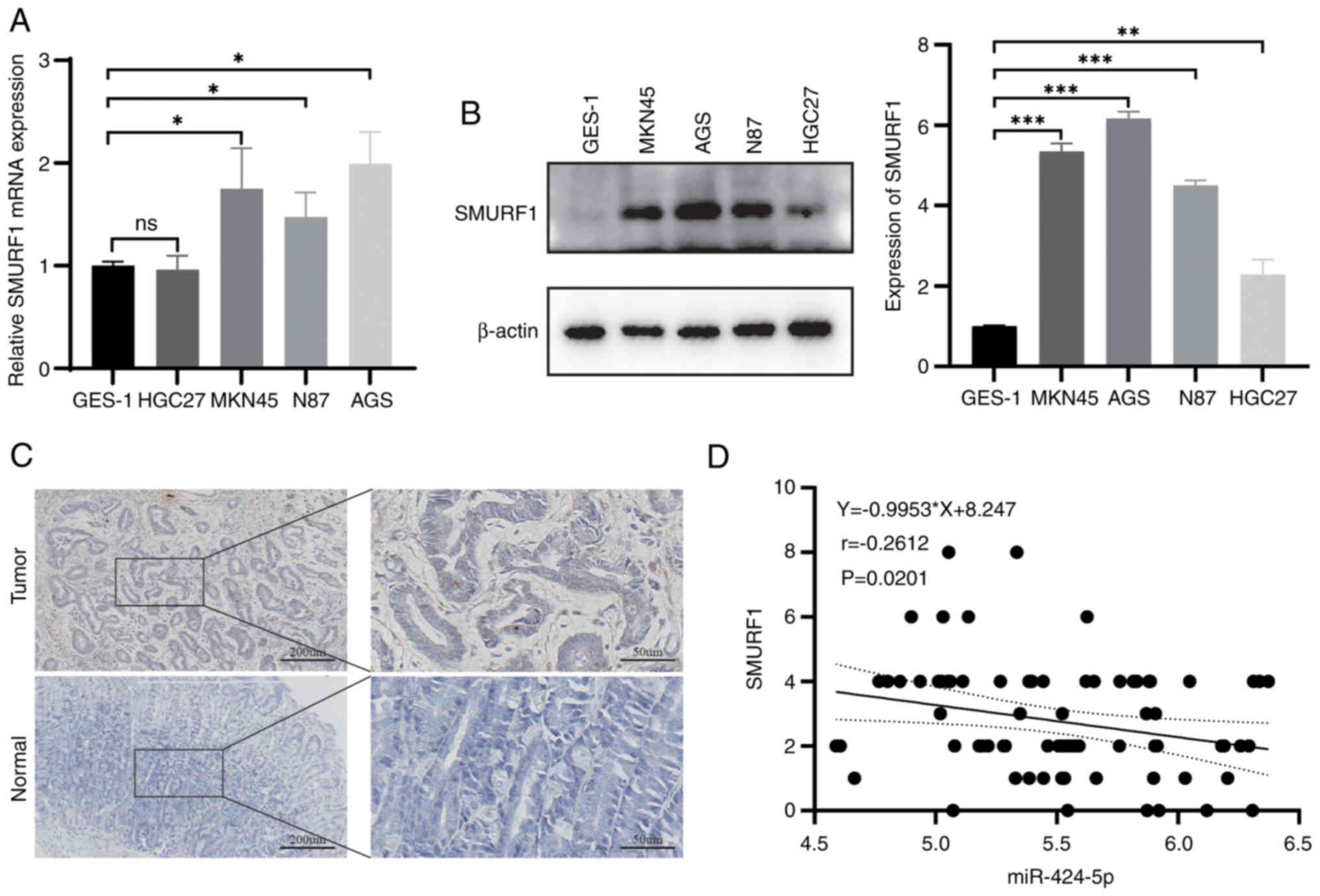
SMURF1 is upregulated in human GC
tissues and cell lines
The present study next assessed the expression of
SMURF1 in GC cell lines and GES-1 cells by RT-qPCR and western
blotting. As shown in Fig. 4A and
B, the mRNA expression levels of SMURF1 in GC cells, such as
NCI-N87, AGS and MKN45, were greater than those in GES-1 cells,
while SMURF1 protein levels were higher in NCI-N87, AGS, MKN45 and
HGC27 cells. IHC analysis revealed that SMURF1 was expressed at
higher levels in GC tissues than in adjacent tissues (Fig. 4C). Furthermore, miR-424-5p was
revealed to be negatively correlated with SMURF1 in GC tissues,
although the correlation was weak (Fig.
