The expression of cytosolic aldehyde dehydrogenases
(ALDHs), which mediate the last step in the synthesis of all-trans
retinoic acid (ATRA), is dysregulated in various types of human
cancer, and is associated with the development of cancer stem cells
(CSCs) in both solid tumors and hematological malignancies
(1,2). CSCs are considered a minor fraction of
cancer cells with the capacity to initiate neoplastic tumors. The
purification of CSCs from patient samples, in almost all cases,
requires antibodies against specific surface markers or the use of
specific culture conditions to promote the enrichment of CSC
populations. Therefore, these methods require identification of
CSC-specific markers that are not available or adequate in a number
of types of cancer (3). The
expression of retinaldehyde dehydrogenases, of which ALDH1A1 is
one, has been included in the biomarkers that are most often used
for breast CSCs (4–6) together with the hyaluronic acid
receptor CD44 (7) and glycoprotein
CD133 (8,9). While ALDH1A1 does not have a uniform
impact on cancer cells, it does nevertheless acquire a crucial role
under certain conditions, which is associated with resistance to
certain aspects of oxidative stress and the generation of RA, as
discussed in the present review. The elucidation of the precise
mode of regulation of this enzyme and of the gene that encodes it
are therefore of paramount importance in biology.
ALDHs are evolutionary well-conserved enzymes in all
living taxonomic groups from bacteria to mammals (10). Previous studies have demonstrated
that ALDH enzymes are involved in various biological processes,
such as proliferation, differentiation and immune system
regulation, by participating in the detoxification of aldehydes
(10–12). Notably, ALDH proteins may function
in the process of cellular UV absorption, and have also been shown
to bind various compounds, such as endobiotics, xenobiotics,
androgens and thyroid hormones (11,12).
Although ALDH activity and expression are generally high in
mitochondria-rich organs, such as the liver and kidney, their
expression is not specific to these organs and ALDH enzymes have a
wider expression profile throughout the body (13).
ALDH family enzymes catalyze the oxidation and
thereby the detoxification of aldehydes, which are highly toxic and
reactive molecules generated from various endogenous or exogenous
sources (14,15). The generation and accumulation of
endogenous aldehydes depends on cellular activities that take place
during various metabolic processes, such as amino acid/alcohol
metabolism and lipid peroxidation in cells (16). Aldehydes are also abundantly present
in the environment and in may be taken up from water, food and air
(17).
Although the substrates of ALDH family members are
generally called ‘aldehydes’, these substrates are diverse aldehyde
molecules that differ according to the substrate-binding
characteristics of the different ALDH subtypes; these binding
characteristics depend on the amino acid sequences and structural
properties of each ALDH protein. Further adding to the subtype
diversity, ALDH family proteins can be localized in different
cellular compartments, such as the cell membrane, cytoplasm,
nucleus, endoplasmic reticulum, mitochondria and in lipid droplets
(18). In humans there are 19 ALDH
proteins, and these proteins can be divided into 11 different
classes, as follows: Class I (ALDH1A1, ALDH1A2, ALDH1A3, ALDH1B1,
ALDH1L1 and ALDH1L2), Class II (ALDH2), Class III (ALDH3A1,
ALDH3A2, ALDH3B1 and ALDH3B2), Class IV (ALDH4A1), Class V
(ALDH5A1), Class VI (ALDH6A1), Class VII (ALDH7A1), Class VIII
(ALDH8A1), Class IX (ALDH9A1), Class X (ALDH16A1) and Class XI
(ALDH18A1) based on their amino acid sequence similarities
(19). Recently Xanthis et
al (10), suggested that the
mitochondrial enzyme ALDH2, which accounts for most acetaldehyde
detoxification, should be included in Class I due to its high amino
acid sequence similarity to the Class I ALDH proteins; notably,
there are important differences between members of this broad class
(Class I) regarding the substrate binding pocket and in the
rate-limiting step (20). The Class
I subclass composed of ALDH1A1, ALDH1A2 and ALDH1A3 has a unique
role; these proteins function as retinaldehyde dehydrogenases, and
are the main enzymes required for the biosynthesis of RA in the
cytosol (21), having a larger
substrate binding cleft that allows them to work more efficiently
on large aldehydes (22,23).
In healthy cells and tissues, the controlled
expression and activities of ALDH family proteins contribute to the
maintenance of homeostasis. In this context, the scavenging of
aldehydes via the activities of ALDH proteins is an important
process in preventing oxidative stress caused by aldehydes in cells
(24). However, the current
approach to cancer treatment is generally based on inducing
oxidative stress in tumor cells via chemotherapy or radiotherapy,
causing substantial cell damage and consequently promoting cell
death (25). Therefore, an increase
in the expression or activity of ALDH family proteins may
negatively affect the success of therapy (26). Notably, it has long been considered
that ALDH family proteins may participate in cancer-related
processes (27,28). Several members of this family,
including ALDH1A1, have been extensively studied for their
contributions to the emergence of the CSC phenotype in malignant
cells (29).
ALDH1A1 has a mostly cytoplasmic and lesser nuclear
localization in cells, and commonly uses aliphatic aldehydes as
substrates, including 4-hydroxynonenal (4-HNE), malondialdehyde
(MDA) and retinaldehyde, which are lipid peroxidation products,
among a number of different compounds (16,30,31).
ALDH1A1 expression and activity are regulated by
various mechanisms. For example, prostate tumor overexpressed 1
(PTOV1) directly binds to the ALDH1A1 promoter and increases its
expression (32). Notably, it is
well known that PTOV1 levels are increased in the tumors of some
patients with breast cancer (BCa), and that PTOV1 upregulation is
associated with disease progression and poor prognosis (33). Although, to the best of our
knowledge, no functional studies have been conducted on this
subject, the tumor-promoting effect of PTOV1 may be related to the
increased expression of ALDH1A1, at least in part.
Mucin 1 (MUC1) expression is increased in various
types of cancer, including BCa, and elevated MUC1 expression can
promote the malignant behavior of cancer cells (34,35).
Furthermore, MUC1 is involved in chemotherapy resistance in cancer
(36). It has been shown that MUC1
induces ALDH1A1 expression via activation of ERK and then
phosphorylation-coupled activation of C/EBPβ. Activated C/EBPβ
directly binds to the ALDH1A1 promoter and increases its expression
(37). Notably, it has been
reported that MUC1 silencing inhibits the CSC phenotypic
manifestation of BCa cells (38).
ALDH1A1 expression may also be regulated in a
Wnt/β-catenin-dependent manner in BCa (39); the β-catenin/TCF complex directly
binds to the ALDH1A1 promoter and increases its expression
(40). In addition, β-catenin
depletion has been shown to decrease ALDH1A1 expression (41).
Although not yet demonstrated in BCa, to the best of
our knowledge, Smad4 has been shown to bind to the ALDH1A1 promoter
and suppress its transcription in TGF-β-treated pancreatic cancer
cells (42). Although TGF-β
inhibits tumorigenesis in normal healthy cells and early-stage
cancer, it mainly promotes invasion, metastasis and therapy
resistance by promoting epithelial-mesenchymal transition (EMT) in
advanced-stage cancer (43–46). In this sense, it will be important
to reveal the effects of the decrease in ALDH1A1 expression that is
mediated by TGF-β/Smad4, by using detailed mechanistic approaches
in terms of identifying and characterizing downstream targets and
analyzing their impacts on cancer biology in diverse model systems,
such as organoids.
NFκB has also been shown to bind directly to the
ALDH1A1 promoter and to positively regulate its expression
(47), although this has not yet
been demonstrated in BCa. NFκB is a well-known pro-inflammatory
transcription factor that is involved in the pathogenesis of BCa
and other types of cancer by controlling the expression of various
genes involved in proliferation, invasion, metastasis and drug
resistance (48–50). However, it is known that NFκB has a
tumor-suppressing role in addition to its tumorigenesis-promoting
role (51); in this sense, the
physiological consequences of NFκB-induced ALDH1A1 expression need
to be studied further in terms of cancer biology.
Post-translational mechanisms are also important in the regulation
of ALDH1A1 activity. It has been shown that acetylation of the K353
amino acid residue is important for ALDH1A1 activity in BCa cells
(52); acetylation of this residue
by P300/CBP-associated factor results in inhibition of ALDH1A1
activity, whereas its de-acetylation by SIRT2 results in ALDH1A1
activation (52). Phosphorylation
of ALDH1A1 by Aurora kinase A on the T267, T442 and T493 amino acid
residues increases both its intracellular stability and activity;
these effects can be attributed to a decrease in ALDH1A1
ubiquitylation, and an increase in the conversion of ALDH1A1 from
an oligomeric to a monomeric form (53). Although this study was conducted in
pancreatic cancer cells, its findings may also apply to other types
of mammalian cells.
Retinaldehyde is an intermediate product in vitamin
A metabolism. Vitamin A is a lipid-soluble molecule that cannot be
synthesized in mammals and therefore needs to be taken up from food
(54,55); however, it has critical roles in
normal cellular physiology and its deficiency may result in various
pathological conditions, including inflammation. Vitamin A exists
in three forms within cells: Retinol, retinaldehyde and RA. RA is
the most active form and its generation from retinol occurs through
two basic enzymatic reactions. In the first step, retinol is
converted to retinaldehyde by the oxidation activities of alcohol
dehydrogenases and in the second step, retinaldehyde is oxidized to
RA by the activities of ALDH family members (56). Retinaldehyde formation by oxidation
of retinol is a reversible process, as retinaldehyde can be reduced
back to retinol by retinal reductases (57). However, the generation of RA by
oxidation of retinaldehyde is an irreversible process and RA is
rapidly degraded by P450 family enzymes after it is generated
(56).
RA signaling generally works in an autocrine and
paracrine manner, and after RA is produced it binds to RA receptor
(RAR) and retinoid X receptor (RXR), which are members of the
nuclear receptor family (58).
Consequently, in the nucleus, ligand-activated receptors (RARα, β
and γ, which form RA-induced heterodimers with RXRα, β and γ) bind
to RA response elements on the promoters of RA responsive genes and
regulate their expression (59).
Generated RA is thereby involved in various cellular processes,
such as development and differentiation. ATRA, 9-cis RA and 13-cis
RA are natural RA isomers, and their receptor preferences may
differ from each other in RA signaling (60). In this context, although ATRA
selectively binds to RAR, 9-cis RA can bind to both receptor types
(RAR and RXR) (61). Although
activated RAR and RXR regulate the expression of target genes by
establishing homo- or hetero-complexes in the canonical pathway, it
is known that these receptors (especially RXR) can affect cell
physiology by forming hetero-complexes with other receptors, such
as estrogen receptor (ER) or peroxisome proliferator-activated
receptor (PPAR) (62–64). PPAR alone can be activated by RA;
however, in contrast to RAR, PPAR signaling supports cell survival
and proliferation, but PPAR requires a higher concentration of RA
to be activated when compared with RAR (63). This multiplicity of downstream
effectors allows RA to affect a number of signaling pathways,
including PI3K/AKT, Notch and Wnt/β-catenin, and to consequently
regulate several cellular functions (65). The effects of RA on cells therefore
depend on the proteome of the cell, and also on the type and
concentration of RA, which determine the subsets of receptors that
will be activated.
Regarding the most common aldehydes that are
products of lipid peroxidation, 4-HNE induces chemical
modifications on DNA, and thereby causes DNA damage and mutations
(66). Notably, it has been shown
that 4-HNE forms an adduct at codon 249 of the p53 encoding gene in
the human genome and promotes liver cancer malignancy (67). In addition, 4-HNE can modify
proteins directly. In the context of DNA damage, it has been shown
that 4-HNE reduces the activities of DNA damage repair proteins,
resulting in more severe damage to DNA, under oxidative stress
conditions (68). Notably,
increased 4-HNE levels are involved in carcinogenesis, and excess
accumulation of 4-HNE has been reported in various types of cancer,
including esophageal, colon and lung cancer (69–71).
Another reactive aldehyde, MDA, is an end product in
the peroxidation of polyunsaturated fatty acids found in the cell
and mitochondrial membranes, and has long been used to monitor
lipid peroxidation (16,72). An increase in MDA levels may reflect
a decrease or insufficient activity of antioxidant systems, and
this event has been associated with various neoplastic (including
BCa) and non-neoplastic diseases (73–75).
MDA activity is pH-dependent and MDA strongly reacts with basic
amino acids, such as lysine and arginine, at lower pH values, to
generate adducts with free amino acids or proteins (16,76).
MDA also reacts with aminophospholipids and generates adducts such
as MDA-phosphatidylethanolamine (77). In addition, MDA reacts with DNA to
generate adducts, and consequently induces DNA damage and mutations
(78). The guanine base in DNA has
the highest susceptibility to the formation of MDA adducts and
therefore the MDA-reacted DNA is generally detected in a form of
MDA-deoxyguanosine (79).
Furthermore, it has been demonstrated that MDA inhibits DNA damage
repair mechanisms (80). In this
context, although MDA is less toxic compared with 4-HNE, which is
the most toxic lipid peroxidation product, it still has potent
mutagenic activity (81).
ALDH1A1 is generally considered to be a marker of
CSCs, and elevated ALDH1A1 expression is generally associated with
increased malignant behaviors and therapy resistance in cancer
(82). Although the antitumor
activities of ALDH1A1 have been demonstrated in some types of
cancer (or some conditions in cancer), it is generally accepted
that it is involved in the regulation of multiple mechanisms to
promote cancer progression (83).
ALDH1A1 promotes stemness and therapeutic resistance
mainly by being involved in RA synthesis (84). In this sense, as aforementioned,
reactive aldehydes are detoxified during RA synthesis. In addition,
synthesized RA induces signaling mechanisms such as PI3K/AKT,
Wnt/β-catenin and Notch, which stimulate the activity of several
transcription factors that promote stem cell behavior, and various
ABC family transporter proteins that are directly involved in drug
resistance (85,86). Consequently, elevated ALDH1A1
activity results in an increase in CSCs, and promotes therapy
resistance and tumor recurrence (87–89).
One important self-limiting factor against ALDH1A1-overexpressing
tumors is the impact of RA on cell physiology; specifically RA
induces differentiation to a number of cell types, such as normal
stem cells or acute promyelocytic leukemia cells (1), thus making ALDH1A1 an attractive
intervention target since in those cell types both exogenous RA and
endogenous ALDH1A1 activity, which generates RA, limit their
proliferation; by contrast, in cells insensitive to RA, ALDH1A1
inhibition decreases CSC frequency (1).
