Introduction
Breast cancer (BC) is the most prevalent malignancy
in female patients worldwide, representing ~25.1% of all new female
cancer cases and remains the leading cause of cancer-related
mortality in female patients across 112 countries, with an
estimated 715,000 deaths annually. Over the past five years
(2019–2024), global incidence rates have risen by 1.2% per year,
attributed to lifestyle transitions and expanded mammography
screening in low-resource regions, despite declining mortality in
high-income countries due to targeted therapies (1). Triple-negative TN(BC) accounts for
~15% of all BC cases (2,3). This type of BC has a poor prognosis
and low survival rate due to its high metastasis rate (4,5).
Metastasis occurs when tumor cells escape the primary site and
travel to distant tissue (6).
Epithelial-mesenchymal transition (EMT) is a main driver of
metastasis and is reportedly a cause of poor prognosis in
metastatic cancer (7,8). BC is primarily treated using surgery;
however, due to its high rate of early metastasis, TNBC is
primarily treated by radiotherapy (5,9).
Targeted therapies may also be used to treat TNBC. However, due to
the shortcomings of targeted drugs (10–14),
the effects of these treatments in patients with advanced
metastasis remain unsatisfactory (15). Therefore, identification of targeted
small-molecule drugs has become an important topic in BC treatment.
Currently, STAT3 is a promising novel target in clinical BC therapy
(16,17).
STAT3 signaling is involved in tumor cell
proliferation, survival, invasion and immunosuppression. STAT3
contributes to cancer development through epigenetic mechanisms
associated with mitochondrial function, inflammation, stem cells
and metastasis (18,19). A growing body of literature has
demonstrated the association between the tumor microenvironment and
the STAT3 signaling pathway (16,20).
Identification of drugs that target pathways inhibiting STAT3
signaling may decrease cancer progression and increase anticancer
immune responses (21). STAT3
signaling in cancer is activated by ligands that bind to cell
surface receptors, leading to the phosphorylation of STAT3. STAT3
regulates cell proliferation and survival through genes such as
cyclin D1, c-MYC, survivin, B cell lymphoma-extra-large and induced
myeloid leukemia cell differentiation protein Mcl-1. It also
promotes vascularization via VEGF and hypoxia-inducible factor 1α
(22–24). In addition, STAT3 regulates tumor
invasion and metastasis by regulating the expression of MMP2, MMP9
and vimentin (25,26). Moreover, STAT3 serves a key role in
immunity and tumor stem cell formation (27,28)
Fibronectin 1 (FN1) is a high molecular weight
glycoprotein of the extracellular matrix (ECM) that mediates
cellular interactions with the ECM and is involved in cell
adhesion, migration, proliferation and differentiation (29,30).
The expression of FN1 is higher in several tumor types including
nasopharyngeal carcinoma, osteosarcoma and esophageal and ovarian
cancer, compared with normal tissue (31–33).
Previously, FN1 was identified as an important tumor-associated
gene that regulates the development of cancer (34). Studies have revealed the role of FN1
in promoting BC invasion and metastasis (32,35,36).
In addition, high FN1 expression levels are associated with
advanced BC and poor prognosis (37).
Bioinformatics analysis can be used to screen vital
disease-associated proteins and provide direction for clinical
research (38). TNBC is
characterized by aggressive metastasis and limited therapeutic
options. The STAT3 signaling pathway has emerged as a critical
driver of TNBC progression, promoting epithelial-mesenchymal
transition (EMT) and extracellular matrix (ECM) remodeling through
downstream effectors such as FN1). Despite preclinical evidence
supporting STAT3 inhibition as a therapeutic strategy, clinical
translation remains hindered by the lack of agents capable of
simultaneously targeting STAT3 and its metastatic mediators. SB939,
a novel hydroxamic acid derivative, has shown pan-histone
deacetylase (HDAC) inhibitory activity in solid tumors (39–41).
However, its potential effects on STAT3/FN1 signaling crosstalk in
TNBC remain unexplored. Clinical studies have demonstrated that it
has good tolerability, efficacy and pharmacokinetics in solid
tumors including non-small cell lung cancer, HER2-negative
metastatic BC and platinum-resistant ovarian cancer (42,43).
It inhibits the metastasis and growth of BC tumors by targeting
histone deacetylase (HDAC) with minimal overall toxicity.
Dysregulation of histone acetylation is associated with
tumorigenesis and cancer progression. SB939 regulates cellular
processes involved in cancer development, such as inducing cell
cycle arrest, promoting apoptosis and suppressing metastasis
(44,45) and also serves a role in cell
differentiation and DNA replication and repair. Acetylation is a
key post-transcriptional modification in epigenetics and is
involved in multiple cellular processes (46,47).
Deacetylation is catalyzed by HDAC; therefore, SB939 is a promising
targeted inhibitor for use in BC treatment.
Materials and methods
RNA isolation and transcriptome
sequencing
Total RNA from MDA-MB-231 breast cancer cells) was
isolated using TRIzol (Catalog #15596026, Invitrogen, USA)
according to the manufacturer's protocol. RNA sequencing libraries
were constructed using the Ion Total RNA-Seq Kit v2 (Catalog
#4475936, Thermo Fisher Scientific, USA), which includes steps for
RNA fragmentation, adapter ligation, and amplification, followed by
template preparation on the Ion Chef system. Libraries were
sequenced on an Ion Torrent S5 XL system (Thermo Fisher Scientific,
Inc.) with a read depth of 10–20 million reads per sample, and raw
data were processed using Torrent Suite software for base calling
and quality control. Library concentrations (2–10 nM) were verified
using a Qubit 4.0 Fluorometer (Thermo Fisher Scientific). RNA
integrity and library fragment size distribution were assessed by
agarose gel electrophoresis (1.5% gel stained with GelRed) and
visualized under UV light.
Culture and pre-treatment of BC
cells
Cell culture
The MDA-MB-231 TNBC cell line (purchased from ATCC)
was cultured in RPMI-1640 medium (MultiCell Technologies, Inc.)
containing 10% fetal bovine serum (MultiCell Technologies, Inc.) at
37°C in a 5% CO2 incubator. MCF-10A, as well as the TNBC
cell lines MDA-MB-468, BT549, BT-20, MCF-7 and MDA-MB-231, were
purchased from American Type Culture Collection. SUM159 cells were
a gift from Dr Yuzhu Zhang (Guangdong Academy of Chinese Medicine,
Guangzhou, China). Cells were cultured in DMEM with 10% fetal
bovine serum (Gibco; Thermo Fisher Scientific) and 1% 100X
penicillin and streptomycin in a 37°C incubator with 5%
CO2. Tanks with liquid nitrogen (−196°C) were used for
long-term storage of the cells.
Pre-treatment of specimens
The control group was three groups of MDA-MB-231
cell samples (C1-C3) treated with DMSO at the same dosage as that
in the experimental group. (Selleck Chemicals LLC). The
experimental group was two groups of MDA-MB-231 cell samples
treated with SB939 [low dose group (T1-T3, 40 µM); high dose group
(T4-T6, 60 µM)]. The SB939 was dissolved in DMSO, and the cells
were treated for 24 h (37°C incubator with 5% CO2)
(44).
Bioinformatics analysis
Screening of differentially expressed genes
GSEA (gsea-msigdb.org/gsea/index.jsp; version
number: 4.3.3) was used to analyze genome-wide expression profiling
microarray data by sorting functionally similar or identical genes
and testing whether differentially expressed genes were enriched in
a predefined set of genes in the control and experimental groups.