4D). In addition, the association between SMURF1 expression
levels and clinicopathological characteristics was analyzed. High
expression of SMURF1 was associated with increased
Tumor-Node-Metastasis stage (Table
I).
miR-424-5p suppresses SMURF1 protein
expression via mRNA degradation
The present study confirmed that miR-424-5p
regulated CDDP resistance in GC cells. To further confirm whether
miR-424-5p regulated drug resistance in GC cells by partially
inhibiting SMURF1 expression, AGS cells overexpressing SMURF1 and
HGC27 cells with SMURF1 knockdown were constructed, and the effects
of its overexpression and knockdown were verified by RT-qPCR and
western blotting (Fig. S3). In
addition, the expression of miR-424-5p was detected by RT-qPCR
following SMURF1 knockdown or overexpression and it was found that
the knockdown or overexpression of SMURF1 did not influence
miR-424-5p expression (Fig. S4).
Then, four groups of AGS cells, namely, the miR-nc + LV-nc,
miR-424-5p mimics + LV-nc, miR-nc + LV-SMURF1 and miR-424-5p mimics
+ LV-SMURF1 groups, were constructed via cotransfection. Similarly,
miR-nc + sh-nc, miR-424-5p inhibitor + sh-nc, miR-nc + sh-SMURF1
and miR-424-5p inhibitor + sh-SMURF1 HGC27 cells were constructed.
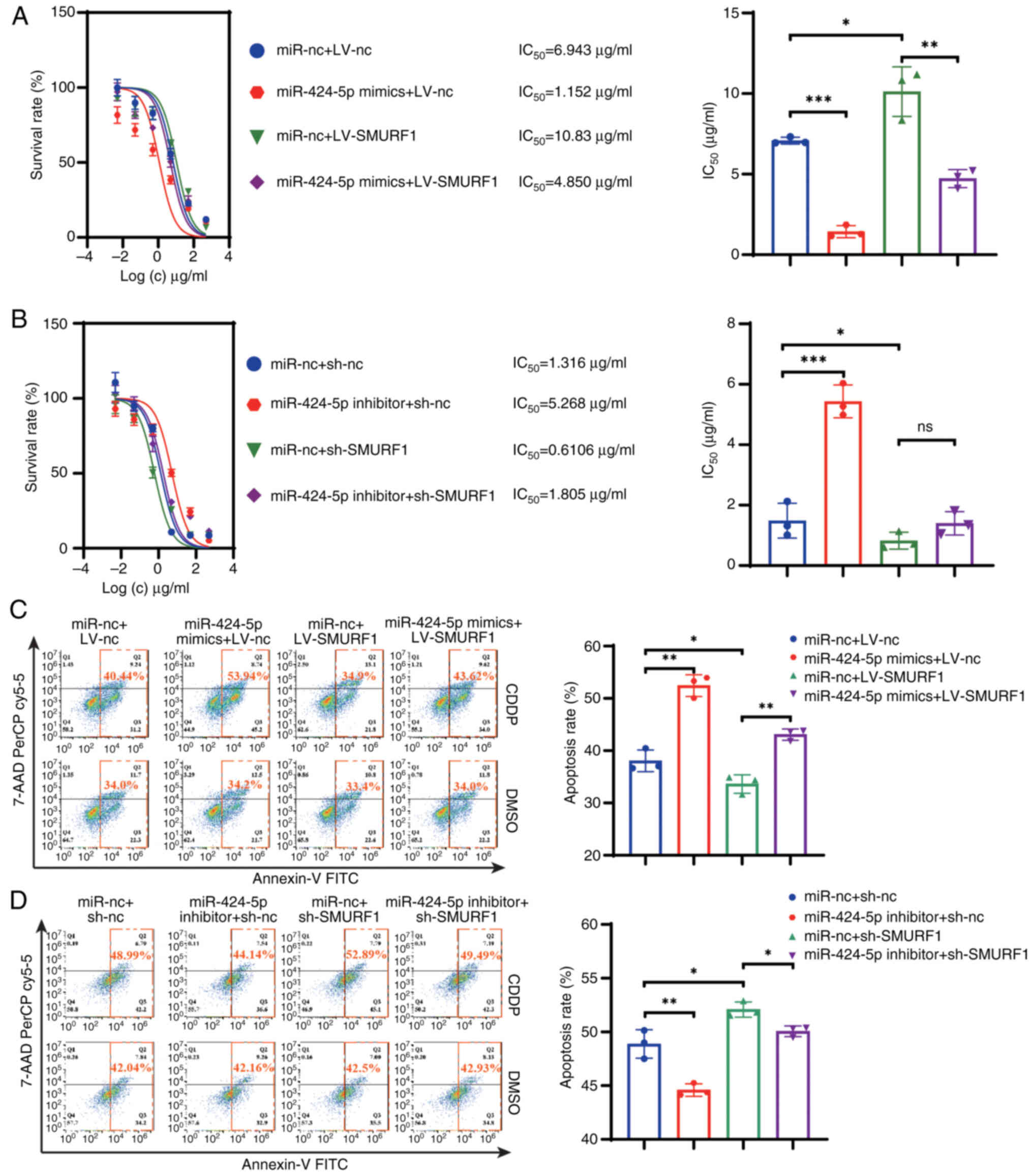
In terms of drug response, the CCK-8 assay results revealed that
CDDP resistance was increased after SMURF1 was upregulated in AGS
cells and decreased after SMURF1 was downregulated in HGC27 cells