ALDH1A1 upregulation has been associated with higher
grade tumors and increased malignancy in patients with BCa
(90). It has been reported that
patients with ALDH1A1 (+) non-triple-negative BCa (TNBC) tumors
have a shorter survival time compared with that of patients with
ALDH1A1 (−) tumors (91). Notably,
ALDH1A1 positivity has been reported as a signature for early
relapse and a more aggressive phenotype in patients with ER
(+)/HER2 (−) BCa (92). In the
context of TNBC, it has been reported that ALDH (+) cells are
enriched in TNBC cell lines compared with in non-TNBC cell lines
(93), and ALDH1A1 positivity is an
independent prognostic factor in TNBC (94). Furthermore, an association between
ALDH1A1 positivity and tumor grade, ER/progesterone receptor (PR)
negativity and HER2 positivity has been reported (90,95),
and it has been shown that ALDH1A1 expression is higher in ER/PR
(−) and HER2 (+) tumors that have high Ki67 levels in patients with
BCa (96). Numerous studies have
reported that ALDH1A1 positivity could be an independent marker of
poor prognosis in patients with luminal or TNBC tumors (97–102);
however, although ALDH1A1 positivity in tumor cells is important
for predicting prognosis in BCa, serum ALDH1A1 has been reported to
be inappropriate as a marker (103). Notably, high ALDH1A1 expression in
stromal cells of TNBC tumors has been reported to predict a
favorable prognosis in BCa (104).
At least in normal epithelial cells, the effects of
ALDH1A1 on stemness can be attributed to RA. The use of
fluorescence-activated cell sorting of primary human mammary
epithelial cells, along with in vitro and in vivo
functional assays to examine the relationship between cells with
ALDH enzymatic activity (ALDH+ cells) and ER+
cells in the normal human breast epithelium, demonstrated that
ALDH1A1 knockdown could significantly reduce the number of primary
and secondary mammospheres formed in suspension culture and that
this effect could be rescued by RA. Notably, this approach
dissociated between the effects of ALDH1A1 on mammospheres and
proliferation, as RA (produced by ALDH1A1) resulted in a block in
proliferation, whereas ALDH1A1 was shown to have an important role
in the formation of mammospheres (105). In this study it was shown that
ER− cells gave rise to ER+ cells; the
ER− cell population contains a subset of cells that can
generate ER+ cells, which are able to proliferate
proving that ALDH1A1 expression is consistent with stem cell
function, since ER− (ALDH1A1 expressing) cells generated
ER+ cells, in the same manner that stem cells generate
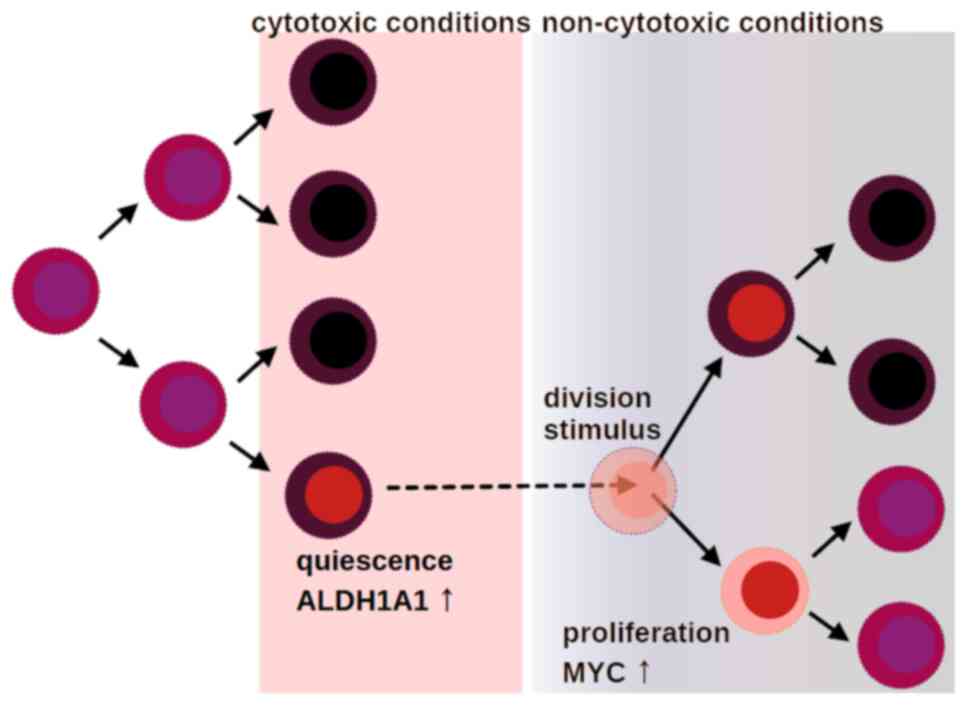
both proliferating and differentiating cells (105). This is noteworthy because in a
later study, ALDH+ BCa CSCs were shown to include both
quiescent as well as proliferating clones, suggesting the role of
ALDH activity as a viability safeguard during the phenotypic
transitions of malignant stem cells, which permits them to generate
diverse subclones with variable adaptation potential; single-cell
RNA profiling previously identified a dormant ALDH+
population that expanded after anti-estrogen treatment (106). Anti-estrogen treatment led to
expansion of the quiescent clones, which supports the hypothesis
that quiescence is a mechanism of malignant cell adaptation to
antineoplastic treatment; under conditions that trigger cell death
or cell cycle arrest, the cells that enter a dormant state in
respect to cell growth and metabolism appear to be protected
(Fig. 1). Exposure to a number of
cytotoxic and cytostatic agents may favor the growth of clones that
have adaptive mechanisms; for example, tamoxifen treatment has been
shown to induce ALDH1A1 expression in breast cancer cells
expressing the ER variant ERα36, and these cells have the capacity
for proliferation and metastasis in BALB/c nude mice (107).
The effect of ALDH1A1 on stemness via RA production
may also be associated with SRC-3, a steroid receptor co-activator,
which is a critical factor in the development and progression of
BCa (108). SRC-3 interacts with
RA-activated RARα and regulates RARα transcriptional activity
(109). Conversely, RA also
promotes the phosphorylation and ensuing ubiquitin-dependent
proteasomal degradation of SRC-3 in a p38/Cul3-dependent manner and
this event contributes to an anti-proliferative effect of RA
(110). In this manner, RARα
transcriptional activity is regulated by SRC-3 under the control of
RA and p38/Cul3 (110,111). However, RA has been reported to
induce SRC-3 phosphorylation and degradation only in
HER2− cells, such as MCF-7, but not in HER2+
cells, such as BT474 and MDA-MB-361 cells (110). Therefore, due to the different
molecular contents of tumor cells, the increase in the amount of RA
produced due to upregulation of ALDH1A1 will not have the same
effect on every cell. Although erbB-2 expression is apparently a
critical factor that determines the mechanism of cell response to
increased ALDH1A1 activity and RA production, there may be more
contributing factors that remain to be defined. In the context of
CSCs, SRC-3 has been implicated in the induction and maintenance of
breast CSCs (112). Notably, SRC-3
(but not SRC-1 or SRC-2) silencing by small interfering (si)RNA or
inhibition using the chemical inhibitor SI-2 in HER2+
and TNBC BCa cell lines has been shown to lead to a decrease in
ALDH activity and in ALDH+ cell populations (112).
While estrogen alone induces BCa cell proliferation,
RA generally inhibits cell proliferation through multiple
mechanisms including interactions between different proteins of the
RA and estrogen signaling pathways (113,114). Notably, the effects of RA on BCa
cells are generally regulated by a crosstalk between ER and RAR
signaling mechanisms (115). In
addition, ATRA inhibits the proliferation of ER (+) BCa cells, but
not of ER (−) cells (116);
however, at the genomic level, the interaction between RAR and ER
signaling mechanisms is complex. RARα is an estrogen-regulated gene
that is associated with ER expression in BCa (117). In addition, RARα and ER share
common cis-regulatory elements in the genome, and RARα interacts
with ER during estrogen stimulation and regulates the transcription
of ER target genes as part of a joint RAR/ER transcriptional
complex (64). However, the
scenario in which ER and RAR interact in the presence of estrogen
and bind to the promoter of target genes as a transcriptional
complex may change when RA is also present. It has been reported
that in some cases of BCa, ER and RAR can compete with each other
to bind to regions of the genome where they recognize common target
DNA sequences, that ER signaling can be inhibited in the presence
of RARα ligands, and conversely that RAR signaling can be
suppressed by the presence of estrogens (118). For example, estradiol (E2)
treatment has been shown to lead to protein kinase A-mediated
lysine-specific histone demethylase 1 (LSD-1) activation and
thereby to demethylation of H3K9me2, resulting in the joint binding
of ERα and RARα to the promoter of target genes such as BCL-2
(119). However, RA inhibits
E2-induced LSD-1 activation, preventing H3K9me2 demethylation and
consequently suppressing this part of estrogen signaling (119). In the context of high ALDH1A1
expression in BCa, high ALDH1A1 levels may result in more RA
production, which could then inhibit cell proliferation by blocking
ER-mediated signaling by binding to regions of the genome where ER
and RAR can bind together. However, the proposed mechanism is based
on the currently available literature and it is possible that other
parameters of this relationship may also emerge due to genomic,
transcriptomic/proteomic and metabolomic differences in different
subsets of BCa.
A critical observation that has been made in BCa is
that CD44 (+)/ALDH1A1 (+)/Ki-67 (−) tumor cells may favor distant
metastasis and predict poor overall survival in patients with
ductal carcinoma in situ (135). In this previous study, quiescence
of breast CSCs was shown to be associated with tumor progression,
treatment resistance and metastatic capacity. Quiescence can
protect stem cells in general by decreasing the generation of
reactive oxygen species (ROS) through a lower metabolic rate, since
it has been shown that quiescence of hematopoietic stem cells
protects them from DNA damage (136–139). Support for this hypothesis was
provided when reviewing information from previously published
studies, including studies conducted with samples from healthy
volunteers, as well as studies with samples from patients with
cancer, including primary cancer samples from the Genomic Data
Commons-deposited data of The Cancer Genome Atlas BRCA study
(Table I).
When the correlation between mRNAs extracted from
healthy volunteers and patients is taken into consideration, three
trends appear regarding the mRNA expression of MYC, ALDH1A1 and
ALDH2: i) The two ALDH genes are correlated at the mRNA level,
indicating that cells tend to express similar levels of ALDH1A1 and
ALDH2; ii) the two ALDH genes are either not correlated, or even
inversely correlated with MYC; and iii) the two ALDH genes are
either not correlated, or even inversely correlated with cellular
DNA damage, as indicated by the homologous recombination deficiency
(HRD) score, whereas the opposite occurs with MYC RNA: MYC RNA is
correlated with the DNA damage index (HRD score), which may reflect
increased metabolic activity of cells that generate ROS to the
extent that leads to DNA damage. Therefore, ALDH1A1 and ALDH2 RNA
tend to be expressed more under conditions not conducive to
cellular DNA damage. This is consistent with their increased
importance for slower cell growth states. Such slower growth may
also correspond to some forms of dormant growth arrest that places
limits to DNA damage (138,140,141).
The platform used for extracting the information shown in Table I was University of California, Santa
Cruz Xena (https://xenabrowser.net/) (142). This concise overview of previous
studies supports the hypothesized role of ALDH1A1 in quiescent stem
cells, and suggests that ALDH1A1 has an important role in quiescent
CSCs, which is consistent with the aforementioned role of ALDH1A1
in mammospheres (105).
Regarding how aggressive cancer cells arise from
quiescent cells, cell quiescence can still result in aggressive
cancer after relapse due to the aberrantly exposed chromatin on
certain key genes in CSCs, such as MYC, which permit rapid
phenotypic changes under favorable conditions (51,143).
A more precise association of the function of each gene can only be
made after considering the impact of the gene product under
different conditions in vitro and in vivo, and after
factoring the hazard ratio of the expression of the gene for
different patient groups.
Any hypothesis that is solely based on the
measurements of RNA steady-state levels can only have a theoretical
value in the absence of mechanistic studies in cultured cells.
Results as those shown in Table I
can easily become irrelevant in datasets obtained from slightly
different sample types. Thus it is important to bear this in mind
until multiple types of analysis support this hypothesis.
A key recent discovery enabled tracking of CSCs in
BCa with a reporter system using a far-red fluorescent protein
under the control of the ALDH1A1 promoter. Positively stained cells
have been shown to exhibit stemness characteristics that include
higher sphere-forming capacity, tumor formation and increased
resistance to anticancer treatments (144). Notably, live tracking of cells in
a microfluidic system has revealed a higher extravasation potential
of CSCs, and for the first time, the live reprogramming of non-CSCs
into CSCs (144). This
reprogramming that facilitates interconversion between CSCs and
non-CSCs can explain why ALDH1A1-positive CSCs may prove far more
elusive than anticipated, especially in light of a recently
discovered variability in the effects of RA-binding proteins on the
capacity for proliferation and drug resistance of BCa cells
(145). One interesting approach
to assess the complexity of RA effects is to design interventions
that selectively target intracellular RARγ; if this has similar
results to knocking out or inhibiting ALDH1A1, it may be
hypothesized that RARγ mediates the effects of ALDH1A1 on CSCs
(146).
In lung cancer ALDH1A1 levels are high in both
non-small cell lung cancer (NSCLC) and SCLC compared with in normal
healthy cells, and much higher in NSCLC compared with in SCLC
(147). ALDH1A1 has been shown to
promote cell cycle arrest by inhibiting the Notch/CDK2/Cyclin E
pathway in lung cancer cells, thus improving clonogenic abilities
and stemness (148). Notably,
ALDH1A1 expression has been reported to be higher in advanced-stage
lung tumors and cisplatin-resistant lung cancer cells compared with
in early-stage tumors and cisplatin-sensitive cells, and
ALDH1A1-depleted cells are sensitive to cisplatin (149,150). In addition, inhibition of ALDH1A1
activity using a disulfiram/copper complex can suppress the
malignant behaviors and relapse of NSCLC (151). Therefore, ALDH1A1 expression has
been proposed to be associated with poor prognosis in patients with
NSCLC (152–154). Furthermore, the S100A9/ALDH1A1/RA
pathway has been reported to promote metastatic brain relapse in
patients with EGFR-mutant lung cancer treated with the EGFR
tyrosine kinase inhibitor osimertinib, whereas targeting of S100A9,
RAR or ALDH1A1 may inhibit brain metastasis in these patients
(155).
It has also been suggested that ALDH1A1 may act as a
tumor suppressor in NSCLC, especially in smokers (156). Although a mechanistic explanation
has not been provided in this study, a recent study revealed that
patients with lung cancer lacking ALDH1A, CD133 and mutant p53 have
a better prognosis (157).
Therefore, the results of Okudela et al (156), which do not comply with the
existing literature, may hint to a tumor-suppressing impact of
ALDH1A1-generated RA. This situation also shows that a number of
molecules may be involved in the downstream effects of ALDH1A1 on
tumorigenesis. Another likely explanation is that the proportion of
ALDH1A1-positive lung cancer stem-like cells is low in aggressive
tumors. As mentioned in the present review, one explanation that
should be considered for solid tumors, is that when ALDH1A1
activity causes an increase in RA, activated RARs confer a positive
disease outcome by leading to the suppression of aggressive tumors
through a number of different mechanisms (158). This suppression can be in part
attributed to an increase in differentiated cell phenotypes
(159). Moreover, ectopic
expression of the RA-induced G gene (also known as IFIT3) has been
shown to lead to a significant decrease in the proliferation of
lung cancer cells, resulting in an inhibition of tumor xenograft
growth in mice (160). Solid tumor
cells derive from non-circulating cell clones that in general do
not adapt to drastic changes in their microenvironment; therefore
the influence of local gradients of RA during primary cancer growth
should be significant and affect the disease course. It must also
be noted that RAR competes with the vitamin D receptor for RXR
binding and for interference with RXR signaling (161), which adds a substantial degree of
flexibility for RA signaling and its downstream effects. In
particular, these interactions may have an important role in
restoration and maintenance of epithelial barrier function; while
this has been suggested for intestinal tissue it is very likely to
apply for other types of epithelial tissue as well (162).