The false discovery rate (FDR) of GSEA was calculated using the
built-in program; FDR <0.25 and a nominal P-value <0.05 were
considered statistically different.
Gene expression profiling interaction
analysis (GEPIA)
GEPIA (gepia.cancer-pku.cn/index.html) was used to
assess differential gene expression of BRCA, disease-associated
gene prognosis and expression of tumor-associated genes in tumor
and normal tissue. GEPIA was also used to analyze disease staging
correlations, prognostic survival curves and correlation between
different proteins.
Kaplan-Meier analysis was performed using R
statistical platform (v4.2.2; R-project.org/) with the survival
(v3.5–3) (https://CRAN.R-project.org/package=survival) and
survminer (v0.4.9) (https://CRAN.R-project.org/package=survminer)
packages. The primary endpoint was overall survival (OS), defined
as the time from initial diagnosis to death from any cause, while
the secondary endpoint, disease-free survival, was calculated as
the time from surgery to recurrence or metastasis. Censoring
criteria included patients lost to follow-up or deceased from
non-cancer-related causes. Survival curves were compared by the
log-rank test with a two-sided significance threshold (α=0.05).
Optimal cut-off thresholds for risk stratification were determined
using X-tile software (v3.6.1, Yale University) through a minimum
P-value approach, validated on the TCGA-BRCA cohort (n=1,091).
Gene expression database of normal and
tumor tissues 2 (GENT2)
GENT2 (gent2.appex.kr) was used to compare gene
expression profiles across normal and tumor tissues, enabling the
identification of genes with significant expression changes for
subsequent genomic and survival analyses in TCGA cohorts via
cBioPortal. Cancer Genomics (cbioportal.org/), a multidimensional
database was used to evaluate tumor genomics in The Cancer Genome
Atlas (TCGA) datasets (ID: syn300013), specifically ‘TCGA Nature
2012 (825 samples)’ and the ‘TCGA Pancancer Atlas (1,084 samples)’.
These datasets were used to study gene copy number alteration,
mutations, survival rates and pathways using the cBioPortal
database (cbioportal.org/). The tabs were selected
based on the default settings of cBioPortal.
Search tool for the retrieval of
interacting genes/proteins (STRING)
STRING (string-db.org/) was used to analyze the
protein-protein interaction (PPI) network of differentially
expressed proteins, and derive proteins with a strong association
with differential genes, which were used to research differential
gene-associated pathways.
Tumor immune estimation resource
(TIMER)
TIMER (cistrome.shinyapps.io/timer/) was used to
evaluate the association between differential gene expression and
immune cell infiltration using the ‘gene module’. The ‘survival
module’ was used to evaluate the association between clinical
prognosis and immune cell infiltration and differential gene
expression.
University of Alabama at Birmingham
cancer data analysis portal (UALCAN)
UALCAN (ualcan.path.uab.edu/.) is a web-based
database of expression levels of specific genes in various cancer
types and conduct grouped comparisons according to clinical
variables such as tumor stage, sex and age. UALAN database was used
to analyze the expression of STAT3 in different types of BC.
LinkedOmics
LinkedOmics (linkedomics.org/) is a publicly
available portal tool that provides comprehensive multi-omics
analysis of data from 32 TCGA cancer types (25 cases). The
‘LinkInterpreter’ module was used to obtain Gene Ontology (GO;
ebi.ac.uk/quickgo/) enrichment for STAT3 in BC and Kyoto
Encyclopedia of Genes and Genomes (KEGG; kegg.jp/) pathway
enrichment. Results were analyzed using Spearman's correlation
test. The P-value cutoff was 0.05.
Western blotting
Cell were seeded into 60-mm cell culture dishes at a
density of 1×105 cells per dish in a cell incubator
maintained at a temperature of 37°C and a carbon dioxide
concentration of 5% for incubation. After 48 h of incubation,
sample collection was performed on the cells were combined with
lysis buffer (KeyGen Total Protein Extraction Kit cat. no. KGP250;
Nanjing KeyGen Biotech Co., Ltd.) containing protease and
phosphatase inhibitors, following the manufacturer's instructions.
The specimens were homogenized on ice to extract total protein. The
lysate was centrifuged at 12,000 g and 4°C for 15 min and the
supernatant was harvested as the protein extract. Protein
concentration was assessed using a BCA kit (Sigma-Aldrich; Merck
KGaA). A polyacrylamide gel was prepared, with the separation gel
at 15% [based on target protein molecular weight (MW)] and the
stacking gel at 5%. Protein samples (30 µg) were separated and a
PVDF membrane was prepared. The membrane was soaked in methanol for
2 min and equilibrated in transfer buffer for transfer at 300 mA
for 120 min. The membrane was incubated in 3% BSA (Sigma)-0.1% TBST
at room temperature for 1 h for blocking. It was incubated with
primary antibodies against STAT3(Abcam; ab119352)and GAPDH(Abcam;
ab8245) (1:500) at 4°C overnight. After washing with TBST 3 times
(10 min each), the membrane was incubated with horseradish
peroxidase-conjugated secondary antibodies(Abcam; cat. no.
ab205719-HRP) of matching species (1:10,000) at room temperature
for 1 h. Following TBST washing (three times, 10 min each), the
membrane was inserted into the Odyssey system for exposure using
ECL kit (PN3300; Beijing GenClone Biotechnology Co., Ltd.) and
protein band image analysis (Image Lab 6.0.1; Bio-Rad).
Statistical analysis
Statistical analysis was performed using GraphPad
Prism 9.3 (GraphPad Software, Inc.). Each group of experiments was
repeated three times. Differences between groups were analyzed by
one-way ANOVA followed by Tukey's post hoc test for multiple
comparisons. Data are expressed as mean ± standard deviation.
P<0.05 was considered to indicate a statistically significant
difference.