(Fig. 5A and B). The CDDP
resistance induced by LV-SMURF1 could be reversed by overexpressing
miR-424-5p in GC cells. By contrast, the CDDP sensitivity induced
by sh-SMURF1 was not be altered by the miR-424-5p inhibitor. In
addition, flow cytometry demonstrated that the overexpression of
SMURF1 in GC cells reduced the number of apoptotic GC cells and
this effect was reversed by the transfection of miR-424-5p mimics
(Fig. 5C). Moreover, the miR-424-5p
inhibitor was found to reduce the apoptosis rate. By contrast,
sh-SMURF1 increased the apoptosis rate, and this could be slightly
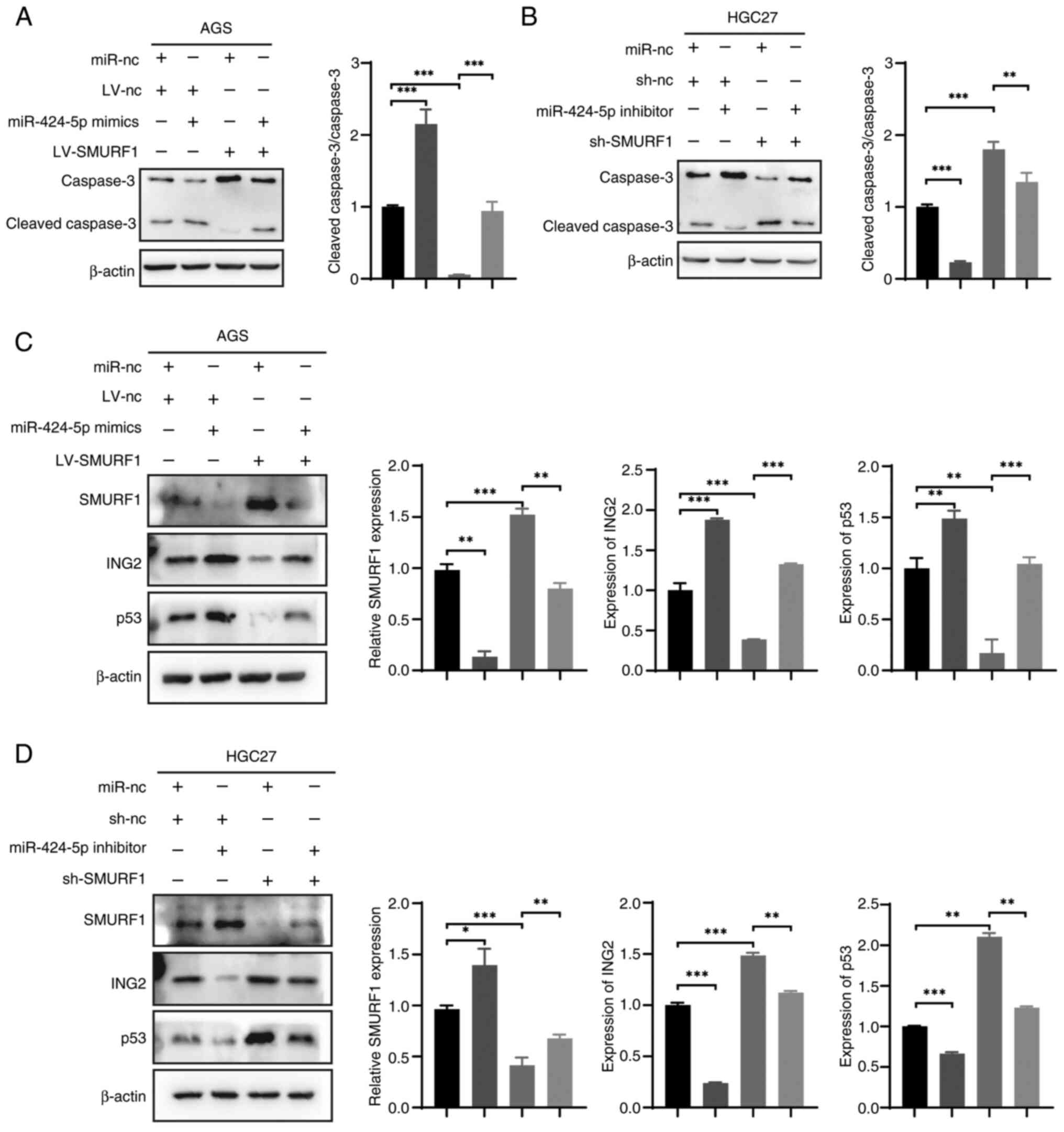
attenuated by the miR-424-5p inhibitor (Fig. 5D). The western blotting results
revealed a consistent trend in caspase 3 changes; the effects of
SMURF1 expression on apoptosis in response to CDDP treatment were
validated by the protein expression levels of cleaved caspase 3.
The results indicated that miR-424-5p mimics enhanced the cleaved
caspase 3/caspase 3 ratio. By contrast, SMURF1 overexpression
significantly reduced this ratio, an effect that could be reversed
by miR-424-5p mimics. Similarly, the miR-424-5p inhibitor decreased
the cleaved caspase 3/caspase 3 ratio, whereas sh-SMURF1 increased
the ratio, with this increase being modestly attenuated by the
miR-424-5p inhibitor (Fig. 6A and
B).
p53 serves an important role in chemosensitivity and
ING2 activates p53 in neoplasms. Moreover, SMURF1 interacts with
and targets ING2 for polyubiquitination and proteasomal degradation