In prostate cancer (PCa), it has been reported that
a relationship exists between ALDH1A1 expression levels and Gleason
score, and that ALDH1A1 expression is higher in
castration-resistant PCa compared with in castration-sensitive PCa
(163). Notably, increased ALDH1A1
expression is associated with metastasis and poor prognosis in PCa
(164); however, increased ALDH1A1
expression in stromal cells adjacent to cancer cells is associated
with a better prognosis, similar to in BCa (165). ALDH1A1 expression has also been
shown to be associated with resistance to radiotherapy (41). In PCa, among other effects, ATRA
treatment suppresses ALDH1A1 expression, activates CDK5 and
increases p27 levels in androgen receptor-negative cells, thereby
inhibiting proliferation (166).
This could make ALDH1A1 expression self-liming under certain
conditions of slow ATRA turnover, particularly in tumors where
oncogenic RA signaling pathways, such as RARγ, do not prevail over
tumor-suppressing mechanisms (146).
In ovarian cancer, high ALDH1A1 levels are
associated with chemotherapy resistance and poor prognosis
(167). Notably, an association
has been identified between high ALDH1A1 expression levels and
shorter overall survival (168).
In this context, ALDH1A1 alters the signaling network in cell cycle
checkpoints and DNA repair processes, and thereby maintains ovarian
CSCs (169). ALDH1A1 levels are
increased after neoadjuvant treatment, and this event is associated
with chemoresistance and poor prognosis (170). The levels of ALDH1A1 and several
drug transporter proteins are high in paclitaxel- and
topotecan-resistant ovarian cancer cells, and ATRA treatment can
decrease both ALDH1A1 and drug transporter protein levels leading
to a decrease in the resistance to chemotherapy agents (171). However, ALDH1A1 depletion in
topotecan- and paclitaxel-resistant ovarian cancer cells results in
an increase in paclitaxel resistance, although it causes a decrease
in topotecan resistance (172).
Inhibition of ALDH1A1 activity can both diminish the CSC population
and inhibit cisplatin-induced senescence that would otherwise
promote stemness via paracrine signaling (173). Consequently, co-expression of
ALDH1A1 and SALL4 in patients with serous ovarian cancer is
associated with an overall unfavorable prognosis (174).
A negative association has been demonstrated between
high ALDH1A1 RNA expression and overall survival in patients with
acute myeloid leukemia (AML) (175). Notably, it has previously been
shown that ALDH1A1 RNA-null patients belong to the AML favorable
prognosis risk group (176). These
findings suggest that ALDH1A1 is a potential target for AML
treatment. One compound that targets ALDH1A1 and possibly other
similar enzymes is DIMATE (177,178), which is currently under study for
AML in the phase 1 clinical trial ‘ODYSSEY’ (NCT05601726) for
patients with relapsed AML (179,180).
Disulfiram is a Food and Drug Administration
(FDA)-approved drug that, among a number of other targets, inhibits
ALDH1A1 at sub-micromolar concentrations (181). Disulfiram specifically targets
CSCs in AML by increasing activity of the ROS-induced JNK pathway
and by inhibiting the NFκB and Nrf2 pathways (182). Furthermore, elevated ALDH1A1
expression is associated with sorafenib resistance in various types
of cancer, including AML (183).
This suggests that, at least for AML, preclinically targeting
ALDH1A1 is an option worth considering. Notably, AML cells can
escape the downstream effects of RA production. In the normal human
bone marrow, mesenchymal cells prevent retinoid-induced
differentiation of hematopoietic stem cells by maintaining a low RA
concentration, via CYP26-mediated degradation (184,185). However aggressive AML cells can be
expected to adapt to a RA-rich microenvironment, and thereby
tolerate high levels of ALDH1A1 expression and activity (82). This aberration renders a substantial
portion of AML cells highly resistant to RA (186). However conversely, this discovery
leads to the expectation that AML cells resistant to RA can be
killed by inhibition of ALDH1A1, since ALDH1A1 protects them from
cytotoxic aldehydes. This RA insensitivity brings AML into sharp
contrast with BCa, since at least a notable part of BCa cells
contains functional RARα and RARγ, and responds to RA; this at
least applies to cytokeratin 5-positive BCa cells, which are the
most aggressive malignant cells (187). ALDH1A1-overexpressing BCa cells
would therefore be expected to thrive mostly in association with
stromal cells that remove RA; stromal cells metabolize RA and
decrease exposure of BCa cells to RA; therefore, interfering with
stromal cells may aid the development of experimental therapeutic
approaches.
Another pathway for the cancer-supporting effects of
ALDH1A1 has been discovered in multiple myeloma cells, where
ALDH1A1-generated 9-cis RA activates RXR to induce NIMA-related
kinase 2 (NEK2) expression; this has been shown to increase
clonogenicity and tumorigenicity, and additionally cause resistance
to two widely used myeloma drugs (bortezomib and doxorubicin) by
enhancing expression of the drug-efflux pump ABCB1 and of survival
proteins, AKT and BCL-2 (188).
NEK2 activation in multiple myeloma is important, since it also
activates autophagy (via the lysosome), which helps malignant cells
survive for several reasons in addition to the most obvious reason,
which is resistance to proteasome inhibition (189). The discovery of the effect of
ALDH1A1 on NEK2 may also be relevant in BCa, since NEK2 has been
shown to control proliferation, migration, invasion and viability
of cultured BCa cells (190).
The positive association between ALDH1A1
expression/activity and tumorigenesis, poor prognosis and therapy
resistance is not limited to the aforementioned types of cancer,
and has also been demonstrated in a number of other types of
cancer, including bladder, colorectal, head and neck, esophageal
and gastric cancer (191–196). In comparison to other CSC markers,
ALDH1A1 expression in adenocarcinoma appears to have a stronger
association with tumor initiation, asymmetric division and
interconversion between cellular phenotypes, properties that are
consistent with increased flexibility during critical phases of
cancer progression (197). The net
effect of the expression of ALDH1A1 in the different cell types of
a given tissue depends on the interactions of its reaction products
with diverse signaling pathways, which include, but are not limited
to, nuclear receptor-activated cascades. Notably, in addition to
the detoxification of aldehydes, ALDH1A1 can contribute to drug
resistance in several other manners, such as via the expression of
drug transporter proteins and of antiapoptotic factors, in addition
to the activation of autophagy, most of these effects are also
paradoxically linked to the generation of RA, due to differential
activation of RA-dependent pathways; therefore increases in ALDH1A1
activity and RA concentration elicit fundamental alterations in
cell signaling mechanisms that affect how the cells respond to
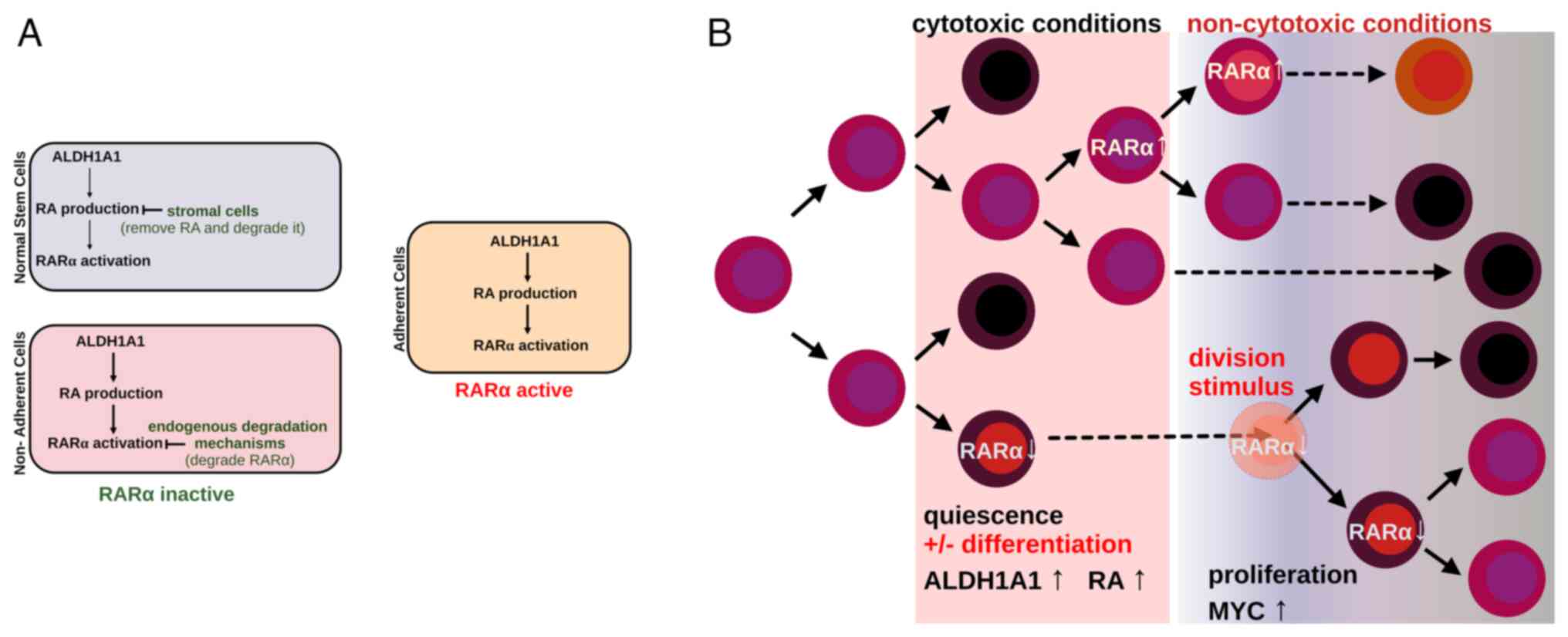
stimuli and whether cells proliferate, differentiate or die.
Specifically, cells that express RARα may differentiate upon
increased ALDH1A1 activity that generates RA, while cells deficient
in RARα are in position to resume proliferation once the cell
microenvironment transitions from cytotoxic to non-cytotoxic
conditions that provide additional stimuli, which induce cell
division (Fig. 2).
There is a clear negative association between
ALDH1A1 levels and treatment success in BCa. Notably, high ALDH1A1
expression results in resistance to numerous chemotherapeutic drugs
that use different cellular mechanisms to exert their
antineoplastic effect. This makes ALDH1A1 an important target in
the treatment of BCa.
ALDH1A1 is involved in cyclophosphamide resistance
and a decrease in ALDH1A1 expression is important in the success of
cyclophosphamide treatment in BCa (13). It has been reported that ALDH1A1
levels are lower in BCa cells that respond to cyclophosphamide
compared with in cells that do not respond to this treatment
(198). Similarly, it has been
shown that ALDH1A1 positivity is associated with poor clinical
outcome and prognosis in cyclophosphamide-treated patients
(199). Furthermore, ALDH1A1
levels are higher in metastatic BCa cells treated with
cyclophosphamide compared with in cells not exposed to
cyclophosphamide (198), and an
increase in cytoplasmic β-catenin levels along with an increase in
ALDH1A1 levels is associated with poor prognosis in patients
receiving cyclophosphamide treatment (200). In a recent study, raloxifene and
bazedoxifene were identified as selective ALDH1A1 inhibitors by
using virtual screening approaches, and it was shown that both
compounds can increase the sensitivity of ALDH1A1-overexpressing
cells to mafosfamide sodium salt, a cyclophosphamide analog
(201). Ifosfamide is another
oxazaphosphorine group chemotherapeutic drug like cyclophosphamide,
and ALDH1A1 has been shown to detoxify it as well (202). In a recent study, telmisartan,
irbesartan and maraviroc were reported as prospective ALDH1A1
inhibitors by the use of computational approaches, although
experimental evidence was not provided (203).
As aforementioned, disulfiram is an FDA-approved
substance for individuals wishing to abstain from alcohol, and it
is also a prospective antineoplastic drug that seems to be a
promising molecule for inhibition of ALDH1A1 (204). Notably, disulfiram/copper
complexes have been reported to decrease NFκB activity, increase
total ROS levels and MAPK signaling activity, and inhibit malignant
behaviors in BCa cells (205). In
addition, disulfiram inhibits HER2/AKT signaling and suppresses
stemness in HER2 (+) BCa cells (206). Disulfiram also inhibits STAT3
signaling, and thereby decreases cyclin D1 and survivin levels, in
addition to inhibiting ALDH1A1 activity in TNBC (207). In this context, STAT3 signaling
may be a critical pathway for the regulation of ALDH1A1-induced
stemness and malignant behaviors in BCa. Notably, STAT3 activity is
higher in ALDH (+) BCa cells compared with in ALDH (−) cells and
inhibition of STAT3 activity using a chemical inhibitor can both
suppress the ALDH (+) cell population and inhibit tumor growth
(208). A novel ferrocene
derivative synthetic compound has been reported to inhibit both
mammosphere formation and stem cell properties, including
downregulation of ALDH1A1 expression, in BCa cells through ROS
production and STAT3 inhibition (209). Dinaciclib, a CDK1/2/5/9 inhibitor,
decreases ALDH1A1 levels along with the levels of
pluripotency-associated transcription factors, including NANOG,
OCT4 and SOX2. This effect of dinaciclib has been attributed to
inhibition of FoxM1 in a sonic hedgehog-dependent manner (210). However, more recently it was shown
that in AML cells dinaciclib inhibits STAT3 activity in an
ERK-dependent manner and consequently decreases Myc expression
(211). Although it is not yet
known whether the effect of dinaciclib causing the decrease in
ALDH1A1 levels is dependent on STAT3, it may be at least a viable
working hypothesis, although AML is a different study system from
BCa. In addition, it has been shown that esculentoside A inhibits
mammosphere formation and the proliferation of breast CSCs,
decreases ALDH1A1, SOX2 and OCT4 levels and STAT3 activity, and
induces apoptosis (212). In
summary, STAT3 activity may be important in the control of ALDH1A1
levels, and ALDH1A1 is associated with malignant behaviors and
therapy resistance in BCa.
It has been shown that although tamoxifen and
fulvestrant decrease total BCa cell proliferation, they
nevertheless increase breast CSC activity in a Notch-dependent
manner (213). In addition, it has
been demonstrated that ALDH1A1 levels are increased in the tumors
of patients with ERα (+) BCa and disease relapse after surgery and
tamoxifen treatment (214).
Notably, ALDH1A1 is a tamoxifen-responsive gene: Tamoxifen induces
ERα-36, a variant of ERα, to translocate to the cell nucleus, where
it directly binds to the ALDH1A1 promoter (107), and consequently, increased ALDH1A1
levels promote metastasis and stemness. In addition, the use of
ALDH1A1 inhibitors or ERα-36 antibodies has been shown to abolish
the effects of tamoxifen-induced malignant behaviors (107).
