Introduction
Cancer of unknown primary (CUP) represents a subset
of metastatic malignancies. It is characterized by the inability to
identify the primary tumor site, despite comprehensive clinical
history taking, systematic physical examinations, imaging studies,
endoscopic evaluations, immunohistochemistry (IHC), genetic testing
and other diagnostic methods (1,2).
According to previous reports, CUP accounts for 2–10% of all newly
diagnosed malignancies from certain regions (3,4). With
advancements in diagnostic techniques, the incidence of CUP has now
decreased to 1–2% (5–7). Nonetheless, the diagnostic process for
CUP remains challenging, requiring meticulous efforts to uncover
any diagnostic clues and identify the primary lesion. The diagnosis
of CUP is often one of exclusion, aimed at minimizing misdiagnosis
and avoiding missed diagnoses of tumors with improved prognoses or
curative options.
Currently, the treatment of CUP primarily relies on
empirical chemotherapy, as targeted therapies tailored to specific
molecular characteristics are typically not feasible. Consequently,
patients with CUP generally have a poor prognosis, with reported
median survival times of <12 months (8). Thus, exploring the pathogenesis of CUP
and optimizing its diagnostic and therapeutic strategies are of
utmost importance for improving the outcomes of patients with
CUP.
Next-generation sequencing (NGS), also known as
second-generation sequencing or high-throughput sequencing, is a
DNA sequencing technology widely used in the diagnosis and
treatment guidance of malignant tumors. NGS testing enables the
identification of adverse prognostic factors such as drug
resistance and toxicity during cancer treatment (9). In particular, in non-small cell lung
cancer, meaningful therapeutic targets, such as EGFR mutations,
anaplastic lymphoma kinase (ALK) gene fusions, ROS-1 and C-MET,
have been established as the gold standard for selecting targeted
therapies, forming the diagnostic cornerstone for precision
treatment (10). Certain research
suggests that tumors originating from different tissues exhibit
distinct gene expression profiles that correspond to their tissue
of origin, and analyzing these gene expression profiles may help to
identify the tumor type (11).
However, there is currently limited data on the genetic testing of
CUP. Therefore, the present study aimed to evaluate the clinical
characteristics, IHC and genetic mutation status, as well as the
survival outcomes of patients with CUP.
Materials and methods
Study design and patients
The present retrospective study included patients
with CUP who attended the Oncology Department of the Fourth
Hospital of Hebei Medical University (Shijiazhuang, China) between
January 2009 and January 2021. Ethical approval was obtained from
the Ethics Committee of the Fourth Hospital of Hebei Medical
University (approval no. 2024KS052). As the present study was
retrospective, it was exempt from requiring informed consent from
the patients. To ensure the accuracy of the study results, the
correctness of the diagnosis of the included population is very
important, which needs rigorous diagnostic screening, and those who
did not have a clear pathological diagnosis and had not undergone
adequate whole-body imaging, including chest and abdominal CT and
cranial imaging examinations to locate the primary lesion, were
excluded. The population included in the present study were
patients diagnosed with CUP after adequate evaluation under
existing examination and laboratory testing techniques according to
European Society for Medical Oncology (ESMO) CUP guidelines
(12). The specific inclusion
criteria were as follows: i) Histopathologically confirmed
malignant tumors; ii) previous thorough evaluation by oncologists
following the diagnostic criteria outlined in the 2015 ESMO
clinical practice guidelines; and iii) complete clinical and
pathological data. The exclusion criteria were as follows: i) Age
of <18 years; ii) pathological diagnosis of mesenchymal-origin
malignant tumors, peritoneal malignant tumors, malignant melanomas,
hematological malignancies or benign tumors; and iii) presence of
comorbidities involving other malignant tumors or suspected primary
tumor sites.
Data collection
The observational indicators for the present study
included the clinical and pathological characteristics, treatment
regimens and genetic mutation spectrum of CUP, as well as treatment
outcomes. Tissues were fixed in formalin and embedded in wax
blocks. Paraffin sections (4 µm thick) were deparaffinized and
rehydrated, followed by treatment with 0.02 M EDTA buffer (pH 9.0)
and EDTA Antigen Retrieval Solution High-Pressure Heat Retrieval
(both from Gene Tech Biotechnology Co., Ltd.) for 2.5 min. IHC and
H&E staining were then performed according to a previously
published protocol (13).