(14). Western blotting data
revealed that the protein expression levels of ING2 and p53 were
partially inhibited or promoted by the miR-424-5p mimics or
inhibitor, respectively (Fig. 6C and
D). It was found that the expression of ING2 and p53 in AGS
cells transfected with LV-SMURF1 decreased and this change nearly
disappeared following transfection with the miR-424-5p mimics.
Similarly, the levels of ING2 and p53 were increased in HGC27 cells
transfected with sh-SMURF1 and were partially restored by
transfection with the miR-424-5p inhibitor. These results indicated
that the overexpression of miR-424-5p may inhibit the expression
levels of SMURF1 and increase the expression levels of p53.
Discussion
CDDP-based chemotherapy is currently the standard of
care for patients with GC; however, consequent chemotherapy
insensitivity and resistance may lead to tumor progression,
treatment failure and poor prognosis (22,23).
The development of treatment resistance is related to the
expression of multidrug resistance genes, and abnormalities in
apoptosis, the cell cycle and autophagy in tumor cells (24–26). A
large amount of evidence has suggested that miRNAs are involved in
the development of cancer and that they also serve important roles
in the resistance of GC to chemotherapy drugs and targeted therapy
drugs (27,28). Various oncogenic miRNAs, such as
miR-20a, miR-193a and miR-30, have been reported to promote the
resistance of GC cells to CDDP (27,29,30).