Results
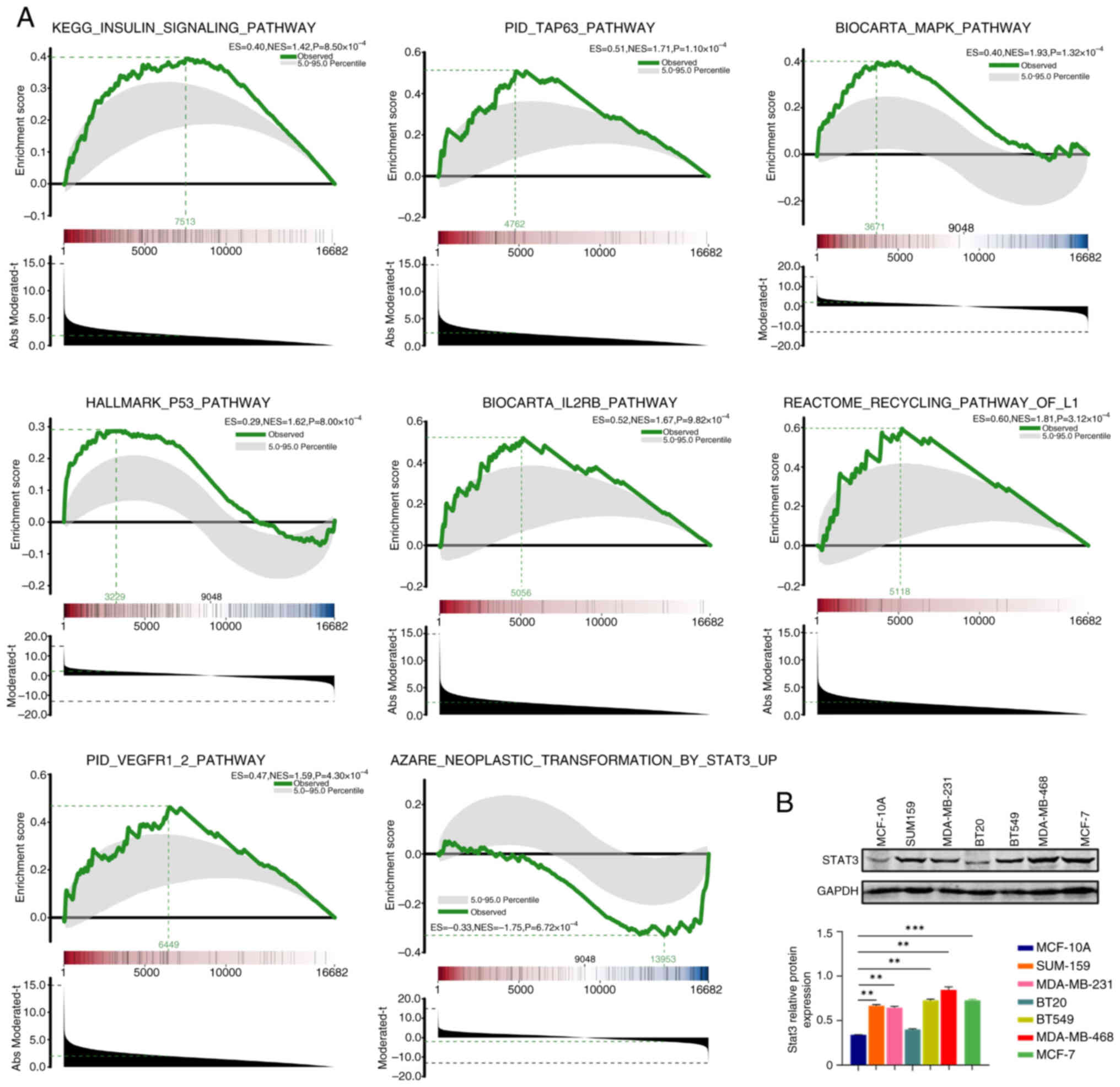
Inhibitory effect of SB939 on the
STAT3-enriched pathway
The results obtained from GSEA of the high-dose
group (T4-T6) treated with SB939 and the untreated control group
(C1-C3) are shown in Fig. 1A (red
represents the upregulated ploidy and blue represents the
downregulated ploidy). There were 16,682 genes associated with
‘insulin signaling pathway’. Furthermore, ‘TAP63 pathway’, ‘MAPK
pathway’, ‘p53 pathway’, ‘IL2RB pathway’, ‘recycling pathway of L1’
and ‘VEGFR1/2 pathway’ had a total of 16,682 genes expressed. These
pathways were enriched in upregulated genes. By contrast, STAT3
target genes were enriched in the downregulated genes, indicating
that the STAT3 signaling pathway was inhibited by high dose SB939,
compared with the control. Expression of the STAT3 protein was also
higher in TNBC cell lines (MDA-MB-231, MDA-MB-468, SUM159, BT549,
MCF-7) compared with a normal breast cell line (MCF-10A; Fig. 1B). Therefore, GSEA results suggest
that SB939 inhibited BC metastasis and invasion by inhibiting
STAT3-associated pathways.
Expression of STAT3
STAT3 was activated and highly expressed in several
cancer types (Fig. 2A). STAT3 is
also highly expressed in ~40% of BC cases. STAT3 expression serves
a vital role in the tumor staging of BC and affects the prognosis
of patients (48). UALCAN
demonstrated that STAT3 expression was increased in the TNBC
basal-like1 phenotype compared with both the luminal and HER2
phenotypes (Fig. 2B and C). STAT3
expression in premenopausal patients was associated with improved
BC survival, with postmenopausal patients having the worst
prognosis (Fig. 2D). GEPIA database
demonstrated that STAT3 was highly expressed in glioblastoma
multiforme, brain low-grade glioma and pancreatic adenocarcinoma
(Fig. 2E). These data suggest that
STAT3 served an essential role in different cancers and markedly
affected BC prognosis. Therefore, there is need to identify a small
molecule compound targeting STAT3 to treat clinical TNBC.
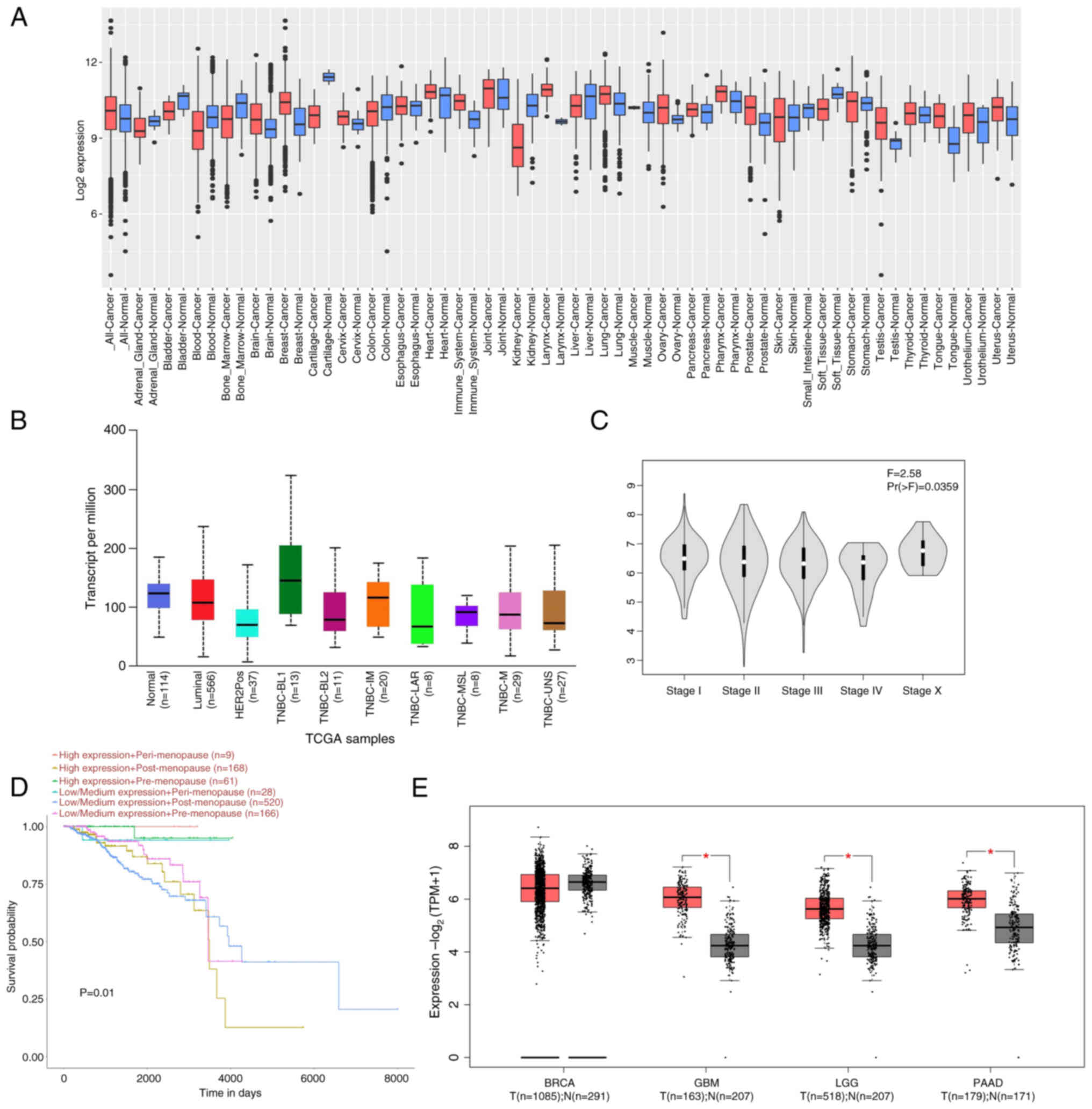 | Figure 2.STAT3 expression in tumors and
prognostic analysis. (A) Expression of STAT3 in pan-cancer obtained
from the Gene Expression Database of Normal and Tumor Tissues 2
database. Red, T; blue, N. (B) STAT3 expression in different types
of breast cancer obtained from the UALCAN database. (C) Effect of
STAT3 in different stages of breast cancer derived from the Gene
Expression Profiling Interaction Analysis database. (D) Prognostic
correlation analysis of STAT3 from the UALCAN database. (E)
Expression levels of STAT3 across varying tumor types. *P<0.05.