Ki-67 protein expression reflects the proliferative
capacity of tumor cells (14,15).
Its expression level can be categorized into three groups: Low
(<30%; Fig. S1A), medium
(30–60%; Fig. S1B) and high
expression (≥60%; Fig. S1C).
Programmed death-ligand 1 (PD-L1) expression, including tumor
proportion score (TPS) or combined positive score (CPS), was
evaluated using methods such as immunohistochemistry (IHC) SP263
pharmDx (Roche Tissue Diagnostics) using the OptiView DAB IHC
detection kit, strictly following the manufacturer's instructions
on a benchmark XT automatic IHC (BenchMark ULTRA; Roche Tissue
Diagnostics), or IHC 22C3 pharmDx (Agilent Technologies, Inc.)
using the Dako Autostainer Link 48 IHC platform (Agilent
Technologies, Inc.) (16). Low
expression was defined as TPS or CPS <10% (Fig. S2A); medium expression as TPS or CPS
(10–50%) (Fig. S2B); and high
expression as TPS or CPS ≥50% (Fig.
S2C).
Patient treatment efficacy and survival follow-up
were assessed using the inpatient medical records system of the
hospital, outpatient visit records and telephone calls. The
follow-up cutoff date was in November 2023. Overall survival (OS)
was defined as the time from the date of surgery to the date of
death or the end of follow-up.
Statistical methods
Statistical analyses were performed using SPSS 23.0
(IBM Corp.). Categorical data are presented as percentages, and
descriptive analyses were performed using frequencies and rates.
Survival curves were plotted using the Kaplan-Meier method, and
differences in survival time were assessed using the log-rank test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Demographic and clinical
characteristics of patients with CUP
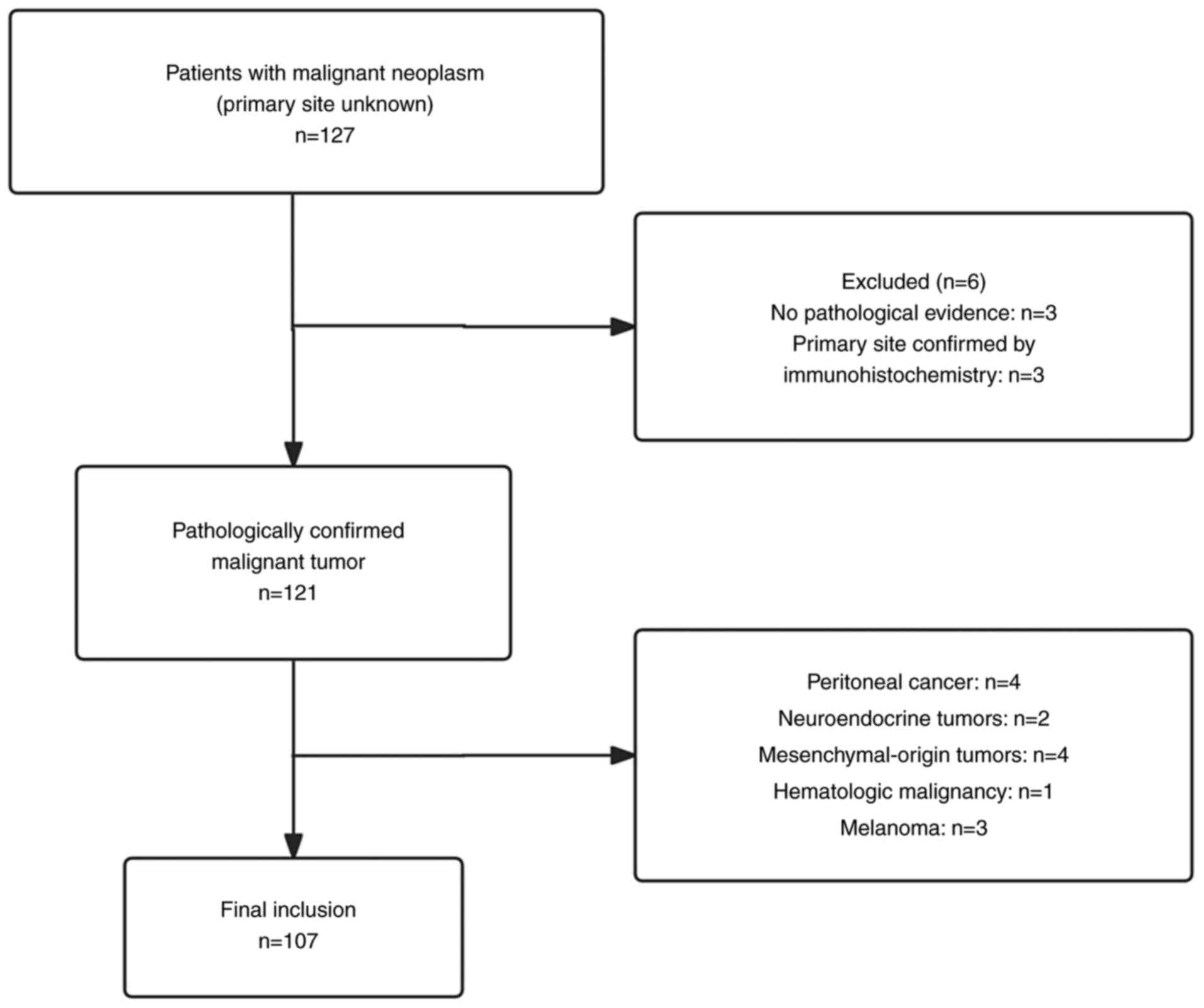
A total of 127 patients with CUP were initially
included in the present study; however, the following were
excluded: Patients without a clear pathological diagnostic basis
(n=3); patients with a confirmed primary lesion post-discharge
(n=3); patients with peritoneal cancer (n=4); patients with
mesenchymal-origin tumors (n=4); patients with neuroendocrine
tumors (n=2); patients with malignant melanoma (n=3); and patients
suspected of having a hematological malignancy (n=1). Ultimately,
107 patients were eligible for analysis (Fig. 1).
Among the 107 patients, 71 were male (66.4%). The
age at onset ranged from 21–76 years, with a mean age of 56.59
years. The median follow-up period was 48.8 months. The most common