A negative association has been reported between
ALDH1A1 expression and neoadjuvant therapy response in BCa
(215,216) and it has been shown that an
increase in ALDH1A1 levels after neoadjuvant therapy may be a
predictor of a weaker therapeutic response (217). Consequently it has been proposed
that ALDH1A1 expression may be used to monitor neoadjuvant
chemotherapy success (199).
CYP2J2-overexpressing BCa cells are generally
resistant to chemotherapy agents and it has been shown that
ALDH1A1s levels are also high in these cells, and that resistance
to chemotherapy agents is mainly regulated by inhibiting the
chemotherapy-mediated ROS production by ALDH1A1, thus protecting
the cells from death (218).
Therefore silencing of ALDH1A1 may be a practical approach to
overcome chemotherapy resistance in CYP2J2-overexpressing BCa
cells.
Various plant-derived molecules have been shown to
reduce ALDH1A1 activity/expression, thereby sensitizing BCa cells
to chemotherapy. For example, curcumin and curcumin derivative
synthetic analogues can decrease ALDH1A1 levels in breast CSCs
(219,220); this effect is dependent on the
sonic hedgehog and Wnt/β-catenin pathways (221). Therefore, curcumin and its
derivatives may be considered as candidate agents for the purpose
of overcoming drug resistance in BCa. A combined curcumin and
vitamin D treatment has been shown to increase sensitivity to
paclitaxel, as well as the apoptotic potential, and to decrease
ALDH1A1 levels in paclitaxel-resistant BCa cells (222). In this context, curcumin-dependent
inhibition of ALDH1A1 may be a useful approach to overcome
paclitaxel and epirubicin resistance, since ALDH1A1 is a reliable
biomarker for paclitaxel and epirubicin resistance in breast CSCs
(223).
Quercetin has also been shown to suppress the
malignant behaviors of breast CSCs and to induce apoptosis via
inhibition of ALDH1A1 along with CXCR4, MUC1 and EpCAM (224). It was also shown that sulforaphane
inhibits TNBC tumor development in an animal model and that it
decreases the expression of various stem cell markers, including
ALDH1A1, via a Cripto-mediated pathway (225). In addition, it has been reported
that sulforaphane decreases the ALDH1A1 (+) cell population in both
TNBC and ER (+)/PR (+) BCa cells (226). Furthermore, 4-vinylphenol
decreases ALDH1A1 levels, and inhibits sphere formation and
malignant behaviors of CSC-enriched BCa cells via inhibition of
EGFR/AKT/β-catenin signaling (227). Although this previous study did
not provide a mechanistic explanation for the association between
the decrease in ALDH1A1 and the inhibition of EGFR/AKT/β-catenin
signaling, a similar mechanism has been observed in esophageal
squamous cell carcinoma cells, where ALDH1A promotes both malignant
behaviors and 5-FU chemotherapeutic resistance by activating AKT
signaling and via interacting with β-catenin (228). Silybin is another plant-derived
complex that inhibits ALDH1A1 expression and thereby inhibits the
malignant behaviors of PCa cells (229). Although its relationship with
ALDH1A1 has not been elucidated, in vitro studies have shown
that silybin inhibits malignant activity in various types of
cancer, including BCa, and that silybin enhances the sensitivity of
BCa and ovarian cancer cells to cisplatin and doxorubicin (230,231).
A recent study has shown that ALDH1A1 inhibits
ferroptosis that is triggered by KRAS inhibitors and thereby leads
to resistance to those agents that target KRAS (232). Although KRAS mutations are not
common in BCa, it is known that mutated KRAS is associated with
metastatic behavior and poor prognosis in BCa (233). Mutated KRAS promotes
chemoresistance via increasing Nrf2 expression; in concordance,
inhibition of the Nrf2 pathway can suppress KRAS-induced
chemoresistance (234). In this
context, it has been shown that ALDH1A1 activates Nrf2 in a
p62-dependent manner (235).
Elevated ALDH1A1 expression has been reported to
facilitate the entry of lysosomal autophagy inhibitors (including
the chloroquine derivative hydroxychloroquine) into cells,
resulting in increased cytotoxicity without affecting lysosome
function or autophagic flux (236). Chloroquine is an anti-malarial
drug and its repurposing as a cancer treatment has been discussed
for years (237,238). Chloroquine targets CSCs by
inducing mitochondrial damage and by impairing DNA break repair, in
addition to inhibiting autophagy (239). In concordance, it has been shown
that chloroquine encapsulated by a
triphenylphosphonium-functionalized hyperbranched polymer results
in a high cytotoxicity in mammospheres in an ATM-dependent manner
(240,241). Therefore, the use of chloroquine
or its derivatives in the treatment of cancer to target cells with
high ALDH1A1 expression (in this case, mostly CSCs) may be a useful
approach.
It may also be possible to specifically target CSCs
by targeting ALDH1A1. Notably, it has been shown that
ALDH1A1-specific CD8+ T cells effectively target and
suppress xenograft tumors and experimental metastases, in a study
conducted for this purpose (242).
This approach may be useful as a means to control the
ALDH1A1-mediated tumor-promoting microenvironment in BCa. In this
context, ALDH1A1 has been shown to trigger a molecular/metabolic
cascade consisting of a decrease in intracellular pH, increased
TAK1 phosphorylation and activation of NFκB signaling in
tumor-initiating breast cells (243). This event results in increased
granulocyte-macrophage colony-stimulating factor secretion from
tumor-initiating cells (TICs) into the tumor microenvironment with
the consequent expansion of myeloid-derived tumor suppressor cells
(MDSCs) (243). Notably, the use
of disulfiram (ALDH1A1 inhibitor) plus gemcitabine may inhibit
tumorigenesis by targeting ALDH1A1 (+) TICs and activating T-cell
immunity (243). The results of
this previous study demonstrated a critical role of ALDH1A1 in the
interaction between BCa-TICs and MDSCs during BCa progression, thus
suggesting that a novel therapeutic approach targeting ALDH1A1 may
be successful by disrupting this interaction in BCa.
The current literature indicates that at least some
bulk tumor cells have the capacity to generate stem-like cells that
act like CSCs, which can contribute to the progression of cancer
and to therapy resistance. In the context of BCa, only a small
fraction of malignant cells exhibit CSC characteristics and these
cells generally have a high ALDH1A1 activity that is critical to
the emergence of the CSC phenotype. As in other types of cancer, in
BCa, decreasing ALDH1A1 expression via gene knockout or
interference with gene expression, or inhibiting ALDH1A1 activity
by using pharmaceutical agents, impedes the malignant behavior of
cancer cells and contributes to overcoming treatment resistance. In
this sense, ALDH1A1 may be an interesting and powerful target for
cancer therapy.
In the context of BCa, it is notable that only a
fraction of the malignant cells are expected to manifest stem-like
features, including increased expression of ALDH1A1. Therefore,
from the angle of disease prognosis, the extent of ALDH1A1
association with increased malignant behavior and drug resistance
remains to be mapped by the application of cutting-edge methods
that define the areas of the expression of biomarkers within
tumors.
Not applicable.
Funding: No funding was received.
Not applicable.
LV, PZ, DAS, VZ and SV contributed to the
conceptualization of the project, to the interpretation and
analysis of data to be included in the review, and wrote and
prepared the draft of the manuscript. Data authentication is not
applicable. All authors read and approved the final version of the
manuscript.
Not applicable.
Not applicable.
DAS is the Editor-in-Chief for the journal, but had
no personal involvement in the reviewing process, or any influence
in terms of adjudicating on the final decision, for this article.
The other authors declare that they have no competing
interests.
|
1
|
Brown G: Targeting the retinoic acid
pathway to eradicate cancer stem cells. Int J Mol Sci. 24:23732023.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Dick JE: Stem cell concepts renew cancer
research. Blood. 112:4793–4807. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hassan G and Seno M: Blood and cancer:
Cancer stem cells as origin of hematopoietic cells in solid tumor
microenvironments. Cells. 9:12932020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liu S, Cong Y, Wang D, Sun Y, Deng L, Liu
Y, Martin-Trevino R, Shang L, McDermott SP, Landis MD, et al:
Breast cancer stem cells transition between epithelial and
mesenchymal states reflective of their normal counterparts. Stem
Cell Reports. 2:78–91. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li W, Ma H, Zhang J, Zhu L, Wang C and
Yang Y: Unraveling the roles of CD44/CD24 and ALDH1 as cancer stem
cell markers in tumorigenesis and metastasis. Sci Rep. 7:138562017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kamalabadi Farahani M, Farjadmehr M,
Atashi A, Momeni A and Behzadifard M: Concise review: Breast cancer
stems cells and their role in metastases. Ann Med Surg (Lond).
86:5266–5275. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhong Y, Shen S, Zhou Y, Mao F, Guan J,
Lin Y, Xu Y and Sun Q: ALDH1 is a better clinical indicator for
relapse of invasive ductal breast cancer than the
CD44+/CD24-phenotype. Med Oncol. 31:8642014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Croker AK, Goodale D, Chu J, Postenka C,
Hedley BD, Hess DA and Allan AL: High aldehyde dehydrogenase and
expression of cancer stem cell markers selects for breast cancer
cells with enhanced malignant and metastatic ability. J Cell Mol
Med. 13:2236–2252. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Brugnoli F, Grassilli S, Al-Qassab Y,
Capitani S and Bertagnolo V: CD133 in breast cancer cells: More
than a stem cell marker. J Oncol. 2019:75126322019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xanthis V, Mantso T, Dimtsi A, Pappa A and
Fadouloglou VE: Human aldehyde dehydrogenases: A superfamily of
similar yet different proteins highly related to cancer. Cancers
(Basel). 15:44192023. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen Y, Thompson DC, Koppaka V, Jester JV
and Vasiliou V: Ocular aldehyde dehydrogenases: Protection against
ultraviolet damage and maintenance of transparency for vision. Prog
Retin Eye Res. 33:28–39. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shortall K, Djeghader A, Magner E and
Soulimane T: Insights into aldehyde dehydrogenase enzymes: A
structural perspective. Front Mol Biosci. 8:6595502021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sládek NE: Human aldehyde dehydrogenases:
Potential pathological, pharmacological, and toxicological impact.
J Biochem Mol Toxicol. 17:7–23. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
O'Brien PJ, Siraki AG and Shangari N:
Aldehyde sources, metabolism, molecular toxicity mechanisms, and
possible effects on human health. Crit Rev Toxicol. 35:609–662.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Marchitti SA, Brocker C, Stagos D and
Vasiliou V: Non-P450 aldehyde oxidizing enzymes: The aldehyde
dehydrogenase superfamily. Expert Opin Drug Metab Toxicol.
4:697–720. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ayala A, Muñoz MF and Argüelles S: Lipid
peroxidation: Production, metabolism, and signaling mechanisms of
malondialdehyde and 4-hydroxy-2-nonenal. Oxid Med Cell Longev.
2014:3604382014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sinharoy P, McAllister SL, Vasu M and
Gross ER: Environmental aldehyde sources and the health
implications of exposure. Adv Exp Med Biol. 1193:35–52. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zanoni M, Bravaccini S, Fabbri F and
Arienti C: Emerging roles of aldehyde dehydrogenase isoforms in
anti-cancer therapy resistance. Front Med (Lausanne). 9:7957622022.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jackson B, Brocker C, Thompson DC, Black
W, Vasiliou K, Nebert DW and Vasiliou V: Update on the aldehyde
dehydrogenase gene (ALDH) superfamily. Hum Genomics. 5:283–303.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Morgan CA, Parajuli B, Buchman CD, Dria K
and Hurley TD: N,N-diethylaminobenzaldehyde (DEAB) as a substrate
and mechanism-based inhibitor for human ALDH isoenzymes. Chem Biol
Interact. 234:18–28. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gomez-Salazar MA, Wang Y, Thottappillil N,
Hardy RW, Alexandre M, Höller F, Martin N, Gonzalez-Galofre ZN,
Stefancova D, Medici D, et al: Aldehyde dehydrogenase, a marker of
normal and malignant stem cells, typifies mesenchymal progenitors
in perivascular niches. Stem Cells Transl Med. 12:474–484. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ambroziak W, Izaguirre G and Pietruszko R:
Metabolism of retinaldehyde and other aldehydes in soluble extracts
of human liver and kidney. J Biol Chem. 274:33366–33373. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bui TBC, Nosaki S, Kokawa M, Xu Y,
Kitamura Y, Tanokura M, Hachimura S and Miyakawa T: Evaluation of
spice and herb as phyto-derived selective modulators of human
retinaldehyde dehydrogenases using a simple in vitro method.
Biosci Rep. 41:BSR202104912021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Vasiliou V, Pappa A and Estey T: Role of
human aldehyde dehydrogenases in endobiotic and xenobiotic
metabolism. Drug Metab Rev. 36:279–299. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Egea J, Fabregat I, Frapart YM, Ghezzi P,
Görlach A, Kietzmann T, Kubaichuk K, Knaus UG, Lopez MG,
Olaso-Gonzalez G, et al: European contribution to the study of ROS:
A summary of the findings and prospects for the future from the
COST action BM1203 (EU-ROS). Redox Biol. 13:94–162. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dinavahi SS, Bazewicz CG, Gowda R and
Robertson GP: Aldehyde dehydrogenase inhibitors for cancer
therapeutics. Trends Pharmacol Sci. 40:774–789. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xia J, Li S, Liu S and Zhang L: Aldehyde
dehydrogenase in solid tumors and other diseases: Potential
biomarkers and therapeutic targets. MedComm (2020). 4:e1952023.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lavudi K, Nuguri SM, Pandey P, Kokkanti RR
and Wang QE: ALDH and cancer stem cells: Pathways, challenges, and
future directions in targeted therapy. Life Sci. 356:1230332024.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Al-Shamma SA, Zaher DM, Hersi F, Abu Jayab
NN and Omar HA: Targeting aldehyde dehydrogenase enzymes in
combination with chemotherapy and immunotherapy: An approach to
tackle resistance in cancer cells. Life Sci. 320:1215412023.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Stagos D, Chen Y, Cantore M, Jester JV and
Vasiliou V: Corneal aldehyde dehydrogenases: multiple functions and
novel nuclear localization. Brain Res Bull. 81:211–218. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhou L, Sheng D, Wang D, Ma W, Deng Q,
Deng L and Liu S: Identification of cancer-type specific expression
patterns for active aldehyde dehydrogenase (ALDH) isoforms in
ALDEFLUOR assay. Cell Biol Toxicol. 35:161–177. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Maggio V, Cánovas V, Félix AJ, Gómez V, de
Torres I, Semidey ME, Morote J, Noé V, Ciudad CJ and Paciucci R: A
novel DNA-binding motif in prostate tumor overexpressed-1 (PTOV1)
required for the expression of ALDH1A1 and CCNG2 in cancer cells.
Cancer Lett. 452:158–167. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lei F, Zhang L, Li X, Lin X, Wu S, Li F
and Liu J: Overexpression of prostate tumor overexpressed 1
correlates with tumor progression and predicts poor prognosis in
breast cancer. BMC Cancer. 14:4572014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Qing L, Li Q and Dong Z: MUC1: An emerging
target in cancer treatment and diagnosis. Bull Cancer.