UALCAN, The University of Alabama at Birmingham Cancer data
analysis portal; TPM, transcripts per million; GBM, glioblastoma;
LGG, low-grade glioma; PAAD, pancreatic adenocarcinoma; TNBC,
triple-negative breast cancer; T, tumor; N, normal; TCGA, The
Cancer Genome Atlas; BL, basal-like 2 subtype); TNBC-IM [Triple
Negative Breast Cancer-Immunomodulatory; TNBC-MSL (Triple Negative
Breast Cancer-Mesenchymal Stem-like subtype); UNS (Triple Negative
Breast Cancer-Unclassified subtype)]. |
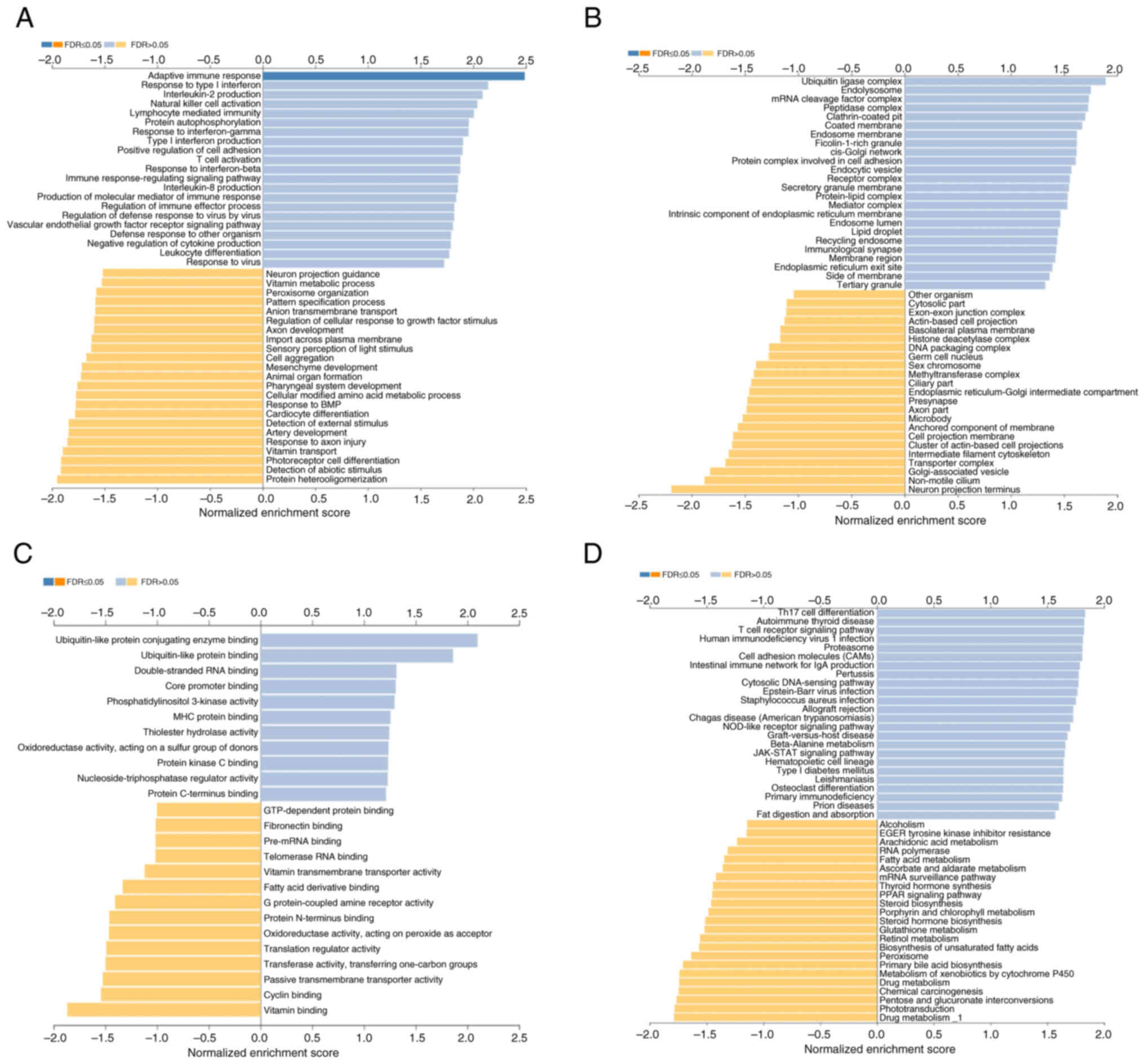
STAT3-associated enriched pathways in
BC
GO enrichment analysis was performed using the GSEA.
Differentially upregulated gene enrichment mediated by STAT3 was
associated with ‘vascular endothelial growth factor receptor
signaling pathway’, ‘adaptive immune response’ and ‘positive
regulation of cell adhesion’ (Fig.
3A). In cellular components, upregulated genes associated with
STAT3 were enriched in ‘ubiquitin ligase complex’, endolysosome and
‘protein complex involved in cell adhesion’ (Fig. 3B). Among the molecular functions,
STAT3-associated upregulated genes were enriched in ‘ubiquitin-like
protein conjugating enzyme binding’, ‘ubiquitin-like protein
binding’ and ‘double-stranded RNA binding’ (Fig. 3C). KEGG pathway analysis was also
performed and STAT3 co-expressed genes were associated with ‘cell
adhesion molecules (CAMs)’, ‘Th17 cell differentiation’ and
‘JAK-STAT signaling pathway’ (Fig.
3D).
FN1 regulates STAT3 pathway enrichment
in tumor tissue
GSEA demonstrated that FN1 had the highest impact
factor and the strongest association (Fig. 4A) in the enrichment heat map of
genes affected by SB939 treatment compared with the control. FN1 is
a glycoprotein molecule widely distributed in cellular structures
such as the smooth muscle cell layer, vascular cell membranes and
the nerve cell layer. It is involved in cell adhesion, migration
and motility. FN1 serves an important role in regulating lung and
colorectal cancer and other malignancies (49). Based on the UALCAN database and
GEPIA data (Fig. 4B and C), FN1
expression was upregulated in tumor tissue including BC, lymphoid
neoplasm diffuse large B cell lymphoma and esophageal carcinoma.