sources of pathological specimens were superficial lymph nodes or
superficial non-visceral tissues (n=78; 72.9%). Adenocarcinoma was
the most prevalent pathological type, accounting for 41 cases
(38.3%), followed by squamous cell carcinoma (n=34; 31.8%) and
neuroendocrine carcinoma (n=18; 16.8%). PET-CT scans were performed
in 63 cases (58.9%) to aid in identifying the primary lesion, and
31 patients (29.0%) were found to have visceral metastases based on
imaging findings. Baseline characteristics of the patients in the
present study are presented in Table
I.
 | Table I.Baseline characteristics of the
patients in the present study (n=107). |
Table I.
Baseline characteristics of the
patients in the present study (n=107).
| Characteristic | n (%) |
|---|
| Sex |
|
| Male | 71 (66.4) |
|
Female | 36 (33.6) |
| Age, years |
|
| ≤40 | 10 (9.3) |
| >40
and ≤65 | 75 (70.1) |
|
>65 | 22 (20.6) |
| Pathological
source |
|
| Lymph
nodes and superficial non-visceral organs | 78 (72.9) |
| Visceral
organs | 16 (15.0) |
| Bone | 9 (8.4) |
| Malignant
pleural effusion | 4 (3.7) |
| Pathological
type |
|
|
Adenocarcinoma | 41 (38.3) |
| Squamous
cell carcinoma | 34 (31.8) |
|
Neuroendocrine carcinoma | 18 (16.8) |
|
Other | 14 (13.1) |
| PET-CT
examination |
|
| Yes | 63 (58.9) |
| No | 44 (41.1) |
| Visceral
metastasis |
|
| Yes | 31 (29.0) |
| No | 76 (71.0) |
Among the 107 patients, 31 (30.0%) received local
treatment, whilst 101 (94.4%) received systemic treatment.
Specifically, 56 patients (52.3%) underwent chemotherapy alone, and
20 patients (18.7%) received a combination of chemotherapy and
immunotherapy, whilst 14 (13.1%) underwent chemotherapy and
anti-angiogenic therapy. A total of two patients received a
combination of chemotherapy, anti-angiogenic drugs and immune
checkpoint inhibitors as first-line treatment, whilst three
patients (2.8%) received a combination of chemotherapy and targeted
therapy as first-line treatment. Additionally, six patients (5.6%)
did not receive any chemotherapy (Table II). Among these patients who did
not undergo chemotherapy, two patients with ALK fusion identified
by genetic testing received oral crizotinib, 2 patients received
anti-angiogenic drugs combined with immune checkpoint inhibitors
and 2 patients were treated with multi-target small molecule
anti-angiogenic agents, such as regorafenib and apatinib.
 | Table II.Treatment modalities of the patients
in the present study (n=107). |
Table II.