The present study found that miR-424-5p was expressed at low levels
in GC cells and inhibited CDDP resistance in GC cells through a
series of experiments. In addition, it was revealed via
clinicopathological data that low miR-424-5p expression was related
to the degree of malignancy of the tumor. These findings provided
new targets for the treatment of chemotherapy resistance in GC.
miRNAs are important noncoding RNAs that regulate
the expression of various genes (4,31).
Notably, miRNAs regulate apoptosis in colorectal tumors (32) and miR-424-5p has been shown to serve
roles in different diseases (33,34).
miR-424-5p is particularly relevant to tumor prognosis and has an
important role in early diagnosis and treatment. Studies have shown
that miR-424-5p can be used as a biomarker to predict the prognosis
of patients with pancreatic cancer, liver cancer or non-small cell
lung cancer (35–37). Accumulating evidence has also shown
that miR-424-5p is involved in regulating cellular functions, such
as cell proliferation, migration and invasion. For example,
miR-424-5p has been shown to inhibit epithelial-mesenchymal
transition in glioma by targeting KIF23 (38). In cervical cancer, miR-424-5p can
target KDM5B through the NOTCH1 signaling pathway to affect
proliferation, and regulate CHK1 to alter migration, invasion,
apoptosis and the cell cycle (39,40).
In addition, miR-424 can increase the sensitivity of ovarian cancer
cells to CDDP by inhibiting the expression of galectin-3 (41), while Geretto et al (42) reported that miR-424 may increase the
sensitivity of tumors to docetaxel. Moreover, miR-424 has been
demonstrated to block the PD-L1 immune checkpoint and reactivate
the T-cell immune response, enhancing immune-killing effects and
attenuating drug resistance (43).
Our previous analysis of the GEO database revealed that miR-424-5p
may regulate CDDP resistance in GC by targeting SMURF1 expression
(11). Therefore, the present study
confirmed this hypothesis using in vitro experiments. It
first studied the effects of miR-424-5p on cell proliferation,
migration and invasion, but the results were not statistically
significant. The present study demonstrated that the expression
levels of miR-424-5p in GC cells were negatively associated with
the IC50 of CDDP and that its sensitivity to CDDP was
affected by the overexpression or inhibition of intracellular
miR-424-5p expression, which could be restored by the
overexpression or knockdown of SMURF1.
In addition, the present study explored the specific
mechanism by which miR-424-5p affects CDDP resistance in GC cells
after SMURF1 is modulated. SMURF1, a ubiquitin ligase, was first
shown to regulate the Smad protein in the TGFβ/BMP signaling
pathway (44). Recently, in-depth
research on ubiquitination-mediated cancer progression and
development has been performed. The function of SMURF1 as a
potential protooncogene has also been identified. SMURF1 promotes
tumor metastasis and inhibits apoptosis in GC, prostate cancer and
ovarian cancer (15,45). In addition, in patient-derived
xenograft models, SMURF1 has been revealed to be associated with
sensitivity to CDDP chemotherapy in colorectal cancer (46). The present study provided further
evidence that the inhibition of SMURF1 may improve the efficacy of
chemotherapy in GC cells. In response to the inhibition of SMURF1
by the overexpression of miR-424-5p, the IC50 of CDDP in
GC cells decreased, whereas CDDP-induced apoptosis increased. A
previous study has shown that SMURF1 regulates apoptosis by
interacting with ING2 through the HECT domain (14). ING2 interacts with p53, enhances the
transcriptional activity of p53 and regulates cellular senescence,
apoptosis and the DNA damage response (47). The present study found that
miR-424-5p inhibited SMURF1 expression, which in turn affected the
expression of ING2 and p53, ultimately regulating CDDP resistance
in GC cells.
In summary, the present study demonstrated that
SMURF1 is a target of miR-424-5p, and contributes to regulating
cell survival and death by regulating apoptosis. In GC, miR-424-5p
inhibited SMURF1 expression by binding to the 3′-UTR of SMURF1,
which may increase p53 expression. p53 is an important regulator of
apoptosis in the CDDP-mediated damage response. Therefore,
miR-424-5p may increase the levels of p53 and thus increase
apoptosis and the sensitivity of GC cells to CDDP treatment.