The BC survival curves demonstrate FN1 exerted similar influence to
STAT3, as a significant difference in survival rates was observed
between pre- and post-menopausal patients and FN1 expression showed
a statistically significant association with menopausal status.
Specifically, high FN1 expression was more prevalent in
pre-menopausal (25.9 vs. 23.1%) and peri-menopausal patients (5.3
vs. 3.4%), whereas post-menopausal patients predominantly exhibited
low/medium FN1 expression (73.5% vs. 68.7%). This pattern suggests
a potential interaction between FN1-mediated extracellular matrix
remodeling and estrogen-driven signaling pathways (Fig. 4D). The aforementioned results
indicate STAT3 may serve an inhibitory role in BC metastasis and
invasion by mediating FN1-associated adhesion proteins.
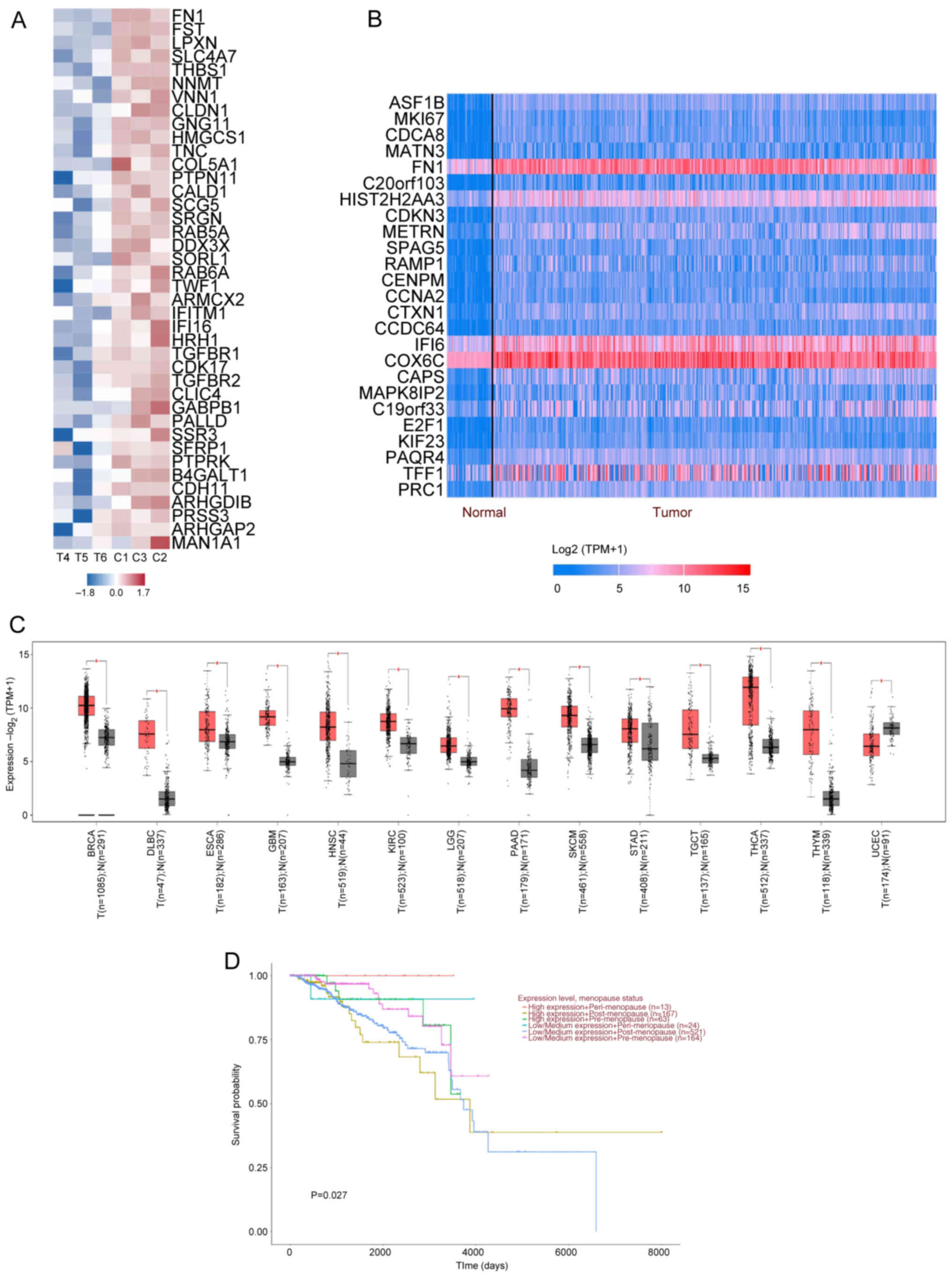 | Figure 4.FN1 expression in T and prognostic
correlation analysis. (A) Gene Set Enrichment Analysis of the top
40 most strongly associated genes. (B) Analysis of the top 51–75
genes overexpressed in BRCA-positive carcinoma using the UALCAN
database. Blue, downregulation; red indicates upregulation. (C)
Gene Expression Profiling Interaction Analysis database yielded
differential expression of FN1 in T. Red represents T; gray, N. (D)
FN1 prognostic association analysis using the UALCAN database.