Treatment modalities of the patients
in the present study (n=107).
| Treatment
modality | n (%) |
|---|
| Local treatment |
|
| None | 76 (71.0) |
|
Radiotherapy | 9 (8.4) |
|
Surgery | 15 (14.0) |
| Surgery
+ radiotherapy | 4 (3.8) |
| Other
local treatments | 3 (2.8) |
| Systemic
treatment |
|
|
None | 6 (5.6) |
|
Chemotherapy | 56 (52.3) |
|
Chemotherapy +
immunotherapy | 20 (18.7) |
|
Chemotherapy + anti-angiogenic
therapy | 14 (13.1) |
|
Chemotherapy + anti-angiogenic
therapy + immunotherapy | 2 (1.9) |
|
Chemotherapy + targeted
therapy | 3 (2.8) |
| No
chemotherapy regimen | 6 (5.6) |
Molecular testing
Among all the included patients, 76 (71.0%)
underwent Ki-67 testing, with 10 (9.4%) demonstrating low
expression, 33 (30.8%) demonstrating medium expression, and 33
(30.8%) demonstrating high expression. PD-L1 testing was performed
in 21 patients (19.6%), with 11 (10.2%) showing low expression, 5
(4.7%) showing medium expression, and 5 (4.7%) showing high
expression. Besides Ki-67 and PD-L1 testing, 37 patients underwent
genetic testing (Table III). A
total of four cases (10.8%) showed no mutations. The most
frequently observed mutations were in tumor protein P53 (TP53;
n=8), human epidermal growth factor receptor 2 (HER-2/ERBB2; n=7)
and SWI/SNF Related BAF Chromatin Remodeling Complex (SMARCA/B;
n=6). Other mutations observed in >1 patient but <15% of the
patients are detailed in Table
IV.
 | Table III.Molecular diagnostic features of the
patients in the present study (n=107). |
Table III.
Molecular diagnostic features of the
patients in the present study (n=107).
| Molecular
diagnostic feature | n (%) |
|---|
| Genetic
testing |
|
|
Yes | 37 (34.6) |
| No | 70 (65.4) |
| Ki-67 expression,
% |
|
|
≥60 | 33 (30.8) |
| ≥30 and
<60 | 33 (30.8) |
|
<30 | 10 (9.4) |
| Not
tested | 31 (29.0) |