However, a limitation of the present study was the absence of
mechanistic experiments in animal models to validate the regulatory
effects of the miR-424-5p/SMURF1/ING2/p53 axis on CDDP resistance.
In the future, the effects of miR-424-5p on the tumor immune
microenvironment will be explored. Subsequently, nanocarriers will
be developed for the targeted delivery of miR-424-5p to overcome
drug resistance in GC.
In conclusion, the present study revealed that
miR-424-5p regulates chemoresistance via the
miR-424-5p/SMURF1/ING2/p53 axis in GC. Thus, the development of
drugs that target miR-424-5p during tumor therapy may ultimately
improve the response of patients with GC to chemotherapeutic
agents.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science
Foundation of China (approval no. 82003301) and the Tianjin
Municipal Education Commission's research project (approval no.
2023KJ117).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
Conceptualization was performed by LL and WF, the
methodology was designed by DW, data analysis was performed by HC,
and formal analysis and investigation by DW. YY participated in the
acquisition and analysis of data. DW was responsible for writing,
reviewing and editing the manuscript and LL was responsible for
visualization. LL and WF confirm the authenticity of all the raw
data. All authors read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
The present study was conducted in accordance with
The Declaration of Helsinki and approved by the Ethics Committee of
Tianjin Medical University General Hospital (approval no.
IRB2020-KY-640). Written informed consent was obtained from all
patients or their relatives for specimen collection.
Patient consent for publication
Written informed consent was obtained from the
patients for their anonymized information to be published in this
article.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Smyth EC, Nilsson M, Grabsch HI, van
Grieken NC and Lordick F: Gastric cancer. Lancet. 396:635–648.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wei L, Sun J, Zhang N, Zheng Y, Wang X, Lv
L, Liu J, Xu Y, Shen Y and Yang M: Noncoding RNAs in gastric
cancer: Implications for drug resistance. Mol Cancer. 19:622020.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rupaimoole R and Slack FJ: MicroRNA
therapeutics: Towards a new era for the management of cancer and
other diseases. Nat Rev Drug Discov. 16:203–222. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang Q, Zhuang J, Deng Y, Yang L, Cao W,
Chen W, Lin T, Lv X, Yu H, Xue Y and Guo H: miR34a/GOLPH3 axis
abrogates urothelial bladder cancer chemoresistance via reduced
cancer stemness. Theranostics. 7:4777–4790. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Peng Y, Hu S, Zhang K, Wang Y, Rouzi M,
Zhou D and Yang R: Downregulation of microRNA-130a inhibits oral
squamous cell carcinoma proliferation and metastasis via the
hippo-YAP pathway. Cancer Manag Res. 13:4829–4840. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wiggins JF, Ruffino L, Kelnar K, Omotola
M, Patrawala L, Brown D and Bader AG: Development of a lung cancer
therapeutic based on the tumor suppressor microRNA-34. Cancer Res.
70:5923–5930. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu C, Kelnar K, Liu B, Chen X,
Calhoun-Davis T, Li H, Patrawala L, Yan H, Jeter C, Honorio S, et
al: The microRNA miR-34a inhibits prostate cancer stem cells and
metastasis by directly repressing CD44. Nat Med. 17:211–215. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Trang P, Wiggins JF, Daige CL, Cho C,
Omotola M, Brown D, Weidhaas JB, Bader AG and Slack FJ: Systemic
delivery of tumor suppressor microRNA mimics using a neutral lipid
emulsion inhibits lung tumors in mice. Mol Ther. 19:1116–1122.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Feng J, Lu SS, Xiao T, Huang W, Yi H, Zhu
W, Fan S, Feng XP, Li JY, Yu ZZ, et al: ANXA1 binds and stabilizes
EphA2 to promote nasopharyngeal carcinoma growth and metastasis.