*P<0.05. FN1, fibronectin 1; N, normal; T, tumor; TPM,
transcripts per million; ARHGDIB, ρ GDP Dissociation Inhibitor
Beta; ARHGAP2, Rho GTPase Activating Protein 2; ARMCX2, armadillo
Repeat Containing X-Linked 2; B4GALT1,
Beta-1,4-Galactosyltransferase 1; CALD1, Caldesmon 1; CDH11,
Cadherin 11; CDK17, cyclin Dependent Kinase 17; CLDN1, Claudin 1;
CLIC4, Chloride Intracellular Channel 4; COL5A1, Collagen Type V
Alpha 1 Chain; DDX3X, DEAD-Box Helicase 3 X-Linked; FN1,
Fibronectin 1; FST, Follistatin; GABPB1, GA Binding Protein
Transcription Factor Beta Subunit 1; GNG11, G Protein Subunit Gamma
11; HMGCS1, 3-Hydroxy-3-Methylglutaryl-CoA Synthase 1; HRH1,
Histamine Receptor H1; IFI16, Interferon Gamma Inducible Protein
16; IFITM1, Interferon Induced Transmembrane Protein 1; LPXN,
Leupaxin; MAN1A1, Mannosidase Alpha Class 1A Member 1; NNMT,
Nicotinamide N-Methyltransferase; PALLD, Palladin, Cytoskeletal
Associated Protein; PRSS3, Protease Serine 3; PTPN11, Protein
Tyrosine Phosphatase Non-Receptor Type 11; PTPRK, Protein Tyrosine
Phosphatase Receptor Type K; SCG5, Secretogranin V; SFRP1, Secreted
Frizzled Related Protein 1; SLC4A7, Solute Carrier Family 4 Member
7; SORL1, Sortilin Related Receptor 1; SRGN, Serglycin; SSR3,
Signal Sequence Receptor Subunit 3; TGFBR1, Transforming Growth
Factor Beta Receptor 1; TGFBR2, Transforming Growth Factor Beta
Receptor 2; THBS1, Thrombospondin 1; TNC, Tenascin C; TWF1,
Twinfilin Actin Binding Protein 1; VNN1, Vanin 1. |
SB939 targets the STAT3 signaling
pathway to downregulate FN1-associated proteins involved in BC
inhibition
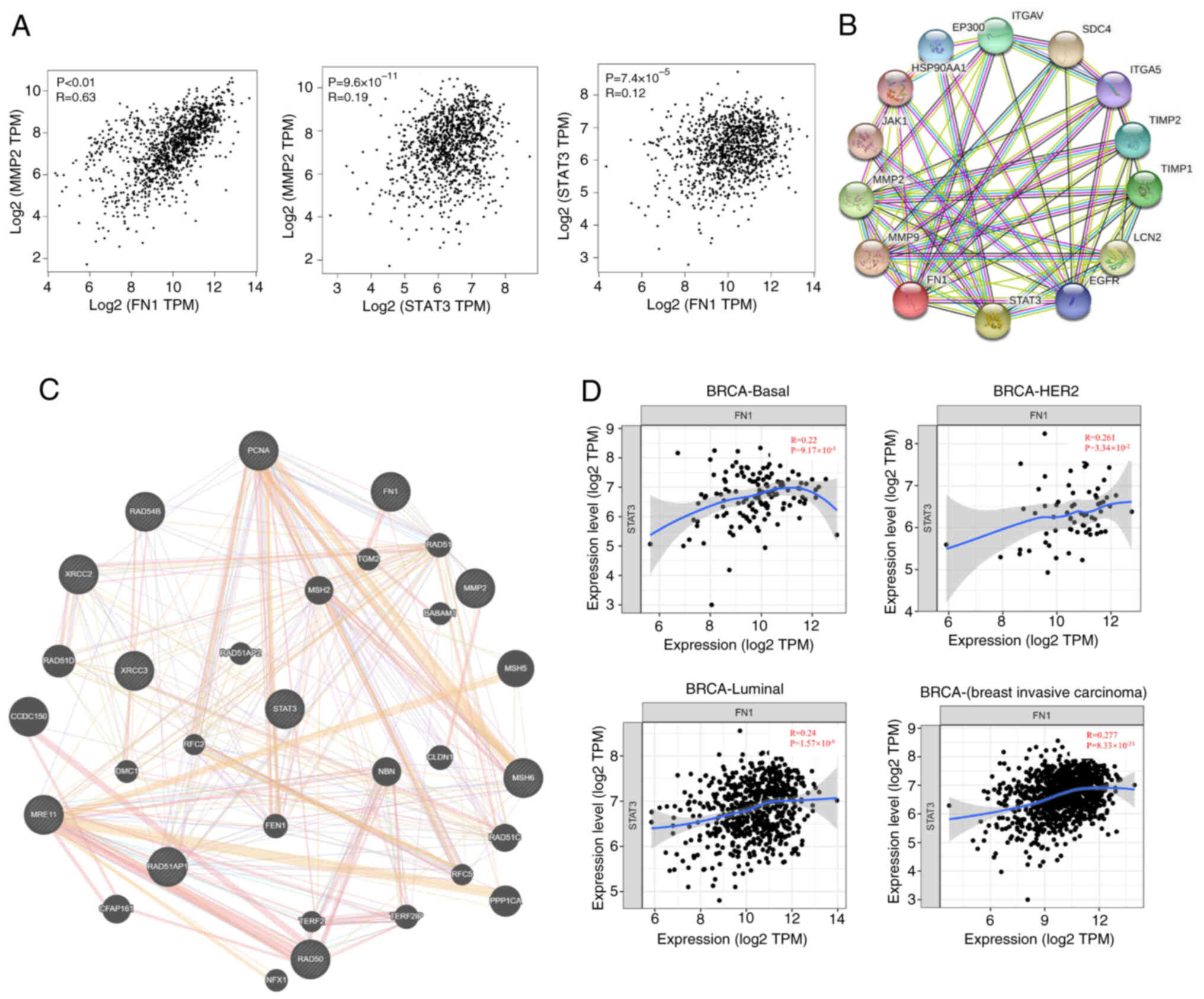
MMP2, a member of the MMP family, serves an
essential role in inflammation regulation, tumor growth and
metastasis (50). Using GEPIA
database correlation analysis, a significant correlation was found
between MMP2 and FN1 (R=0.63; Fig.
5A) and between MMP2 and STAT3 (R=0.19). In addition, the
correlation coefficient between STAT3 and FN1 was 0.12. There was a
significant degree of correlation between STAT3, FN1 and MMP2. PPIs
serve a key role in cellular functions and biological signaling,
which may reveal interactions and pathways. Therefore, the protein
interaction analysis database STRING was used for the association
analysis of the STAT3, FN1 and MMP2 proteins (Fig. 5B). The combined score of STAT3 and
FN1 was 0.651, STAT3 and MMP2 was 0.975 and MMP2 and FN1 was 0.607.
Therefore, SB939 may serve a role in regulating the expression of
STAT3 and the metastasis-associated protein MMP2 through
downregulation of FN1 (Fig. 5C).
TIMER database was used to evaluate the degree of association of
FN1 in types of BC (Fig. 5D). The
association between FN1 and STAT3 was most pronounced in the
invasive BC classification (R=0.277). In summary, SB939 inhibition
of BC invasion and metastasis may be mediated by targeting FN1 to
regulate STAT3 signaling.