| PD-L1 expression,
% |
|
|
<10 | 11 (10.2) |
| ≥10 and
<50 | 5 (4.7) |
|
≥50 | 5 (4.7) |
| Not
tested | 86 (80.4) |
 | Table IV.Spectrum of gene mutations in the
present study (n=37). |
Table IV.
Spectrum of gene mutations in the
present study (n=37).
| Gene mutation | n (%) |
|---|
| TP53 | 8 (21.6) |
| HER-2/ERBB2 | 7 (18.9) |
| SMARCA/SMARCB | 6 (16.2) |
| RAS | 5 (13.5) |
| BRAF | 4 (10.8) |
| CDK | 4 (10.8) |
| NTRK | 2 (5.4) |
| NF | 3 (8.1) |
| MET | 2 (5.4) |
| JAK2 | 2 (5.4) |
| NOTCH1 | 2 (5.4) |
| PIK3CIB | 2 (5.4) |
| POLE | 2 (5.4) |
| PTEN | 2 (5.4) |
| RET | 2 (5.4) |
| RB1 | 2 (5.4) |
| TNF | 2 (5.4) |
| STK11 | 2 (5.4) |
Survival analysis
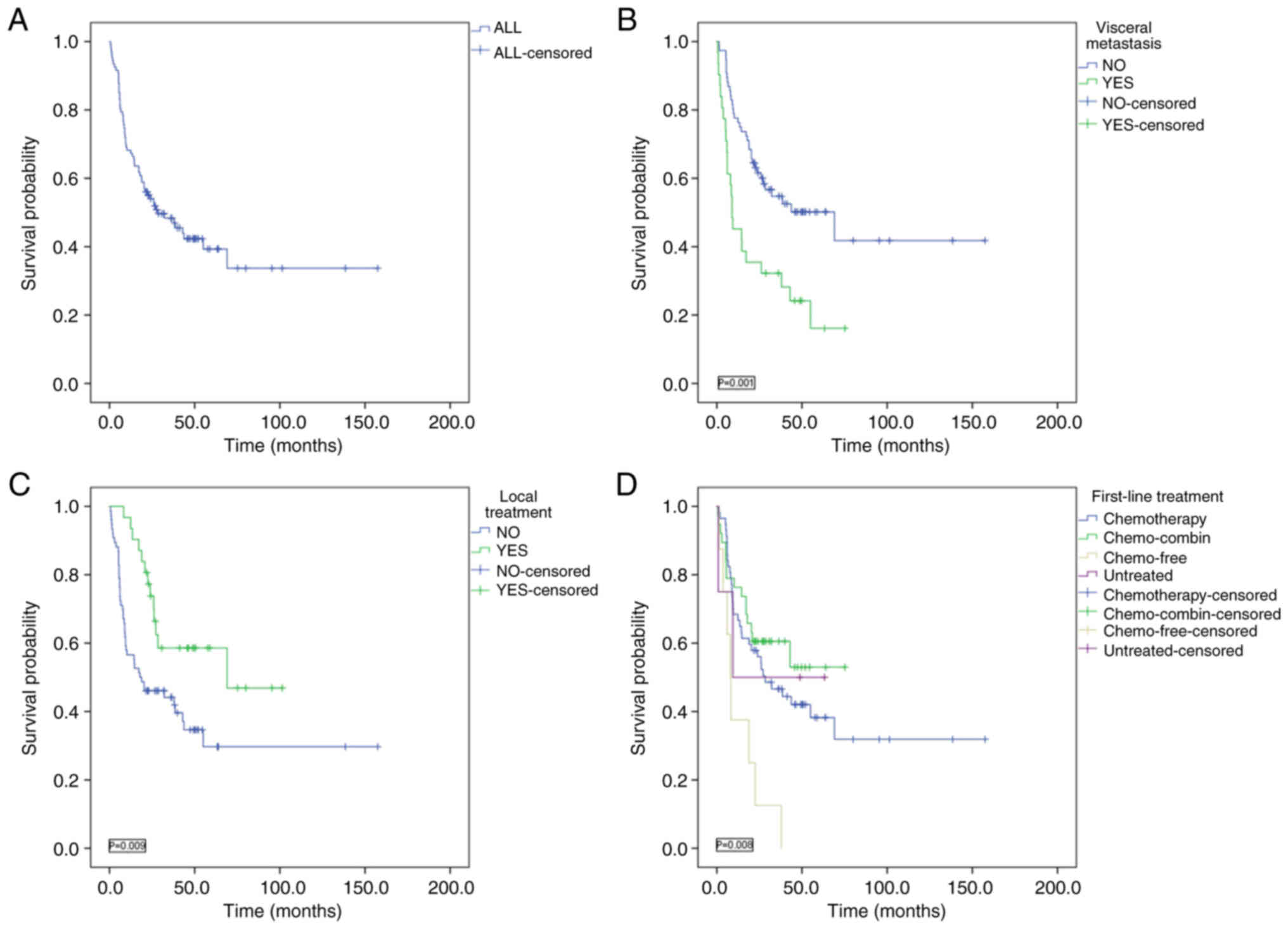
Among the 107 patients, the median OS was 28.4
months, with 1-, 2-, 3- and 4-year OS rates of 68.2, 54.1, 48.4 and
42.3%, respectively. Patients with visceral metastasis had a
significantly shorter median OS compared with those without
visceral metastasis (8.9 vs. 69 months; P=0.001). According to the
stratified analysis of whether local therapy was applied, the
median OS of patients with local treatment was 69 months, and that
of patients without local treatment was 17.9 months (P=0.009). The
survival time of patients given local treatment was significantly
longer compared with those without local treatment. Patients were
stratified into the following four groups based on first-line
treatment: i) Chemotherapy alone; ii) chemotherapy combined with
immune checkpoint inhibitors, anti-angiogenic drugs or targeted
therapies; iii) single-agent targeted therapy or surgery without
chemotherapy; and iv) untreated. The median OS times for these
groups were 28.4 months, not reached, 8 months and 9.4 months,
respectively, with significant differences in survival observed
among the four groups (P=0.008; Fig.