Cancer Res. 80:4386–4398. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lu L, Wu M, Lu Y, Zhao Z, Liu T, Fu W and
Li W: MicroRNA-424 regulates cisplatin resistance of gastric cancer
by targeting SMURF1 based on GEO database and primary validation in
human gastric cancer tissues. Onco Targets Ther. 12:7623–7636.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Narayanan S, Cai CY, Assaraf YG, Guo HQ,
Cui Q, Wei L, Huang JJ, Ashby CR Jr and Chen ZS: Targeting the
ubiquitin-proteasome pathway to overcome anti-cancer drug
resistance. Drug Resist Updat. 48:1006632020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xia Q, Li Y, Han D and Dong L: SMURF1, a
promoter of tumor cell progression? Cancer Gene Ther. 28:551–565.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nie J, Liu L, Wu M, Xing G, He S, Yin Y,
Tian C, He F and Zhang L: HECT ubiquitin ligase Smurf1 targets the
tumor suppressor ING2 for ubiquitination and degradation. FEBS
Lett. 584:3005–3012. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fu L, Cui CP, Zhang X and Zhang L: The
functions and regulation of Smurfs in cancers. Semin Cancer Biol.
67:102–116. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xia Q, Zhang H, Zhang P, Li Y, Xu M, Li X,
Li X and Dong L: Oncogenic Smurf1 promotes PTEN wild-type
glioblastoma growth by mediating PTEN ubiquitylation. Oncogene.
39:5902–5915. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Khammanivong A, Gopalakrishnan R and
Dickerson EB: SMURF1 silencing diminishes a CD44-high cancer stem
cell-like population in head and neck squamous cell carcinoma. Mol
Cancer. 13:2602014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rich JT, Neely JG, Paniello RC, Voelker
CC, Nussenbaum B and Wang EW: A practical guide to understanding
Kaplan-Meier curves. Otolaryngol Head Neck Surg. 143:331–336. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Heid CA, Stevens J, Livak KJ and Williams
PM: Real time quantitative PCR. Genome Res. 6:986–894. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2-ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Guo Z, Zhang X, Zhu H, Zhong N, Luo X,
Zhang Y, Tu F, Zhong J, Wang X, He J and Huang L: TELO2 induced
progression of colorectal cancer by binding with RICTOR through
mTORC2. Oncol Rep. 45:523–534. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
An Y, Wang B, Wang X, Dong G, Jia J and
Yang Q: SIRT1 inhibits chemoresistance and cancer stemness of
gastric cancer by initiating an AMPK/FOXO3 positive feedback loop.
Cell Death Dis. 11:1152020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yang W, Ma J, Zhou W, Cao B, Zhou X, Yang
Z, Zhang H, Zhao Q, Fan D and Hong L: Molecular mechanisms and
theranostic potential of miRNAs in drug resistance of gastric
cancer. Expert Opin Ther Targets. 21:1063–1075. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Czabotar PE, Lessene G, Strasser A and
Adams JM: Control of apoptosis by the BCL-2 protein family:
Implications for physiology and therapy. Nat Rev Mol Cell Biol.
15:49–63. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hammond WA, Swaika A and Mody K:
Pharmacologic resistance in colorectal cancer: A review. Ther Adv
Med Oncol. 8:57–84. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nies AT, Magdy T, Schwab M and Zanger UM:
Role of ABC transporters in fluoropyrimidine-based chemotherapy
response. Adv Cancer Res. 125:217–243. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lee SD, Yu D, Lee DY, Shin HS, Jo JH and
Lee YC: Upregulated microRNA-193a-3p is responsible for cisplatin
resistance in CD44(+) gastric cancer cells. Cancer Sci.
110:662–673. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhu C, Huang Q and Zhu H: miR-383
inhibited the cell cycle progression of gastric cancer cells via
targeting cyclin E2. DNA Cell Biol. 38:849–856. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhu M, Zhou X, Du Y, Huang Z, Zhu J, Xu J,
Cheng G, Shu Y, Liu P, Zhu W and Wang T: miR-20a induces cisplatin
resistance of a human gastric cancer cell line via targeting CYLD.
Mol Med Rep. 14:1742–1750. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Du X, Liu B, Luan X, Cui Q and Li L:
miR-30 decreases multidrug resistance in human gastric cancer cells
by modulating cell autophagy. Exp Ther Med. 15:599–605.
2018.PubMed/NCBI
|
|
31
|
Pillai RS, Bhattacharyya SN and Filipowicz
W: Repression of protein synthesis by miRNAs: How many mechanisms?