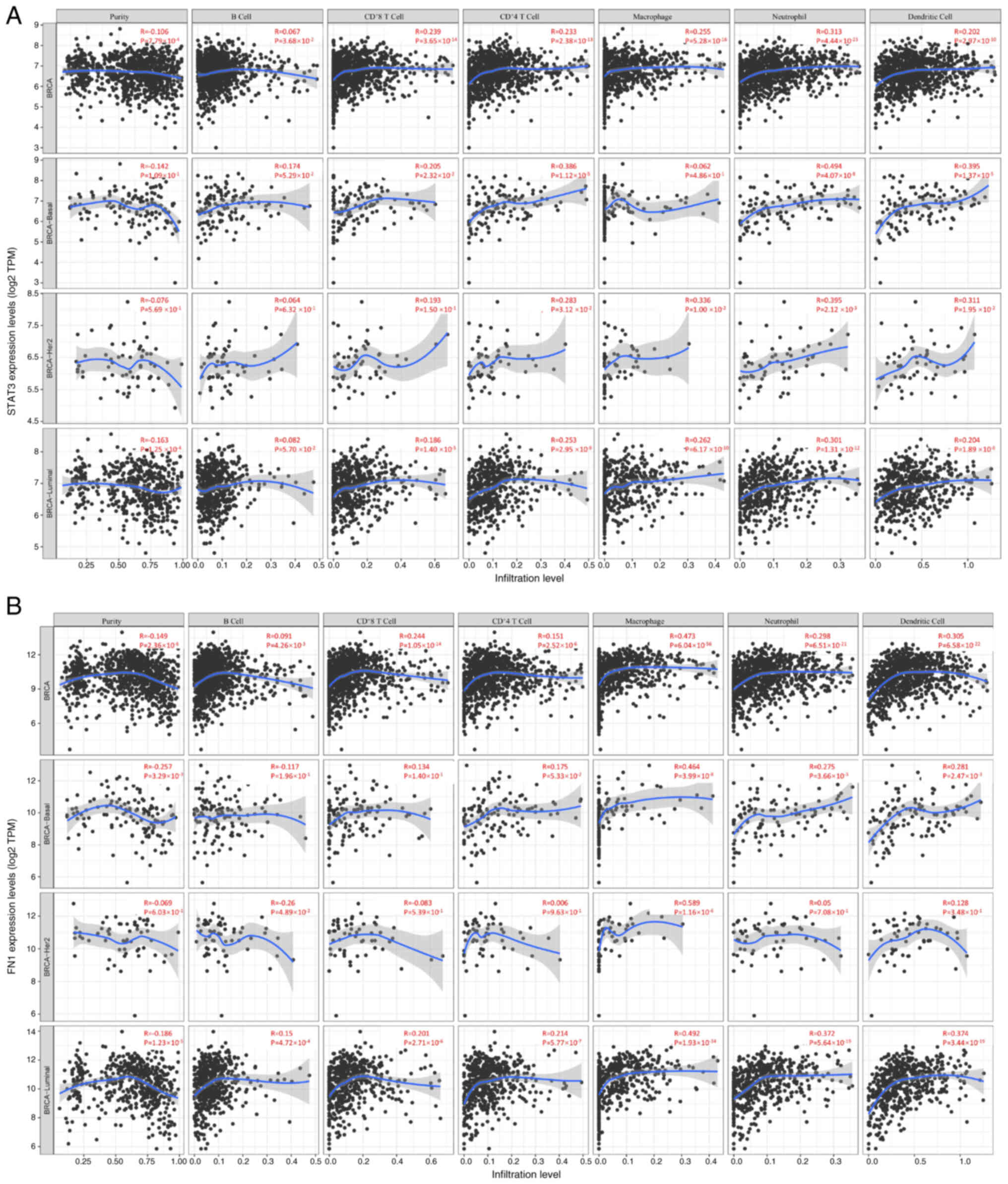
Correlation analysis of STAT3 and FN1
in immune infiltration
STAT3 chemokines are involved in inflammatory
responses and immune cell infiltration, thus affecting the clinical
outcome of patients with BC (50).
Therefore, evaluation of the correlation between differentially
expressed STAT3 chemokines and immune cell infiltration was
performed using TIMER database. In different BC types (basal;
BRCA-Her2; BRCA-Luminal), there was a negative correlation between
STAT3 expression and tumor purity (R=−0.106), and a positive
correlation between STAT3 expression and the infiltration of B
cells (R=0.067), CD8+ (R=0.239) and CD4+ T
cells (R=0.233), macrophages (R=0.255), neutrophils (R=0.313) and
dendritic cells (R=0.202; Fig. 6A).
In basal types, STAT3 expression was negatively associated with the
purity (R=−0.142) and positively associated with the infiltration
of B (R=0.174) and CD8+ (R=0.205) and CD4+ T
cells (R=0.386), macrophages (R=0.062), neutrophils (R=0.107) and
dendritic cells (R=0.305). In HER2 BC types, STAT3 expression was
negatively associated with the purity (R=−0.076) and there was a
positive correlation between STAT3 expression and the infiltration
of B (R=0.064) and CD8+ (R=0.193) and CD4+ T
cells (R=0.283), macrophages (R=0.336), neutrophils (R=0.395) and
dendritic cells (R=0.311). Similarly, the expression of STAT3 was
positively associated with the infiltration of B (R=0.082) and
CD8+ (R=0.186) and CD4+ T cells (R=0.253),
macrophages (R=0.262), neutrophils (R=0.301) and dendritic cells
(R=0.204). There was a negative correlation between STAT3
expression and purity (R=−0.163) in luminal BC types (Fig. 6A). The correlation between
differentially expressed FN1 chemokines with immune cell
infiltration was similar to that of STAT3. FN1 expression is
generally positively correlated with macrophage infiltration across
various breast cancer subtypes, with differing correlation
strengths among other immune cells. These patterns suggest a
potential role for FN1 in modulating immune cell infiltration
within tumor microenvironments. (Fig.
6B). Taken together, these data suggested that STAT3 and FN1
not only served a role in regulating BC invasion and metastasis but
also influenced immune cell infiltration.
Discussion
The 2024 global cancer statistics show that BC is
the top malignant tumor endangering the lives and health of female
patients worldwide (51). Its
incidence and mortality are greater than those of lung cancer, and
it has been the most common malignant cancer in female patients for
the past 5 years (1,52). TNBC accounts for 15–20% of all BC
cases (53). It is highly malignant
and aggressive and has a poor prognosis. Chemotherapy is typically
the main treatment for TNBC; however, this often has poor results
(54,55). Therefore, the identification of a
novel, targeted drug for TNBC is needed to decrease the incidence
of metastasis and improve prognosis for patients. Targeted drugs
often do not result in side effects exhibited by standard
chemotherapeutic drugs (56).
Distal metastasis occurs when tumor cells lose
epithelial adhesion and gain mesenchymal cell motility,
subsequently colonizing distal vital organs (57). SB939 is an effective oral HDAC
inhibitor with high selectivity and low toxicity compared with
traditional drugs (such as vorinostat, romidepsin and Panobinostat)
(51). It has efficacy in
inhibiting the proliferation of numerous cancer cell lines and has
shown marked efficacy and low toxicity in seven clinical studies
(42,43,58–62).
However, its effectiveness in TNBC is unknown. Therefore, the
potential efficacy of SB939 in TNBC was explored. STAT3, one of the
seven STAT family members, is responsible for transcriptional
regulation of the cell cycle in normal cells. However, it is
activated in BC, highly expressed in TNBC and promotes tumor
growth, metastasis and invasion. STAT3 is activated by upstream
signaling, leading to phosphorylation of tyrosine and serine
residues and dimerization. The activated dimer translocates to the
nucleus and binds target genes. Receptors such as EGFR and VEGFR
can activate STAT3 directly or indirectly to regulate downstream
target genes and stimulate tumor progression. These target genes
control tumor cell proliferation, angiogenesis and
epithelial-mesenchymal transition. STAT3 regulation is multi-modal,
serving as a central link between signaling processes, and is a
popular target in clinical research (63–66).
MMPs are involved in cellular phenotypical
responses, such as the degradation of ECM proteins and cleavage of
cell surface receptors (67,68).
When STAT3 promoter activity increases, MMP-associated proteins are
upregulated. MMP2 is a Zn2+-dependent MMP associated
with cancer and angiogenesis (69).
FN1 is a glycoprotein distributed in cellular structures and
involved in cell adhesion, migration and motility, serving a key
role in numerous malignancies (BC and Lung cancer) (70). Decreasing FN1 expression can
alleviate chemotherapy resistance in TNBC. Studies have reported
that inhibition of metastasis and invasion can be achieved by
regulating the expression of MMP2 and FN1 (71,72).
In MDA-MB-231 TNBC cell lines with and without SB939
treatment, GSEA demonstrated that STAT3 expression was inhibited in
the treatment group. Therefore, it was hypothesized that SB939 may
inhibit the metastatic invasion of TNBC by downregulating the STAT3
signaling pathway. Through heat map enrichment analysis, FN1 was
identified as a key protein in the downregulation of the STAT3
signaling pathway. The expression of STAT3 and FN1 in tumors and
the prognostic survival curves of patients were analyzed. Protein
networks were used to study the association between STAT3, FN1 and
other associated proteins. In TNBC, STAT3 promotes tumor metastasis
and invasion mainly by regulating EMT (73,74).