2).
Discussion
The findings of the present study indicate that the
survival outcomes of patients with CUP differ based on the presence
of visceral metastasis and the administered treatment modalities.
Systemic therapies, particularly those combining chemotherapy with
immune checkpoint inhibitors, anti-angiogenic drugs or targeted
therapies, show promise in improving prognosis.
Currently, advancements in imaging technologies,
improved tissue acquisition methods for pathology and progress in
molecular genetic testing have led to a more rigorous diagnostic
approach for CUP. In the present study, PET-CT scans were performed
on 58.9% and aggressive IHC and genetic testing accounting for
one-third of patients to assist in diagnosing the primary lesion,
but the diagnosis of the primary lesion was still not definitive.
Furthermore, the present study explored the current methods for the
detection of primary lesions and the reduction in the misdiagnosis
rate of metastatic CUP lesions. However, despite these
advancements, determining the primary site remains challenging. The
following factors may contribute to this issue: i) Metastasis to
distant sites may cause the primary tumor to regress or disappear,
making it difficult to identify, with 20–50% of patients failing to
have a primary tumor identified even upon autopsy; ii) current
detection methods may be limited by inadequate sampling or by the
removal of the primary tumor during the sampling process; iii)
certain primary tumors may be small or slow-growing, rendering them
undetectable through current imaging and detection technologies;
iv) the primary tumor may have been eradicated by the immune
system; and v) metastatic tumor cells may undergo phenotypic
changes, causing them to appear distinct from the primary tumor
(such as, poorly differentiated, undifferentiated or heterogeneous)
(17). This is consistent with the
findings of the present study, as nearly 60% of patients underwent
PET-CT examination but still failed to have their primary tumor
site identified.
Although IHC is frequently used to predict the
primary site in CUP, it is not typically considered a treatment
guideline due to the inherent ambiguity of pathology reports, which
often employ terms such as ‘favor’ or ‘consistent with’ rather than
providing a definitive diagnosis (18–20).
In a cohort of 252 patients with CUP, the Cancer TYPE ID assay
successfully predicted the primary site in 98% of cases (11). However, like IHC, the accuracy of
these predictions is difficult to ascertain due to the lack of
identified anatomical primary sites. Nevertheless, in a subset of
24 patients with CUP with primary sites identified months after
diagnosis, the Molecular Cancer Classifier Assay correctly
predicted the primary site in 75% of cases, provided that
sufficient tissue for analysis was available (13,21).
CUP often presents with widespread metastases, most commonly
involving lymph nodes, lungs, bones and liver (22). In the present study, most specimens
were obtained from lymph nodes, followed by visceral organs, bone
tissue and pleural effusion.
Apart from the primary tumor site, the pathological
type of CUP is another critical factor influencing treatment
decisions. A previous study indicated that adenocarcinomas,
squamous cell carcinomas, neuroendocrine carcinomas and poorly
differentiated carcinomas accounted for 60, 5, 2 and 30% of all
pathological types, respectively. Another study by Meijer et
al (23) reported that
adenocarcinoma represented ~41% of all CUP cases. In the present
study, adenocarcinoma, squamous cell carcinoma and neuroendocrine
carcinoma constituted 38.3, 31.8 and 16.8% of the CUP cases,
respectively. Given the unknown primary site, applying limited
tissue specimens to refine IHC and genetic testing at key
diagnostic points is essential for guiding clinical treatment
decisions (24).
Regarding genetic testing, the present study
identified TP53, HER-2 and SMARCA as the most commonly mutated
genes. The TP53 mutation, known for its association with cancer, is
observed in carcinomas originating from several tissues, including
lung, colorectal and gynecological cancers. Consequently, TP53
detection generally lacks specificity in determining the primary
site (25). Other studies have also
reported TP53 as the most frequently mutated gene in CUP. Bochtler
et al (26) reported a TP53
mutation rate of 49.6%, whereas Gerard et al (27) reported an even higher positive rate
of 87.5%. These discrepancies highlight differences in baseline
characteristics between the cohort in the present study and the
aforementioned populations, although the underlying cause of this
variation requires further analysis. Median survival varied by
histological type, with squamous cell carcinoma demonstrating the
longest median survival at 25.1 months and poorly differentiated
carcinoma the shortest at 3.0 months. Survival rates for each
histological type were generally consistent across sex and physical
status, with women showing higher survival rates than men for
well-differentiated and moderately differentiated adenocarcinomas.