109:1202–1216. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chen W, Zhang Z, Zhang S, Zhu P, Ko JK and
Yung KK: MUC1: Structure, function, and clinic application in
epithelial cancers. Int J Mol Sci. 22:65672021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ren J, Agata N, Chen D, Li Y, Yu WH, Huang
L, Raina D, Chen W, Kharbanda S and Kufe D: Human MUC1
carcinoma-associated protein confers resistance to genotoxic
anticancer agents. Cancer Cell. 5:163–175. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Alam M, Ahmad R, Rajabi H, Kharbanda A and
Kufe D: MUC1-C oncoprotein activates ERK-C/EBPβ signaling and
induction of aldehyde dehydrogenase 1A1 in breast cancer cells. J
Biol Chem. 288:30892–30903. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Alam M, Rajabi H, Ahmad R, Jin C and Kufe
D: Targeting the MUC1-C oncoprotein inhibits self-renewal capacity
of breast cancer cells. Oncotarget. 5:2622–2634. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Jang GB, Hong IS, Kim RJ, Lee SY, Park SJ,
Lee ES, Park JH, Yun CH, Chung JU, Lee KJ, et al: Wnt/β-Catenin
Small-molecule inhibitor CWP232228 preferentially inhibits the
growth of breast cancer Stem-like cells. Cancer Res. 75:1691–1702.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
King TD, Suto MJ and Li Y: The
Wnt/β-catenin signaling pathway: A potential therapeutic target in
the treatment of triple negative breast cancer. J Cell Biochem.
113:13–18. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Cojoc M, Peitzsch C, Kurth I, Trautmann F,
Kunz-Schughart LA, Telegeev GD, Stakhovsky EA, Walker JR, Simin K,
Lyle S, et al: Aldehyde dehydrogenase is regulated by β-Catenin/TCF
and promotes radioresistance in prostate cancer progenitor cells.
Cancer Res. 75:1482–1494. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hoshino Y, Nishida J, Katsuno Y, Koinuma
D, Aoki T, Kokudo N, Miyazono K and Ehata S: Smad4 decreases the
population of pancreatic Cancer-initiating cells through
transcriptional repression of ALDH1A1. Am J Pathol. 185:1457–1470.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Varisli L and Vlahopoulos S:
Epithelial-Mesenchymal transition in acute leukemias. Int J Mol
Sci. 25:21732024. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kuburich NA, Sabapathy T, Demestichas BR,
Maddela JJ, den Hollander P and Mani SA: Proactive and reactive
roles of TGF-β in cancer. Semin Cancer Biol. 95:120–139. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Baba AB, Rah B, Bhat GR, Mushtaq I,
Parveen S, Hassan R, Hameed Zargar M and Afroze D: Transforming
growth Factor-Beta (TGF-β) signaling in Cancer-A betrayal within.
Front Pharmacol. 13:7912722022. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Varisli L, Tolan V, Cen JH, Vlahopoulos S
and Cen O: Dissecting the effects of androgen deprivation therapy
on cadherin switching in advanced prostate cancer: A molecular
perspective. Oncol Res. 30:137–155. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Yu Q, Biswas S, Ma G, Zhao P, Li B and Li
J: Canonical NF-κB signaling maintains corneal epithelial integrity
and prevents corneal aging via retinoic acid. Elife. 10:e673152021.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Pavitra E, Kancharla J, Gupta VK, Prasad
K, Sung JY, Kim J, Tej MB, Choi R, Lee JH, Han YK, et al: The role
of NF-κB in breast cancer initiation, growth, metastasis, and
resistance to chemotherapy. Biomed Pharmacother. 163:1148222023.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Vlahopoulos SA, Cen O, Hengen N, Agan J,
Moschovi M, Critselis E, Adamaki M, Bacopoulou F, Copland JA,
Boldogh I, et al: Dynamic aberrant NF-κB spurs tumorigenesis: A new
model encompassing the microenvironment. Cytokine Growth Factor
Rev. 26:389–403. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Lambrou GI, Hatziagapiou K and Vlahopoulos
S: Inflammation and tissue homeostasis: The NF-κB system in
physiology and malignant progression. Mol Biol Rep. 47:4047–4063.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Vlahopoulos SA: Divergent processing of
cell stress signals as the basis of cancer progression: Licensing
NFκB on chromatin. Int J Mol Sci. 25:86212024. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhao D, Mo Y, Li MT, Zou SW, Cheng ZL, Sun
YP, Xiong Y, Guan KL and Lei QY: NOTCH-induced aldehyde
dehydrogenase 1A1 deacetylation promotes breast cancer stem cells.
J Clin Invest. 124:5453–5465. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Wang J, Nikhil K, Viccaro K, Chang L,
White J and Shah K: Phosphorylation-dependent regulation of ALDH1A1
by Aurora kinase A: Insights on their synergistic relationship in
pancreatic cancer. BMC Biol. 15:102017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Ross AC and Moran NE: Our current dietary
reference intakes for vitamin A-Now 20 years old. Curr Dev Nutr.
4:nzaa0962020. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Surman SL, Penkert RR, Sealy RE, Jones BG,
Marion TN, Vogel P and Hurwitz JL: Consequences of vitamin a
deficiency: Immunoglobulin dysregulation, squamous cell metaplasia,
infectious disease, and death. Int J Mol Sci. 21:55702020.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Kedishvili NY: Enzymology of retinoic acid
biosynthesis and degradation. J Lipid Res. 54:1744–1760. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Belyaeva OV, Adams MK, Popov KM and
Kedishvili NY: Generation of retinaldehyde for retinoic acid
biosynthesis. Biomolecules. 10:52019. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Giguère V and Evans RM: Chronicle of a
discovery: The retinoic acid receptor. J Mol Endocrinol. 69:T1–T11.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Bastien J and Rochette-Egly C: Nuclear
retinoid receptors and the transcription of retinoid-target genes.
Gene. 328:1–16. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Jin Y, Teh SS, Lau HLN, Xiao J and Mah SH:
Retinoids as anti-cancer agents and their mechanisms of action. Am
J Cancer Res. 12:938–960. 2022.PubMed/NCBI
|
|
61
|
di Masi A, Leboffe L, De Marinis E, Pagano
F, Cicconi L, Rochette-Egly C, Lo-Coco F, Ascenzi P and Nervi C:
Retinoic acid receptors: From molecular mechanisms to cancer
therapy. Mol Aspects Med. 41:1–115. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Rastinejad F: Retinoic acid receptor
structures: The journey from single domains to full-length complex.
J Mol Endocrinol. 69:T25–T36. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Wolf G: Retinoic acid as cause of cell
proliferation or cell growth inhibition depending on activation of
one of two different nuclear receptors. Nutr Rev. 66:55–59. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Ross-Innes CS, Stark R, Holmes KA, Schmidt
D, Spyrou C, Russell R, Massie CE, Vowler SL, Eldridge M and
Carroll JS: Cooperative interaction between retinoic acid
receptor-alpha and estrogen receptor in breast cancer. Genes Dev.
24:171–182. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Piskunov A, Al Tanoury Z and Rochette-Egly
C: Nuclear and extra-nuclear effects of retinoid acid receptors:
How they are interconnected. Subcell Biochem. 70:103–127. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Sharma S, Sharma P, Bailey T, Bhattarai S,
Subedi U, Miller C, Ara H, Kidambi S, Sun H, Panchatcharam M and
Miriyala S: Electrophilic aldehyde 4-Hydroxy-2-nonenal mediated
signaling and mitochondrial dysfunction. Biomolecules. 12:15552022.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Hu W, Feng Z, Eveleigh J, Iyer G, Pan J,
Amin S, Chung FL and Tang MS: The major lipid peroxidation product,
trans-4-hydroxy-2-nonenal, preferentially forms DNA adducts at
codon 249 of human p53 gene, a unique mutational hotspot in
hepatocellular carcinoma. Carcinogenesis. 23:1781–1789. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Suman S, Kumar S, N'Gouemo P and Datta K:
Increased DNA double-strand break was associated with
downregulation of repair and upregulation of apoptotic factors in
rat hippocampus after alcohol exposure. Alcohol. 54:45–50. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Zhang Y, Wang H, Wu K and Liu Z:
Expression of 4-hydroxynonenal in esophageal squamous cell
carcinoma. Oncol Lett. 14:35–40. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Yang Y, Huycke MM, Herman TS and Wang X:
Glutathione S-transferase alpha 4 induction by activator protein 1
in colorectal cancer. Oncogene. 35:5795–5806. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Gęgotek A, Nikliński J, Žarković N,
Žarković K, Waeg G, Łuczaj W, Charkiewicz R and Skrzydlewska E:
Lipid mediators involved in the oxidative stress and antioxidant
defence of human lung cancer cells. Redox Biol. 9:210–219. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Fritz KS and Petersen DR: An overview of
the chemistry and biology of reactive aldehydes. Free Radic Biol
Med. 59:85–91. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Sener DE, Gönenç A, Akinci M and Torun M:
Lipid peroxidation and total antioxidant status in patients with
breast cancer. Cell Biochem Funct. 25:377–382. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Hassan W, Noreen H, Rehman S, Kamal MA and
da Rocha JBT: Association of oxidative stress with neurological
disorders. Curr Neuropharmacol. 20:1046–1072. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Menon B, Ramalingam K and Kumar R:
Evaluating the role of oxidative stress in acute ischemic stroke. J
Neurosci Rural Pract. 11:156–159. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Jové M, Mota-Martorell N, Pradas I,
Martín-Gari M, Ayala V and Pamplona R: The advanced lipoxidation
End-product Malondialdehyde-lysine in aging and longevity.
Antioxidants (Basel). 9:11322020. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Naudí A, Jové M, Ayala V, Cabré R,
Portero-Otín M and Pamplona R: Non-enzymatic modification of
aminophospholipids by carbonyl-amine reactions. Int J Mol Sci.
14:3285–3313. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Plastaras JP, Dedon PC and Marnett LJ:
Effects of DNA structure on oxopropenylation by the endogenous
mutagens malondialdehyde and base propenal. Biochemistry.
41:5033–5042. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Wauchope OR, Mitchener MM, Beavers WN,
Galligan JJ, Camarillo JM, Sanders WD, Kingsley PJ, Shim HN,
Blackwell T, Luong T, et al: Oxidative stress increases M1dG, a
major peroxidation-derived DNA adduct, in mitochondrial DNA.
Nucleic Acids Res. 46:3458–3467. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Feng Z, Hu W, Marnett LJ and Tang M:
Malondialdehyde, a major endogenous lipid peroxidation product,
sensitizes human cells to UV- and BPDE-induced killing and
mutagenesis through inhibition of nucleotide excision repair. Mutat
Res. 601:125–136. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Ramana KV, Srivastava S and Singhal SS:
Lipid peroxidation products in human health and disease 2016. Oxid
Med Cell Longev. 2017:21632852017. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Dancik GM, Varisli L and Vlahopoulos SA:
The molecular context of oxidant stress response in cancer
establishes ALDH1A1 as a critical target: What this means for acute
myeloid leukemia. Int J Mol Sci. 24:93722023. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Yue H, Hu Z, Hu R, Guo Z, Zheng Y, Wang Y
and Zhou Y: ALDH1A1 in cancers: Bidirectional function, drug
resistance, and regulatory mechanism. Front Oncol. 12:9187782022.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Tomita H, Tanaka K, Tanaka T and Hara A:
Aldehyde dehydrogenase 1A1 in stem cells and cancer. Oncotarget.
7:11018–11032. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Stromskaya TP, Rybalkina EY, Zabotina TN,
Shishkin AA and Stavrovskaya AA: Influence of RARalpha gene on MDR1
expression and P-glycoprotein function in human leukemic cells.
Cancer Cell Int. 5:152005. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Poturnajova M, Kozovska Z and Matuskova M:
Aldehyde dehydrogenase 1A1 and 1A3 isoforms-mechanism of activation
and regulation in cancer. Cell Signal. 87:1101202021. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Wei Y, Li Y, Chen Y, Liu P, Huang S, Zhang
Y, Sun Y, Wu Z, Hu M, Wu Q, et al: ALDH1: A potential therapeutic
target for cancer stem cells in solid tumors. Front Oncol.
12:10262782022. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Yang L, Shi P, Zhao G, Xu J, Peng W, Zhang
J, Zhang G, Wang X, Dong Z, Chen F and Cui H: Targeting cancer stem
cell pathways for cancer therapy. Signal Transduct Target Ther.
5:82020. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Ciccone V, Morbidelli L, Ziche M and
Donnini S: How to conjugate the stemness marker ALDH1A1 with tumor
angiogenesis, progression, and drug resistance. Cancer Drug Resist.
3:26–37. 2020.PubMed/NCBI
|
|
90
|
Zhou L, Jiang Y, Yan T, Di G, Shen Z, Shao
Z and Lu J: The prognostic role of cancer stem cells in breast
cancer: A meta-analysis of published literatures. Breast Cancer Res
Treat. 122:795–801. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Kim YS, Jung MJ, Ryu DW and Lee CH:
Clinicopathologic characteristics of breast cancer stem cells
identified on the basis of aldehyde dehydrogenase 1 expression. J
Breast Cancer. 17:121–128. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Miyoshi Y, Shien T, Ogiya A, Ishida N,
Yamazaki K, Horii R, Horimoto Y, Masuda N, Yasojima H, Inao T, et
al: Differences in expression of the cancer stem cell marker
aldehyde dehydrogenase 1 among estrogen receptor-positive/human
epidermal growth factor receptor type 2-negative breast cancer
cases with early, late, and no recurrence. Breast Cancer Res.
18:732016. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Li H, Ma F, Wang H, Lin C, Fan Y, Zhang X,
Qian H and Xu B: Stem cell marker aldehyde dehydrogenase 1
(ALDH1)-expressing cells are enriched in triple-negative breast
cancer. Int J Biol Markers. 28:e357–e364. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Ma F, Li H, Li Y, Ding X, Wang H, Fan Y,
Lin C, Qian H and Xu B: Aldehyde dehydrogenase 1 (ALDH1) expression
is an independent prognostic factor in triple negative breast
cancer (TNBC). Medicine (Baltimore). 96:e65612017. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Liu Y, Lv DL, Duan JJ, Xu SL, Zhang JF,
Yang XJ, Zhang X, Cui YH, Bian XW and Yu SC: ALDH1A1 expression
correlates with clinicopathologic features and poor prognosis of
breast cancer patients: A systematic review and meta-analysis. BMC
Cancer. 14:4442014. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Morimoto K, Kim SJ, Tanei T, Shimazu K,
Tanji Y, Taguchi T, Tamaki Y, Terada N and Noguchi S: Stem cell
marker aldehyde dehydrogenase 1-positive breast cancers are
characterized by negative estrogen receptor, positive human
epidermal growth factor receptor type 2, and high Ki67 expression.
Cancer Sci. 100:1062–1068. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Althobiti M, El Ansari R, Aleskandarany M,
Joseph C, Toss MS, Green AR and Rakha EA: The prognostic
significance of ALDH1A1 expression in early invasive breast cancer.