Trends Cell Biol. 17:118–126. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang H: MicroRNAs and apoptosis in
colorectal cancer. Int J Mol Sci. 21:53532020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Min KH, Yang WM and Lee W: Saturated fatty
acids-induced miR-424-5p aggravates insulin resistance via
targeting insulin receptor in hepatocytes. Biochem Biophys Res
Commun. 503:1587–1593. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cheng D, Zhu C, Liang Y, Xing Y and Shi C:
MiR-424 overexpression protects alveolar epithelial cells from
LPS-induced apoptosis and inflammation by targeting FGF2 via the
NF-kappaB pathway. Life Sci. 242:1172132020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang Y, Lv Z, Fu J, Wang Z, Fan Z and Lei
T: Endogenous microRNA-424 predicts clinical outcome and its
inhibition acts as cancer suppressor in human non-small cell lung
cancer. Biomed Pharmacother. 89:208–214. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wu L, Yang F, Lin B, Chen X, Yin S, Zhang
F, Xie H, Zhou L and Zheng S: MicroRNA-424 expression predicts
tumor recurrence in patients with hepatocellular carcinoma
following liver transplantation. Oncol Lett. 15:9126–9132.
2018.PubMed/NCBI
|
|
37
|
Wang ZX, Deng TX and Ma Z: Identification
of a 4-miRNA signature as a potential prognostic biomarker for
pancreatic adenocarcinoma. J Cell Biochem. 120:16416–16426. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhao C, Wang XB, Zhang YH, Zhou YM, Yin Q
and Yao WC: MicroRNA-424 inhibits cell migration, invasion and
epithelial-mesenchymal transition in human glioma by targeting
KIF23 and functions as a novel prognostic predictor. Eur Rev Med
Pharmacol Sci. 22:6369–6378. 2018.PubMed/NCBI
|
|
39
|
Xu J, Li Y, Wang F, Cheng B, Ye F, Xie X,
Zhou C and Lu W: Suppressed miR-424 expression via upregulation of
target gene Chk1 contributes to the progression of cervical cancer.
Oncogene. 32:976–987. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhou Y, An Q, Guo RX, Qiao YH, Li LX,
Zhang XY and Zhao XL: miR-424-5p functions as an anti-oncogene in
cervical cancer cell growth by targeting KDM5B via the Notch
signaling pathway. Life Sci. 171:9–15. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Bieg D, Sypniewski D, Nowak E and Bednarek
I: MiR-424-3p suppresses galectin-3 expression and sensitizes
ovarian cancer cells to cisplatin. Arch Gynecol Obstet.
299:1077–1087. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Geretto M, Pulliero A, Rosano C, Zhabayeva
D, Bersimbaev R and Izzotti A: Resistance to cancer
chemotherapeutic drugs is determined by pivotal microRNA
regulators. Am J Cancer Res. 7:1350–1371. 2017.PubMed/NCBI
|
|
43
|
Xu S, Tao Z, Hai B, Liang H, Shi Y, Wang
T, Song W, Chen Y, OuYang J, Chen J, et al: miR-424(322) reverses
chemoresistance via T-cell immune response activation by blocking
the PD-L1 immune checkpoint. Nat Commun. 7:114062016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Song MK, Lee JH, Ryoo IG, Lee SH, Ku SK
and Kwak MK: Bardoxolone ameliorates TGF-beta1-associated renal
fibrosis through Nrf2/Smad7 elevation. Free Radic Biol Med.
138:33–42. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Tao Y, Sun C, Zhang T and Song Y: SMURF1
promotes the proliferation, migration and invasion of gastric
cancer cells. Oncol Rep. 38:1806–1814. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Guo J, Xu G, Mao C and Wei R: Low
expression of smurf1 enhances the chemosensitivity of human
colorectal cancer to gemcitabine and cisplatin in patient-derived
xenograft models. Transl Oncol. 13:1008042020. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Gozani O, Karuman P, Jones DR, Ivanov D,
Cha J, Lugovskoy AA, Baird CL, Zhu H, Field SJ, Lessnick SL, et al:
The PHD finger of the chromatin-associated protein ING2 functions
as a nuclear phosphoinositide receptor. Cell. 114:99–111. 2003.
View Article : Google Scholar : PubMed/NCBI
|