Younger patients aged 18–64 years also tended to have longer
survival compared with those aged ≥65 years (28).
The present study has several limitations. First, it
is a single-center study with a small sample size of only 107
patients; thus, the findings should be validated in larger
multi-center cohorts. Second, not all patients underwent genetic
testing, leading to certain gene mutations being identified in only
one or two patients, indicating the need for a larger sample size.
Third, the data from the PD-L1 expression assay and counting
criteria are inconsistent and the detection rate is low in the
observed population, and there was a variation in IHC protocols
used for different individuals from the cohort, so the data may be
biased. Finally, the follow-up period was not long enough to
capture long-term outcomes. Future research should focus on
expanding the sample size, extending the follow-up duration and
ensuring comprehensive genetic testing and PD-L1 for all patients
to enhance the robustness of the findings.
In conclusion, the results of the present study
support the use of NGS in patients with CUP. The findings also
demonstrate the feasibility of integrating genomic analysis with
diagnostic histopathology and IHC in a community practice setting.
Future research should consider combining diagnostic algorithms
with genomic analysis to better characterize unknown primary
cancers.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present work was supported by the National Natural Science
Foundation of China (grant no. 82103497) and the Medical Science
Research Project of Hebei (grant no. 20210832).
Availability of data and materials
The data of genetic testing reports generated in the
present study may be found in the figshare database under accession
no. 10.6084/m9.figshare.28012967 or at the following URL:
https://figshare.com/s/6111ff335415093eb404. All other
data generated in the present study may be requested from the
corresponding author.
Authors' contributions
MM and BG performed the experiments and participated
in data collection. PJ and JB drafted the manuscript. JW and QD
performed the statistical analysis and contributed to the study
design. SW, YX, FZ and MH participated in data acquisition,
analysis or interpretation. PZ and JJ were responsible for tissue
management and preparation. MM and BG confirm the authenticity of
all the raw data. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the Fourth Hospital of Hebei Medical University
(approval no. 2024KS052). Informed consent was not required due to
the retrospective nature of the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Levi F, Te VC, Erler G, Randimbison L and
La Vecchia C: Epidemiology of unknown primary tumours. Eur J
Cancer. 38:1810–1812. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lee MS and Sanoff HK: Cancer of unknown
primary. BMJ. 371:m40502020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pavlidis N, Briasoulis E, Hainsworth J and
Greco FA: Diagnostic and therapeutic management of cancer of an
unknown primary. Eur J Cancer. 39:1990–2005. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pavlidis N and Fizazi K: Carcinoma of
unknown primary (CUP). Crit Rev Oncol Hematol. 69:271–278. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Brustugun OT and Helland A: Rapid
reduction in the incidence of cancer of unknown primary. A
population-based study. Acta Oncol. 53:134–137. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Boo YK, Park D, Lim J, Lim HS and Won YJ:
Descriptive epidemiology of cancer of unknown primary in South
Korea, 1999–2017. Cancer Epidemiol. 74:1020002021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rassy E and Pavlidis N: The currently
declining incidence of cancer of unknown primary. Cancer Epidemiol.
61:139–141. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ren M, Cai X, Jia L, Bai Q, Zhu X, Hu X,
Wang Q, Luo Z and Zhou X: Comprehensive analysis of cancer of
unknown primary and recommendation of a histological and
immunohistochemical diagnostic strategy from China. BMC Cancer.
23:11752023. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sunami K, Takahashi H, Tsuchihara K,
Takeda M, Suzuki T, Naito Y, Sakai K, Dosaka-Akita H, Ishioka C,
Kodera Y, et al: Clinical practice guidance for next-generation
sequencing in cancer diagnosis and treatment (Edition 1.0). Cancer
Sci. 109:2980–2985. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Thai AA, Solomon BJ, Sequist LV, Gainor JF
and Heist RS: Lung cancer. Lancet. 398:535–554. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Erlander MG, Ma XJ, Kesty NC, Bao L,
Salunga R and Schnabel CA: Performance and clinical evaluation of
the 92-gene real-time PCR assay for tumor classification. J Mol
Diagn. 13:493–503. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fizazi K, Greco FA, Pavlidis N and
Pentheroudakis G; ESMO Guidelines Working Group, : Cancers of
unknown primary site: ESMO Clinical Practice Guidelines for
diagnosis, treatment and follow-up. Ann Oncol. 26 Suppl
5:vi33–vi38. 2011.
|
|
13
|
Greco FA, Spigel DR, Yardley DA, Erlander
MG, Ma XJ and Hainsworth JD: Molecular profiling in unknown primary
cancer: Accuracy of tissue of origin prediction. Oncologist.