Histopathology. 77:437–448. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Kang EJ, Jung H, Woo OH, Park KH, Woo SU,
Yang DS, Kim AR, Lee JB, Kim YH, Kim JS and Seo JH: Association of
aldehyde dehydrogenase 1 expression and biologically aggressive
features in breast cancer. Neoplasma. 61:352–362. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Yao J, Jin Q, Wang XD, Zhu HJ and Ni QC:
Aldehyde dehydrogenase 1 expression is correlated with poor
prognosis in breast cancer. Medicine (Baltimore). 96:e71712017.
View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Ohi Y, Umekita Y, Yoshioka T, Souda M, Rai
Y, Sagara Y, Sagara Y, Sagara Y and Tanimoto A: Aldehyde
dehydrogenase 1 expression predicts poor prognosis in
triple-negative breast cancer. Histopathology. 59:776–780. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Nogami T, Shien T, Tanaka T, Nishiyama K,
Mizoo T, Iwamto T, Ikeda H, Taira N, Doihara H and Miyoshi S:
Expression of ALDH1 in axillary lymph node metastases is a
prognostic factor of poor clinical outcome in breast cancer
patients with 1–3 lymph node metastases. Breast Cancer. 21:58–65.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Dong Y, Bi LR, Xu N, Yang HM, Zhang HT,
Ding Y, Shi AP and Fan ZM: The expression of aldehyde dehydrogenase
1 in invasive primary breast tumors and axillary lymph node
metastases is associated with poor clinical prognosis. Pathol Res
Pract. 209:555–561. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Kong Y, Lyu N, Wu J, Tang H and Xie X,
Yang L, Li X, Wei W and Xie X: Breast cancer stem cell markers CD44
and ALDH1A1 in serum: Distribution and prognostic value in patients
with primary breast cancer. J Cancer. 9:3728–3735. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Resetkova E, Reis-Filho JS, Jain RK, Mehta
R, Thorat MA, Nakshatri H and Badve S: Prognostic impact of ALDH1
in breast cancer: A story of stem cells and tumor microenvironment.
Breast Cancer Res Treat. 123:97–108. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Honeth G, Lombardi S, Ginestier C, Hur M,
Marlow R, Buchupalli B, Shinomiya I, Gazinska P, Bombelli S,
Ramalingam V, et al: Aldehyde dehydrogenase and estrogen receptor
define a hierarchy of cellular differentiation in the normal human
mammary epithelium. Breast Cancer Res. 16:R522014. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Sarmiento-Castro A, Caamaño-Gutiérrez E,
Sims AH, Hull NJ, James MI, Santiago-Gómez A, Eyre R, Clark C,
Brown ME, Brooks MD, et al: Increased expression of Interleukin-1
receptor characterizes Anti-estrogen-Resistant ALDH+ breast cancer
stem cells. Stem Cell Reports. 15:307–316. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Wang Q, Jiang J, Ying G, Xie XQ, Zhang X,
Xu W, Zhang X, Song E, Bu H, Ping YF, et al: Tamoxifen enhances
stemness and promotes metastasis of ERα36+ breast cancer by
upregulating ALDH1A1 in cancer cells. Cell Res. 28:336–358. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Varisli L, Dancik GM, Tolan V and
Vlahopoulos S: Critical roles of SRC-3 in the development and
progression of breast cancer, rendering it a prospective clinical
target. Cancers (Basel). 15:52422023. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Brown K, Chen Y, Underhill TM, Mymryk JS
and Torchia J: The coactivator p/CIP/SRC-3 facilitates retinoic
acid receptor signaling via recruitment of GCN5. J Biol Chem.
278:39402–39412. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Ferry C, Gaouar S, Fischer B, Boeglin M,
Paul N, Samarut E, Piskunov A, Pankotai-Bodo G, Brino L and
Rochette-Egly C: Cullin 3 mediates SRC-3 ubiquitination and
degradation to control the retinoic acid response. Proc Natl Acad
Sci USA. 108:20603–20608. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Giannì M, Parrella E, Raska I Jr, Gaillard
E, Nigro EA, Gaudon C, Garattini E and Rochette-Egly C:
P38MAPK-dependent phosphorylation and degradation of SRC-3/AIB1 and
RARalpha-mediated transcription. EMBO J. 25:739–751. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Rohira AD, Yan F, Wang L, Wang J, Zhou S,
Lu A, Yu Y, Xu J, Lonard DM and O'Malley BW: Targeting SRC
coactivators blocks the Tumor-initiating capacity of cancer
stem-like cells. Cancer Res. 77:4293–4304. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Fitzgerald P, Teng M, Chandraratna RA,
Heyman RA and Allegretto EA: Retinoic acid receptor alpha
expression correlates with retinoid-induced growth inhibition of
human breast cancer cells regardless of estrogen receptor status.
Cancer Res. 57:2642–2650. 1997.PubMed/NCBI
|
|
114
|
Garattini E, Bolis M, Garattini SK,
Fratelli M, Centritto F, Paroni G, Gianni' M, Zanetti A, Pagani A,
Fisher JN, et al: Retinoids and breast cancer: From basic studies
to the clinic and back again. Cancer Treat Rev. 40:739–749. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Liu Y, Lee MO, Wang HG, Li Y, Hashimoto Y,
Klaus M, Reed JC and Zhang X: Retinoic acid receptor beta mediates
the growth-inhibitory effect of retinoic acid by promoting
apoptosis in human breast cancer cells. Mol Cell Biol.
16:1138–1149. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Centritto F, Paroni G, Bolis M, Garattini
SK, Kurosaki M, Barzago MM, Zanetti A, Fisher JN, Scott MF, Pattini
L, et al: Cellular and molecular determinants of all-trans retinoic
acid sensitivity in breast cancer: Luminal phenotype and RARα
expression. EMBO Mol Med. 7:950–972. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Roman SD, Ormandy CJ, Manning DL, Blamey
RW, Nicholson RI, Sutherland RL and Clarke CL: Estradiol induction
of retinoic acid receptors in human breast cancer cells. Cancer
Res. 53:5940–5945. 1993.PubMed/NCBI
|
|
118
|
Hua S, Kittler R and White KP: Genomic
antagonism between retinoic acid and estrogen signaling in breast
cancer. Cell. 137:2009. View Article : Google Scholar
|
|
119
|
Ombra MN, Di Santi A, Abbondanza C,
Migliaccio A, Avvedimento EV and Perillo B: Retinoic acid impairs
estrogen signaling in breast cancer cells by interfering with
activation of LSD1 via PKA. Biochim Biophys Acta. 1829:480–486.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Charafe-Jauffret E, Ginestier C, Iovino F,
Tarpin C, Diebel M, Esterni B, Houvenaeghel G, Extra JM, Bertucci
F, Jacquemier J, et al: Aldehyde dehydrogenase 1-positive cancer
stem cells mediate metastasis and poor clinical outcome in
inflammatory breast cancer. Clin Cancer Res. 16:45–55. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Croker AK, Rodriguez-Torres M, Xia Y,
Pardhan S, Leong HS, Lewis JD and Allan AL: Differential functional
roles of ALDH1A1 and ALDH1A3 in mediating metastatic behavior and
therapy resistance of human breast cancer cells. Int J Mol Sci.
18:20392017. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Pan H, Wu N, Huang Y, Li Q, Liu C, Liang
M, Zhou W, Liu X and Wang S: Aldehyde dehydrogenase 1 expression
correlates with the invasion of breast cancer. Diagn Pathol.
10:662015. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Sakakibara M, Fujimori T, Miyoshi T,
Nagashima T, Fujimoto H, Suzuki HT, Ohki Y, Fushimi K, Yokomizo J,
Nakatani Y and Miyazaki M: Aldehyde dehydrogenase 1-positive cells
in axillary lymph node metastases after chemotherapy as a
prognostic factor in patients with lymph node-positive breast
cancer. Cancer. 118:3899–3910. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Liang L and Kaufmann AM: The significance
of cancer stem cells and Epithelial-Mesenchymal transition in
metastasis and Anti-cancer therapy. Int J Mol Sci. 24:25552023.
View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Papadaki MA, Stoupis G, Theodoropoulos PA,
Mavroudis D, Georgoulias V and Agelaki S: Circulating tumor cells
with stemness and Epithelial-to-Mesenchymal transition features are
chemoresistant and predictive of poor outcome in metastatic breast
cancer. Mol Cancer Ther. 18:437–447. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Hashemi M, Arani HZ, Orouei S, Fallah S,
Ghorbani A, Khaledabadi M, Kakavand A, Tavakolpournegari A, Saebfar
H, Heidari H, et al: EMT mechanism in breast cancer metastasis and
drug resistance: Revisiting molecular interactions and biological
functions. Biomed Pharmacother. 155:1137742022. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Francou A and Anderson KV: The
Epithelial-to-Mesenchymal transition (EMT) in development and
cancer. Annu Rev Cancer Biol. 4:197–220. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Varisli L and Tolan V: Increased ROS
alters E-/N-cadherin levels and promotes migration in prostate
cancer cells. Bratisl Lek Listy. 123:752–757. 2022.PubMed/NCBI
|
|
129
|
Raimondi C, Gradilone A, Naso G, Vincenzi
B, Petracca A, Nicolazzo C, Palazzo A, Saltarelli R, Spremberg F,
Cortesi E and Gazzaniga P: Epithelial-mesenchymal transition and
stemness features in circulating tumor cells from breast cancer
patients. Breast Cancer Res Treat. 130:449–455. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Kallergi G, Papadaki MA, Politaki E,
Mavroudis D, Georgoulias V and Agelaki S: Epithelial to mesenchymal
transition markers expressed in circulating tumour cells of early
and metastatic breast cancer patients. Breast Cancer Res.
13:R592011. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Papadaki MA, Kallergi G, Zafeiriou Z,
Manouras L, Theodoropoulos PA, Mavroudis D, Georgoulias V and
Agelaki S: Co-expression of putative stemness and
epithelial-to-mesenchymal transition markers on single circulating
tumour cells from patients with early and metastatic breast cancer.
BMC Cancer. 14:6512014. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Vesuna F, Lisok A, Kimble B and Raman V:
Twist modulates breast cancer stem cells by transcriptional
regulation of CD24 expression. Neoplasia. 11:1318–1328. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Ito M, Shien T, Omori M, Mizoo T, Iwamoto
T, Nogami T, Motoki T, Taira N, Doihara H and Miyoshi S: Evaluation
of aldehyde dehydrogenase 1 and transcription factors in both
primary breast cancer and axillary lymph node metastases as a
prognostic factor. Breast Cancer. 23:437–444. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Ciccone V, Terzuoli E, Donnini S,
Giachetti A, Morbidelli L and Ziche M: Stemness marker ALDH1A1
promotes tumor angiogenesis via retinoic acid/HIF-1α/VEGF
signalling in MCF-7 breast cancer cells. J Exp Clin Cancer Res.
37:3112018. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
DA Cruz Paula A, Marques O, Sampaio R,
Rosa A, Garcia J, Rêma A, DE Fátima Faria M, Silva P, Vizcaíno R
and Lopes C: Characterization of CD44+ALDH1+Ki-67-Cells in
Non-malignant and neoplastic lesions of the breast. Anticancer Res.
36:4629–4638. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Walter D, Lier A, Geiselhart A, Thalheimer
FB, Huntscha S, Sobotta MC, Moehrle B, Brocks D, Bayindir I,
Kaschutnig P, et al: Exit from dormancy provokes DNA-damage-induced
attrition in haematopoietic stem cells. Nature. 520:549–552. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Lynch J, Troadec E, Fung TK, Gladysz K,
Virely C, Lau PNI, Cheung N, Zeisig B, Wong JWH, Lopes M, et al:
Hematopoietic stem cell quiescence and DNA replication dynamics
maintained by the resilient β-catenin/Hoxa9/Prmt1 axis. Blood.
143:1586–1598. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Xu J, Fei P, Simon DW, Morowitz MJ, Mehta
PA and Du W: Crosstalk between DNA damage repair and metabolic
regulation in hematopoietic stem cells. Cells. 13:7332024.
View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Becker F, Ouzin M, Liedtke S, Raba K and
Kogler G: DNA damage response after treatment of cycling and
quiescent cord blood hematopoietic stem cells with distinct
genotoxic noxae. Stem Cells. 42:158–171. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Pallis M, Burrows F, Whittall A, Boddy N,
Seedhouse C and Russell N: Efficacy of RNA polymerase II inhibitors
in targeting dormant leukaemia cells. BMC Pharmacol Toxicol.
14:322013. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Min HY and Lee HY: Cellular dormancy in
cancer: Mechanisms and potential targeting strategies. Cancer Res
Treat. 55:720–736. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Goldman MJ, Craft B, Hastie M, Repečka K,
McDade F, Kamath A, Banerjee A, Luo Y, Rogers D, Brooks AN, et al:
Visualizing and interpreting cancer genomics data via the Xena
platform. Nat Biotechnol. 38:675–678. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Vlahopoulos S, Pan L, Varisli L, Dancik
GM, Karantanos T and Boldogh I: OGG1 as an epigenetic reader
affects NFκB: What this means for cancer. Cancers (Basel).
16:1482023. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Bidan N, Bailleul-Dubois J, Duval J,
Winter M, Denoulet M, Hannebicque K, El-Sayed IY, Ginestier C,
Forissier V, Charafe-Jauffret E, et al: Transcriptomic analysis of
breast cancer stem cells and development of a pALDH1A1:mNeptune
reporter system for live tracking. Proteomics. 19:e18004542019.
View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Enikeev AD, Abramov PM, Elkin DS, Komelkov
AV, Beliaeva AA, Silantieva DM and Tchevkina EM: Opposite effects
of CRABP1 and CRABP2 homologs on proliferation of breast cancer
cells and their sensitivity to retinoic acid. Biochemistry (Mosc).
88:2107–2124. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Brown G: Deregulation of All-trans
retinoic acid signaling and development in cancer. Int J Mol Sci.
24:120892023. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Patel M, Lu L, Zander DS, Sreerama L, Coco
D and Moreb JS: ALDH1A1 and ALDH3A1 expression in lung cancers:
Correlation with histologic type and potential precursors. Lung
Cancer. 59:340–349. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Li Z, Xiang Y, Xiang L, Xiao Y, Li F and
Hao P: ALDH maintains the stemness of lung adenoma stem cells by
suppressing the Notch/CDK2/CCNE pathway. PLoS One. 9:e926692014.
View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Yassin Fel-Z: Aldehyde dehyderogenase
(ALDH1A1) delineating the normal and cancer stem cells in spectral
lung lesions: An immunohistochemical appraisal. Pathol Res Pract.
212:398–409. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Wei Y, Wu S, Xu W, Liang Y, Li Y, Zhao W
and Wu J: Depleted aldehyde dehydrogenase 1A1 (ALDH1A1) reverses
cisplatin resistance of human lung adenocarcinoma cell A549/DDP.
Thorac Cancer. 8:26–32. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Liu X, Wang L, Cui W, Yuan X, Lin L, Cao
Q, Wang N, Li Y, Guo W, Zhang X, et al: Targeting ALDH1A1 by
disulfiram/copper complex inhibits non-small cell lung cancer
recurrence driven by ALDH-positive cancer stem cells. Oncotarget.
7:58516–58530. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Gao F, Zhou B, Xu JC, Gao X, Li SX, Zhu
GC, Zhang XG and Yang C: The role of LGR5 and ALDH1A1 in non-small
cell lung cancer: Cancer progression and prognosis. Biochem Biophys
Res Commun. 462:91–98. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Alamgeer M, Ganju V, Szczepny A, Russell
PA, Prodanovic Z, Kumar B, Wainer Z, Brown T, Schneider-Kolsky M,
Conron M, et al: The prognostic significance of aldehyde
dehydrogenase 1A1 (ALDH1A1) and CD133 expression in early stage
non-small cell lung cancer. Thorax. 68:1095–1104. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Li D, Cao Y, Luo CW, Zhang LP and Zou YB:
The clinical significance and prognostic value of ALDH1 expression
in non-small cell lung cancer: A systematic review and
meta-analysis. Recent Pat Anticancer Drug Discov. 19:599–609. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Biswas AK, Han S, Tai Y, Ma W, Coker C,
Quinn SA, Shakri AR, Zhong TJ, Scholze H, Lagos GG, et al:
Targeting S100A9-ALDH1A1-Retinoic acid signaling to suppress brain
relapse in EGFR-mutant lung cancer. Cancer Discov. 12:1002–1021.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Okudela K, Woo T, Mitsui H, Suzuki T,
Tajiri M, Sakuma Y, Miyagi Y, Tateishi Y, Umeda S, Masuda M and
Ohashi K: Downregulation of ALDH1A1 expression in non-small cell
lung carcinomas-its clinicopathologic and biological significance.
Int J Clin Exp Pathol. 6:1–12. 2013.PubMed/NCBI
|
|
157
|
Yamashita N, So T, Miyata T, Yoshimatsu T,
Nakano R, Oyama T, Matsunaga W and Gotoh A: Triple-negative
expression (ALDH1A1-/CD133-/mutant p53-) cases in lung
adenocarcinoma had a good prognosis. Sci Rep. 12:14732022.
View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Pelos G, Riester M, Pal J, Myacheva K,
Moneke I, Rotondo JC, Lübbert M and Diederichs S: Fast
proliferating and slowly migrating non-small cell lung cancer cells
are vulnerable to decitabine and retinoic acid combinatorial
treatment. Int J Cancer. 154:1029–1042. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Zito G, Naselli F, Saieva L, Raimondo S,
Calabrese G, Guzzardo C, Forte S, Rolfo C, Parenti R and Alessandro
R: Retinoic Acid affects Lung adenocarcinoma growth by inducing
differentiation via GATA6 activation and EGFR and Wnt inhibition.
Sci Rep. 7:47702017. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Li D, Sun J, Liu W, Wang X, Bals R, Wu J,
Quan W, Yao Y, Zhang Y, Zhou H and Wu K: Rig-G is a growth
inhibitory factor of lung cancer cells that suppresses STAT3 and
NF-κB. Oncotarget. 7:66032–66050. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
161
|
Rehó B, Fadel L, Brazda P, Benziane A,
Hegedüs É, Sen P, Gadella TWJ, Tóth K, Nagy L and Vámosi G:
Agonist-controlled competition of RAR and VDR nuclear receptors for
heterodimerization with RXR is manifested in their DNA binding. J
Biol Chem. 299:1028962023. View Article : Google Scholar : PubMed/NCBI
|
|
162
|
López-Fandiño R, Molina E and
Lozano-Ojalvo D: Intestinal factors promoting the development of
RORγt+ cells and oral tolerance. Front Immunol. 14:12942922023.
View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Le Magnen C, Bubendorf L, Rentsch CA,
Mengus C, Gsponer J, Zellweger T, Rieken M, Thalmann GN, Cecchini
MG, Germann M, et al: Characterization and clinical relevance of
ALDHbright populations in prostate cancer. Clin Cancer Res.
19:5361–5371. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
164
|
Gorodetska I, Offermann A, Püschel J,
Lukiyanchuk V, Gaete D, Kurzyukova A, Freytag V, Haider MT, Fjeldbo
CS, Di Gaetano S, et al: ALDH1A1 drives prostate cancer metastases
and radioresistance by interplay with AR- and RAR-dependent
transcription. Theranostics. 14:714–737. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Nastały P, Filipska M, Morrissey C, Eltze
E, Semjonow A, Brandt B, Pantel K and Bednarz-Knoll N:
ALDH1-positive intratumoral stromal cells indicate differentiated
epithelial-like phenotype and good prognosis in prostate cancer.
Transl Res. 203:49–56. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Liu Z, Ren G, Shangguan C, Guo L, Dong Z,
Li Y, Zhang W, Zhao L, Hou P, Zhang Y, et al: ATRA inhibits the
proliferation of DU145 prostate cancer cells through reducing the
methylation level of HOXB13 gene. PLoS One. 7:e409432012.
View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Landen CN Jr, Goodman B, Katre AA, Steg
AD, Nick AM, Stone RL, Miller LD, Mejia PV, Jennings NB, Gershenson
DM, et al: Targeting aldehyde dehydrogenase cancer stem cells in
ovarian cancer. Mol Cancer Ther. 9:3186–3199. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Izycka N, Rucinski M, Andrzejewska M,
Szubert S, Nowak-Markwitz E and Sterzynska K: The prognostic value
of cancer stem cell markers (CSCs) Expression-ALDH1A1, CD133,
CD44-For survival and long-term follow-up of ovarian cancer
patients. Int J Mol Sci. 24:24002023. View Article : Google Scholar : PubMed/NCBI
|
|
169
|
Meng E, Mitra A, Tripathi K, Finan MA,
Scalici J, McClellan S, Madeira da Silva L, Reed E, Shevde LA,
Palle K and Rocconi RP: ALDH1A1 maintains ovarian cancer stem
cell-like properties by altered regulation of cell cycle checkpoint
and DNA repair network signaling. PLoS One. 9:e1071422014.
View Article : Google Scholar : PubMed/NCBI
|
|
170
|
Kaipio K, Chen P, Roering P, Huhtinen K,
Mikkonen P, Östling P, Lehtinen L, Mansuri N, Korpela T, Potdar S,
et al: ALDH1A1-related stemness in high-grade serous ovarian cancer
is a negative prognostic indicator but potentially targetable by
EGFR/mTOR-PI3K/aurora kinase inhibitors. J Pathol. 250:159–169.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
171
|
Januchowski R, Wojtowicz K, Sterzyſska K,
Sosiſska P, Andrzejewska M, Zawierucha P, Nowicki M and Zabel M:
Inhibition of ALDH1A1 activity decreases expression of drug
transporters and reduces chemotherapy resistance in ovarian cancer
cell lines. Int J Biochem Cell Biol. 78:248–259. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
172
|
Nowacka M, Ginter-Matuszewska B,
Świerczewska M, Sterzyńska K, Nowicki M and Januchowski R: Effect
of ALDH1A1 gene knockout on drug resistance in paclitaxel and
topotecan resistant human ovarian cancer cell lines in 2D and 3D
model. Int J Mol Sci. 23:30362022. View Article : Google Scholar : PubMed/NCBI
|
|
173
|
Muralikrishnan V, Fang F, Given TC,
Podicheti R, Chtcherbinine M, Metcalfe TX, Sriramkumar S, O'Hagan
HM, Hurley TD and Nephew KP: A novel ALDH1A1 inhibitor blocks
platinum-induced senescence and stemness in ovarian cancer. Cancers
(Basel). 14:34372022. View Article : Google Scholar : PubMed/NCBI
|
|
174
|
Sharbatoghli M, Shamshiripour P, Fattahi
F, Kalantari E, Habibi Shams Z, Panahi M, Totonchi M, Asadi-Lari Z,
Madjd Z and Saeednejad Zanjani L: Co-expression of cancer stem cell
markers, SALL4/ALDH1A1, is associated with tumor aggressiveness and
poor survival in patients with serous ovarian carcinoma. J Ovarian
Res. 15:172022. View Article : Google Scholar : PubMed/NCBI
|
|
175
|
Dancik GM, Voutsas IF and Vlahopoulos S:
Lower RNA expression of ALDH1A1 distinguishes the favorable risk
group in acute myeloid leukemia. Mol Biol Rep. 49:3321–3331. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
176
|
Gasparetto M, Pei S, Minhajuddin M, Khan
N, Pollyea DA, Myers JR, Ashton JM, Becker MW, Vasiliou V,
Humphries KR, et al: Targeted therapy for a subset of acute myeloid
leukemias that lack expression of aldehyde dehydrogenase 1A1.
Haematologica. 102:1054–1065. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
177
|
Venton G, Pérez-Alea M, Baier C, Fournet
G, Quash G, Labiad Y, Martin G, Sanderson F, Poullin P, Suchon P,
et al: Aldehyde dehydrogenases inhibition eradicates leukemia stem
cells while sparing normal progenitors. Blood Cancer J. 6:e4692016.
View Article : Google Scholar : PubMed/NCBI
|
|
178
|
Rebollido-Rios R, Venton G,
Sánchez-Redondo S, Iglesias I, Felip C, Fournet G, González E,
Romero Fernández W, Borroto Escuela DO, Di Stefano B,
Penarroche-Díaz R, et al: Dual disruption of aldehyde
dehydrogenases 1 and 3 promotes functional changes in the
glutathione redox system and enhances chemosensitivity in nonsmall
cell lung cancer. Oncogene. 39:2756–2771. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
179
|
Law R: Advanced BioDesign releases
positive data from ODYSSEY AML study. Clinical Trials Arena. June
6–2024.
|
|
180
|
Venton G, Colle J, Tichadou A, Quessada J,
Baier C, Labiad Y, Perez M, De Lassus L, Loosveld M, Arnoux I, et
al: Reactive oxygen species and aldehyde dehydrogenase 1A as
prognosis and theragnostic biomarker in acute myeloid leukaemia
patients. J Cell Mol Med. 28:e700112024. View Article : Google Scholar : PubMed/NCBI
|
|
181
|
Yasgar A, Titus SA, Wang Y, Danchik C,
Yang SM, Vasiliou V, Jadhav A, Maloney DJ, Simeonov A and Martinez
NJ: A High-content assay enables the automated screening and
identification of small molecules with specific ALDH1A1-inhibitory
activity. PLoS One. 12:e01709372017. View Article : Google Scholar : PubMed/NCBI
|
|
182
|
Xu B, Wang S, Li R, Chen K, He L, Deng M,
Kannappan V, Zha J, Dong H and Wang W: Disulfiram/copper
selectively eradicates AML leukemia stem cells in vitro and in vivo
by simultaneous induction of ROS-JNK and inhibition of NF-κB and
Nrf2. Cell Death Dis. 8:e27972017. View Article : Google Scholar : PubMed/NCBI
|
|
183
|
Dancik GM, Voutsas IF and Vlahopoulos S:
Aldehyde dehydrogenase enzyme functions in acute leukemia stem
cells. Front Biosci (Schol Ed). 14:82022. View Article : Google Scholar : PubMed/NCBI
|
|
184
|
Ghiaur G, Yegnasubramanian S, Perkins B,
Gucwa JL, Gerber JM and Jones RJ: Regulation of human hematopoietic
stem cell self-renewal by the microenvironment's control of
retinoic acid signaling. Proc Natl Acad Sci USA. 110:16121–16126.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
185
|
Alonso S, Jones RJ and Ghiaur G: Retinoic
acid, CYP26, and drug resistance in the stem cell niche. Exp
Hematol. 54:17–25. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
186
|
Bunaciu RP, MacDonald RJ, Gao F, Johnson
LM, Varner JD, Wang X, Nataraj S, Guzman ML and Yen A: Potential
for subsets of wt-NPM1 primary AML blasts to respond to retinoic
acid treatment. Oncotarget. 9:4134–4149. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
187
|
McGinn O, Riley D, Finlay-Schultz J, Paul
KV, Kabos P and Sartorius CA: Cytokeratins 5 and 17 maintain an
aggressive epithelial state in basal-like breast cancer. Mol Cancer
Res. 20:1443–1455. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
188
|
Yang Y, Zhou W, Xia J, Gu Z, Wendlandt E,
Zhan X, Janz S, Tricot G and Zhan F: NEK2 mediates
ALDH1A1-dependent drug resistance in multiple myeloma. Oncotarget.
5:11986–11997. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
189
|
Xia J, He Y, Meng B, Chen S, Zhang J, Wu
X, Zhu Y, Shen Y, Feng X, Guan Y, et al: NEK2 induces
autophagy-mediated bortezomib resistance by stabilizing Beclin-1 in
multiple myeloma. Mol Oncol. 14:763–778. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
190
|
Xing Z, Zhang M and Wang X, Liu J, Liu G,
Feng K and Wang X: Silencing of Nek2 suppresses the proliferation,
migration and invasion and induces apoptosis of breast cancer cells
by regulating ERK/MAPK signaling. J Mol Histol. 52:809–821. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
191
|
Szafarowski T, Sierdziński J, Ludwig N,
Głuszko A, Filipowska A and Szczepański MJ: Assessment of cancer
stem cell marker expression in primary head and neck squamous cell
carcinoma shows prognostic value for aldehyde dehydrogenase
(ALDH1A1). Eur J Pharmacol. 867:1728372020. View Article : Google Scholar : PubMed/NCBI
|
|
192
|
Gupta V, Maurya MK, Agarwal P, Kumar M,
Sagar M, Raghuvanshi S and Gupta S: Expression of aldehyde
dehydrogenase 1A1 in oral squamous cell carcinoma and its
correlation with clinicopathological parameters. Natl J Maxillofac
Surg. 13:208–215. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
193
|
Namekawa T, Ikeda K, Horie-Inoue K, Suzuki
T, Okamoto K, Ichikawa T, Yano A, Kawakami S and Inoue S: ALDH1A1
in patient-derived bladder cancer spheroids activates retinoic acid
signaling leading to TUBB3 overexpression and tumor progression.
Int J Cancer. 146:1099–1113. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
194
|
Li X, Xu Q, Fu X and Luo W: ALDH1A1
overexpression is associated with the progression and prognosis in
gastric cancer. BMC Cancer. 14:7052014. View Article : Google Scholar : PubMed/NCBI
|
|
195
|
van der Waals LM, Borel Rinkes IHM and
Kranenburg O: ALDH1A1 expression is associated with poor
differentiation, ‘right-sidedness’ and poor survival in human
colorectal cancer. PLoS One. 13:e02055362018. View Article : Google Scholar : PubMed/NCBI
|
|
196
|
Yang L, Ren Y, Yu X, Qian F, Bian BS, Xiao
HL, Wang WG, Xu SL, Yang J, Cui W, et al: ALDH1A1 defines invasive
cancer stem-like cells and predicts poor prognosis in patients with
esophageal squamous cell carcinoma. Mod Pathol. 27:775–783. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
197
|
Yehya A, Youssef J, Hachem S, Ismael J and
Abou-Kheir W: Tissue-specific cancer stem/progenitor cells:
Therapeutic implications. World J Stem Cells. 15:323–341. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
198
|
Sládek NE, Kollander R, Sreerama L and
Kiang DT: Cellular levels of aldehyde dehydrogenases (ALDH1A1 and
ALDH3A1) as predictors of therapeutic responses to
cyclophosphamide-based chemotherapy of breast cancer: A
retrospective study. Rational individualization of
oxazaphosphorine-based cancer chemotherapeutic regimens. Cancer
Chemother Pharmacol. 49:309–321. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
199
|
Khoury T, Ademuyiwa FO, Chandrasekhar R,
Jabbour M, Deleo A, Ferrone S, Wang Y and Wang X: Aldehyde
dehydrogenase 1A1 expression in breast cancer is associated with
stage, triple negativity, and outcome to neoadjuvant chemotherapy.