15:500–506. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li Z, Li F, Pan C, He Z, Pan X, Zhu Q, Wu
W and Chen L: Tumor cell proliferation (Ki-67) expression and its
prognostic significance in histological subtypes of lung
adenocarcinoma. Lung Cancer. 154:69–75. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sun X and Kaufman PD: Ki-67: More than a
proliferation marker. Chromosoma. 127:175–186. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wei B, Kang J, Kibukawa M, Arreaza G,
Maguire M, Chen L, Qiu P, Lang L, Aurora-Garg D, Cristescu R and
Levitan D: Evaluation of the trusight oncology 500 assay for
routine clinical testing of tumor mutational burden and clinical
utility for predicting response to pembrolizumab. J Mol Diagn.
24:600–608. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Society of Cancer of Multiple and Unknown
Primary of China Anti-Cancer Association, . China Anti-Cancer
Association guideline for diagnosis and treatment of cancer of
multiple and unknown primaries (2023 edition). Chin Oncol.
33:403–422. 2023.
|
|
18
|
Greco FA, Lennington WJ, Spigel DR and
Hainsworth JD: Molecular profiling diagnosis in unknown primary
cancer: Accuracy and ability to complement standard pathology. J
Natl Cancer Inst. 105:782–790. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Weiss LM, Chu P, Schroeder BE, Singh V,
Zhang Y, Erlander MG and Schnabel CA: Blinded comparator study of
immunohistochemical analysis versus a 92-gene cancer classifier in
the diagnosis of the primary site in metastatic tumors. J Mol
Diagn. 15:263–269. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Morawietz L, Floore A, Stork-Sloots L,
Folprecht G, Buettner R, Rieger A, Dietel M and Huebner G:
Comparison of histopathological and gene expression-based typing of
cancer of unknown primary. Virchows Arch. 456:23–29. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Meiri E, Mueller WC, Rosenwald S, Zepeniuk
M, Klinke E, Edmonston TB, Werner M, Lass U, Barshack I, Feinmesser
M, et al: A second-generation microRNA-based assay for diagnosing
tumor tissue origin. Oncologist. 17:801–812. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Rassy E and Pavlidis N: Progress in
refining the clinical management of cancer of unknown primary in
the molecular era. Nat Rev Clin Oncol. 17:541–554. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Meijer L, Verhoeven RHA, de Hingh IHJT,
van de Wouw AJ, van Laarhoven HWM, Lemmens VEPP and Loef C:
Extensive diagnostic work-up for patients with carcinoma of unknown
primary. Clin Exp Metastasis. 38:231–238. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Beauchamp K, Moran B, O'Brien T, Brennan
D, Crown J, Sheahan K and Cotter MB: Carcinoma of unknown primary
(CUP): An update for histopathologists. Cancer Metastasis Rev.
42:1189–1200. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Joerger AC, Stiewe T and Soussi T: TP53:
The unluckiest of genes? Cell Death Differ. Oct 23–2024.(Epub ahead
of print). PubMed/NCBI
|
|
26
|
Bochtler T, Reiling A, Endris V, Hielscher
T, Volckmar AL, Neumann O, Kirchner M, Budczies J, Heukamp LC,
Leichsenring J, et al: Integrated clinicomolecular characterization
identifies RAS activation and CDKN2A deletion as independent
adverse prognostic factors in cancer of unknown primary. Int J
Cancer. 146:3053–3064. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gerard L, Garcia J, Gauthier A, Lopez J,
Durand A, Hervieu V, Lemelin A, Chardon L, Landel V, Gibert B, et
al: ctDNA in neuroendocrine carcinoma of gastroenteropancreatic
origin or of unknown primary: The CIRCAN-NEC pilot study.
Neuroendocrinology. 111:951–964. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bytnar JA, Lin J, Moncur JT, Shriver CD
and Zhu K: Cancers of unknown primary: Survival by histologic type,
demographic features, and treatment in the U.S. Military Health
System. Cancer Epidemiol. 82:1023162023. View Article : Google Scholar : PubMed/NCBI
|