Mod Pathol. 25:388–397. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
200
|
Sun M, Zhao H, Xiao Q, Yu Z, Song Z, Yao
W, Tang H, Guan S, Jin F and Wei M: Combined expression of aldehyde
dehydrogenase 1A1 and β-catenin is associated with lymph node
metastasis and poor survival in breast cancer patients following
cyclophosphamide treatment. Oncol Rep. 34:3163–3173. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
201
|
Narendra G, Raju B, Verma H, Kumar M, Jain
SK, Tung GK, Thakur S, Kaur R, Kaur S, Sapra B and Silakari O:
Scaffold hopping based designing of selective ALDH1A1 inhibitors to
overcome cyclophosphamide resistance: Synthesis and biological
evaluation. RSC Med Chem. 15:309–321. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
202
|
Wang D and Wang H: Oxazaphosphorine
bioactivation and detoxification The role of xenobiotic receptors.
Acta Pharm Sin B. 2:10.1016/j.apsb.2012.02.004. 2012. View Article : Google Scholar
|
|
203
|
Paul SK, Guendouzi A, Banerjee A,
Guendouzi A and Haldar R: Identification of approved drugs with
ALDH1A1 inhibitory potential aimed at enhancing chemotherapy
sensitivity in cancer cells: An in-silico drug repurposing
approach. J Biomol Struct Dyn. 1–15. 2024.doi:
10.1080/07391102.2023.2300127 (Epub ahead of print). View Article : Google Scholar
|
|
204
|
Lu C, Li X, Ren Y and Zhang X: Disulfiram:
A novel repurposed drug for cancer therapy. Cancer Chemother
Pharmacol. 87:159–172. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
205
|
Yip NC, Fombon IS, Liu P, Brown S,
Kannappan V, Armesilla AL, Xu B, Cassidy J, Darling JL and Wang W:
Disulfiram modulated ROS-MAPK and NFκB pathways and targeted breast
cancer cells with cancer stem cell-like properties. Br J Cancer.
104:1564–1574. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
206
|
Kim JY, Cho Y, Oh E, Lee N, An H, Sung D,
Cho TM and Seo JH: Disulfiram targets cancer stem-like properties
and the HER2/Akt signaling pathway in HER2-positive breast cancer.
Cancer Lett. 379:39–48. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
207
|
Kim YJ, Kim JY, Lee N, Oh E, Sung D, Cho
TM and Seo JH: Disulfiram suppresses cancer stem-like properties
and STAT3 signaling in triple-negative breast cancer cells. Biochem
Biophys Res Commun. 486:1069–1076. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
208
|
Lin L, Hutzen B, Lee HF, Peng Z, Wang W,
Zhao C, Lin HJ, Sun D, Li PK, Li C, et al: Evaluation of STAT3
signaling in ALDH+ and ALDH+/CD44+/CD24-subpopulations of breast
cancer cells. PLoS One. 8:e828212013. View Article : Google Scholar : PubMed/NCBI
|
|
209
|
Nourbakhsh M, Farzaneh S, Taghikhani A,
Zarghi A and Noori S: The effect of a newly synthesized ferrocene
derivative against MCF-7 breast cancer cells and spheroid stem
cells through ROS production and inhibition of JAK2/STAT3 signaling
pathway. Anticancer Agents Med Chem. 20:875–886. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
210
|
Tsao AN, Chuang YS, Lin YC, Su Y and Chao
TC: Dinaciclib inhibits the stemness of two subtypes of human
breast cancer cells by targeting the FoxM1 and Hedgehog signaling
pathway. Oncol Rep. 47:1052022. View Article : Google Scholar : PubMed/NCBI
|
|
211
|
Teng CJ, Cheng PT, Cheng YC, Tsai JR, Chen
MC and Lin H: Dinaciclib inhibits the growth of acute myeloid
leukemia cells through either cell cycle-related or ERK1/STAT3/MYC
pathways. Toxicol In Vitro. 96:1057682024. View Article : Google Scholar : PubMed/NCBI
|
|
212
|
Liu C, Dong L, Sun Z, Wang L, Wang Q, Li
H, Zhang J and Wang X: Esculentoside A suppresses breast cancer
stem cell growth through stemness attenuation and apoptosis
induction by blocking IL-6/STAT3 signaling pathway. Phytother Res.
32:2299–2311. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
213
|
Simões BM, O'Brien CS, Eyre R, Silva A, Yu
L, Sarmiento-Castro A, Alférez DG, Spence K, Santiago-Gómez A,
Chemi F, et al: Anti-estrogen resistance in human breast tumors is
driven by JAG1-NOTCH4-dependent cancer stem cell activity. Cell
Rep. 12:1968–1977. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
214
|
Notas G, Pelekanou V, Kampa M, Alexakis K,
Sfakianakis S, Laliotis A, Askoxilakis J, Tsentelierou E, Tzardi M,
Tsapis A and Castanas E: Tamoxifen induces a pluripotency signature
in breast cancer cells and human tumors. Mol Oncol. 9:1744–1759.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
215
|
Li J, Zhang B, Yang YF, Jin J and Liu YH:
Aldehyde dehydrogenase 1 as a predictor of the neoadjuvant
chemotherapy response in breast cancer: A meta-analysis. Medicine
(Baltimore). 97:e120562018. View Article : Google Scholar : PubMed/NCBI
|
|
216
|
Alamgeer M, Ganju V, Kumar B, Fox J, Hart
S, White M, Harris M, Stuckey J, Prodanovic Z, Schneider-Kolsky ME
and Watkins DN: Changes in aldehyde dehydrogenase-1 expression
during neoadjuvant chemotherapy predict outcome in locally advanced
breast cancer. Breast Cancer Res. 16:R442014. View Article : Google Scholar : PubMed/NCBI
|
|
217
|
Lee A, Won KY, Lim SJ, Cho SY, Han SA,
Park S and Song JY: ALDH1 and tumor infiltrating lymphocytes as
predictors for neoadjuvant chemotherapy response in breast cancer.
Pathol Res Pract. 214:619–624. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
218
|
Allison SE, Chen Y, Petrovic N, Zhang J,
Bourget K, Mackenzie PI and Murray M: Activation of ALDH1A1 in
MDA-MB-468 breast cancer cells that over-express CYP2J2 protects
against paclitaxel-dependent cell death mediated by reactive oxygen
species. Biochem Pharmacol. 143:79–89. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
219
|
Kakarala M, Brenner DE, Korkaya H, Cheng
C, Tazi K, Ginestier C, Liu S, Dontu G and Wicha MS: Targeting
breast stem cells with the cancer preventive compounds curcumin and
piperine. Breast Cancer Res Treat. 122:777–785. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
220
|
Kesharwani RK, Srivastava V, Singh P,
Rizvi SI, Adeppa K and Misra K: A novel approach for overcoming
drug resistance in breast cancer chemotherapy by targeting new
synthetic curcumin analogues against aldehyde dehydrogenase 1
(ALDH1A1) and glycogen synthase kinase-3 β (GSK-3β). Appl Biochem
Biotechnol. 176:1996–2017. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
221
|
Li X, Wang X, Xie C, Zhu J, Meng Y, Chen
Y, Li Y, Jiang Y, Yang X, Wang S, et al: Sonic hedgehog and
Wnt/β-catenin pathways mediate curcumin inhibition of breast cancer
stem cells. Anticancer Drugs. 29:208–215. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
222
|
Attia YM, El-Kersh DM, Ammar RA, Adel A,
Khalil A, Walid H, Eskander K, Hamdy M, Reda N, Mohsen NE, et al:
Inhibition of aldehyde dehydrogenase-1 and p-glycoprotein-mediated
multidrug resistance by curcumin and vitamin D3 increases
sensitivity to paclitaxel in breast cancer. Chem Biol Interact.
315:1088652020. View Article : Google Scholar : PubMed/NCBI
|
|
223
|
Tanei T, Morimoto K, Shimazu K, Kim SJ,
Tanji Y, Taguchi T, Tamaki Y and Noguchi S: Association of breast
cancer stem cells identified by aldehyde dehydrogenase 1 expression
with resistance to sequential Paclitaxel and epirubicin-based
chemotherapy for breast cancers. Clin Cancer Res. 15:4234–4241.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
224
|
Wang R, Yang L, Li S, Ye D, Yang L, Liu Q,
Zhao Z, Cai Q, Tan J and Li X: Quercetin inhibits breast cancer
stem cells via downregulation of aldehyde dehydrogenase 1A1
(ALDH1A1), chemokine receptor type 4 (CXCR4), Mucin 1 (MUC1), and
epithelial cell adhesion molecule (EpCAM). Med Sci Monit.
24:412–420. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
225
|
Castro NP, Rangel MC, Merchant AS,
MacKinnon G, Cuttitta F, Salomon DS and Kim YS: Sulforaphane
suppresses the growth of triple-negative breast cancer stem-like
cells in vitro and in vivo. Cancer Prev Res (Phila). 12:147–158.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
226
|
Li Y, Zhang T, Korkaya H, Liu S, Lee HF,
Newman B, Yu Y, Clouthier SG, Schwartz SJ, Wicha MS and Sun D:
Sulforaphane, a dietary component of broccoli/broccoli sprouts,
inhibits breast cancer stem cells. Clin Cancer Res. 16:2580–2590.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
227
|
Leung HW, Ko CH, Yue GL, Herr I and Lau
CS: The natural agent 4-vinylphenol targets metastasis and stemness
features in breast cancer stem-like cells. Cancer Chemother
Pharmacol. 82:185–197. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
228
|
Wang W, He S, Zhang R, Peng J, Guo D,
Zhang J, Xiang B and Li L: ALDH1A1 maintains the cancer stem-like
cells properties of esophageal squamous cell carcinoma by
activating the AKT signal pathway and interacting with β-catenin.
Biomed Pharmacother. 125:1099402020. View Article : Google Scholar : PubMed/NCBI
|
|
229
|
Jiang Y, Song H, Jiang L, Qiao Y, Yang D,
Wang D and Li J: Silybin prevents prostate cancer by inhibited the
ALDH1A1 expression in the retinol metabolism pathway. Front Cell
Dev Biol. 8:5743942020. View Article : Google Scholar : PubMed/NCBI
|
|
230
|
Scambia G, De Vincenzo R, Ranelletti FO,
Panici PB, Ferrandina G, D'Agostino G, Fattorossi A, Bombardelli E
and Mancuso S: Antiproliferative effect of silybin on
gynaecological malignancies: Synergism with cisplatin and
doxorubicin. Eur J Cancer. 32A:877–882. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
231
|
Bhatia N, Zhao J, Wolf DM and Agarwal R:
Inhibition of human carcinoma cell growth and DNA synthesis by
silibinin, an active constituent of milk thistle: Comparison with
silymarin. Cancer Lett. 147:77–84. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
232
|
Bian Y, Shan G, Bi G, Liang J, Hu Z, Sui
Q, Shi H, Zheng Z, Yao G, Wang Q, et al: Targeting ALDH1A1 to
enhance the efficacy of KRAS-targeted therapy through ferroptosis.
Redox Biol. 77:1033612024. View Article : Google Scholar : PubMed/NCBI
|
|
233
|
Galiè M: RAS as supporting actor in breast
cancer. Front Oncol. 9:11992019. View Article : Google Scholar : PubMed/NCBI
|
|
234
|
Tao S, Wang S, Moghaddam SJ, Ooi A,
Chapman E, Wong PK and Zhang DD: Oncogenic KRAS confers
chemoresistance by upregulating NRF2. Cancer Res. 74:7430–7441.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
235
|
Kim D, Choi BH, Ryoo IG and Kwak MK: High
NRF2 level mediates cancer stem cell-like properties of aldehyde
dehydrogenase (ALDH)-high ovarian cancer cells: Inhibitory role of
all-trans retinoic acid in ALDH/NRF2 signaling. Cell Death Dis.
9:8962018. View Article : Google Scholar : PubMed/NCBI
|
|
236
|
Piao S, Ojha R, Rebecca VW, Samanta A, Ma
XH, Mcafee Q, Nicastri MC, Buckley M, Brown E, Winkler JD, et al:
ALDH1A1 and HLTF modulate the activity of lysosomal autophagy
inhibitors in cancer cells. Autophagy. 13:2056–2071. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
237
|
Varisli L, Cen O and Vlahopoulos S:
Dissecting pharmacological effects of chloroquine in cancer
treatment: Interference with inflammatory signaling pathways.
Immunology. 159:257–278. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
238
|
Vlahopoulos S, Critselis E, Voutsas IF,
Perez SA, Moschovi M, Baxevanis CN and Chrousos GP: New use for old
drugs? Prospective targets of chloroquines in cancer therapy. Curr
Drug Targets. 15:843–851. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
239
|
Liang DH, Choi DS, Ensor JE, Kaipparettu
BA, Bass BL and Chang JC: The autophagy inhibitor chloroquine
targets cancer stem cells in triple negative breast cancer by
inducing mitochondrial damage and impairing DNA break repair.
Cancer Lett. 376:249–258. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
240
|
Stagni V, Kaminari A, Sideratou Z,
Sakellis E, Vlahopoulos SA and Tsiourvas D: Targeting breast cancer
stem-like cells using chloroquine encapsulated by a
triphenylphosphonium-functionalized hyperbranched polymer. Int J
Pharm. 585:1194652020. View Article : Google Scholar : PubMed/NCBI
|
|
241
|
Panagiotaki KN, Sideratou Z, Vlahopoulos
SA, Paravatou-Petsotas M, Zachariadis M, Khoury N, Zoumpourlis V
and Tsiourvas D: A Triphenylphosphonium-functionalized
mitochondriotropic nanocarrier for efficient co-delivery of
doxorubicin and chloroquine and enhanced antineoplastic activity.
Pharmaceuticals (Basel). 10:912017. View Article : Google Scholar : PubMed/NCBI
|
|
242
|
Visus C, Wang Y, Lozano-Leon A, Ferris RL,
Silver S, Szczepanski MJ, Brand RE, Ferrone CR, Whiteside TL,
Ferrone S, et al: Targeting ALDH(bright) human carcinoma-initiating
cells with ALDH1A1-specific CD8+ T cells. Clin Cancer
Res. 17:6174–6184. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
243
|
Liu C, Qiang J, Deng Q, Xia J, Deng L,
Zhou L, Wang D, He X, Liu Y, Zhao B, et al: ALDH1A1 activity in
tumor-initiating cells remodels myeloid-derived suppressor cells to
promote breast cancer progression. Cancer Res. 81:5919–5934. 2021.
View Article : Google Scholar : PubMed/NCBI
|