MMP2 and FN1 have the same association with tumor angiogenesis.
Suppression of FN1 and MMP2 expression leads to decreased tumor
metastasis and invasion (75,76)
This suggests that SB939 may serve an inhibitory role in tumor
metastasis and invasion by regulating STAT3 signaling and
downregulating MMP2 and FN1. STAT3 affects multiple intracellular
signal transductions through associated pathways such as JAK/STAT3,
growth factor receptor-mediated and G-protein-coupled receptor
(77,78). SB939 inhibits activity of HDAC,
leading to an increase in histone acetylation, which may alter the
accessibility of the STAT3 gene promoter region, thereby regulating
the expression of the STAT3 protein (59,79).
FN1 expression is associated with cell adhesion, migration,
proliferation and differentiation (54,80).
SB939 inhibits STAT3, changing the activity of HDAC
in the cell, which affects the expression or modification state of
FN1 and ultimately has an impact on cell migration or
proliferation. Future studies should assess STAT3 activity and FN1
expression in SB939-treated cells. Silencing and overexpression of
STAT3 and FN1, respectively, should be performed to assess
phenotypical changes of cells and the alterations in associated
signaling pathways in the presence or absence of SB939. These in
vitro experiments will more accurately analyze the complex
mechanism by which SB939 and STAT3 affect FN1. Cellular and animal
experiments are required to determine the mechanism of SB939 in
vivo.
Although TNBC differs from other BC subtypes in the
expression of key receptors [estrogen receptor (ER), progesterone
receptor (PR) and HER2], they may share key signaling pathways. For
example, the STAT3 pathway serves a role in the proliferation,
survival and metastasis of several types of cancer cell, including
BC. If SB939 mediates FN1 expression in TNBC by inhibiting the
STAT3 pathway, the same signaling pathway may also be partially
involved in the development of other BC subtypes. However, due to
the signal interference of receptors such as ER, PR or HER2 in
non-TNBC subtypes, the role of the STAT3 pathway may be less
prominent or it may be regulated by other factors.
SB939 shares both similarities and differences with
other HDAC inhibitors in terms of efficacy, toxicity and mechanism
of action. The antitumor activity of SB939 and its combinatorial
effects exhibit tumor type-specific mechanisms. In acute myeloid
leukemia (AML), SB939 suppresses proliferation and induces
apoptosis by downregulating JAK/FLT3 signaling in JAK2V617F and
FLT3-ITD mutant cell lines. Synergy with the JAK2/FLT3 inhibitor
pacritinib further reduces tumor growth and metastases while
normalizing tumor-induced dysregulation of plasma cytokines, growth
factors, and chemokines. In contrast, in colorectal cancer, SB939
demonstrates dose-dependent growth inhibition in HCT-116 ×enografts
and significant antitumor efficacy in the
Apc<sup>min</sup> mouse model, with selective
accumulation observed in tumor tissues. These differential
manifestations highlight its context-dependent therapeutic
potential across malignancies (42,43,81).
It exhibits common gastrointestinal and hematological toxicity
similar to those of other inhibitors, as well as low hepatotoxicity
(82). Inhibition of HDAC and the
regulation of downstream signaling pathways have distinct
epigenetic and non-epigenetic mechanisms: HDAC inhibition directly
modulates chromatin accessibility through histone acetylation,
while concurrently disrupting non-histone protein interactions
critical for STAT3 activation. Unlike broad-spectrum epigenetic
modifiers, HDAC-targeted regulation selectively amplifies feedback
loops between FN1-mediated extracellular matrix remodeling and
STAT3-dependent transcriptional programs.) in influencing key
factors such as STAT3 and FN1. Understanding of these differences
is conducive to a more precise selection and application of HDAC
inhibitors in cancer treatment, providing a theoretical basis for
the development of more effective tumor treatment strategies. In
future, more clinical and basic research is needed to explore the
characteristics and application potentials of SB939 as well as
other HDAC inhibitors.
The present study primarily relied on bioinformatics
tools and database analyses, which may have limitations in terms of
accuracy and representativeness and there is a possibility of bias
or inaccuracies. In addition, the present study was not a
large-scale clinical trial, which is needed to validate the
efficacy and safety of SB939. Future studies should include more
comprehensive clinical evaluations to understand its potential in
BC treatment. SB939 may show value in cancer treatment and underpin
future HDAC inhibitor investigation, drug structure optimization,
target expansion and delivery system innovation. Whether used alone
or in combination with endocrine therapy drugs (such as exemestane,
letrozole, anastrozole, etc.) and cyclin-dependent kinase 4/6
(CDK4/6) inhibitors), it may offer patients with cancer more
choices and improved survival, enabling precise treatment based on
individual differences. As a drug, SB939 has a complex in
vivo mechanism, affecting multiple organs (42,43).
The strict regulatory approval safety evaluations, such as
toxicology studies and adverse reaction monitoring in animal and
clinical trials, demonstrate risks, ensure clinical safety and
protect the health of patients. To provide a more comprehensive
understanding of the effects of SB939 across a wider range of
biological systems, further study should include different cell
lines and animal models.
In conclusion, SB939 downregulated MMP2 and FN1
protein expression by regulating STAT3 expression. Western blotting
demonstrated that STAT3 was highly expressed in BC, and TCGA
database revealed that STAT3 was strongly associated with the
development of BC. In summary, the present study provides new
guidance for the potential use of SB939 in BC treatment.
Acknowledgements
Not applicable.
Funding
The present study was supported by Natural Science Foundation of
Shanxi (grant no. 202403021212224).
Availability of data and materials
The data generated in the present study may be found
in the Figshare database under accession number 28032146/28032149
or at the following URLs: figshare.com/articles/dataset/Groups_xlsx__/28032146;
figshare.com/articles/dataset/Go_/28032149.
Authors' contributions
CQ and JL designed the experiments. CQ, JL and RS
performed the experiments. CQ, SZ, XH analyzed the data and wrote
the manuscript. CQ and JL confirm the authenticity of all the raw
data. All authors have read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Giaquinto AN and Jemal A:
Cancer statistics, 2024. CA Cancer J Clin. 74:12–49. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Dent R, Trudeau M, Pritchard KI, Hanna WM,
Kahn HK, Sawka CA, Lickley LA, Rawlinson E, Sun P and Narod SA:
Triple-negative breast cancer: Clinical features and patterns of
recurrence. Clin Cancer Res. 13:4429–4434. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cancer Genome Atlas Network, .
Comprehensive molecular portraits of human breast tumours. Nature.
490:61–70. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yin L, Duan JJ, Bian XW and Yu SC:
Triple-negative breast cancer molecular subtyping and treatment
progress. Breast Cancer Res. 22:612020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Karim AM, Eun Kwon J, Ali T, Jang J, Ullah
I, Lee YG, Park DW, Park J, Jeang JW and Kang SC: Triple-negative
breast cancer: Epidemiology, molecular mechanisms, and modern
vaccine-based treatment strategies. Biochem Pharmacol.
212:1155452023. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gerstberger S, Jiang Q and Ganesh K:
Metastasis. Cell. 186:1564–1579. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Manfioletti G and Fedele M:
Epithelial-mesenchymal transition (EMT). Int J Mol Sci.
24:113862023. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Saitoh M: Transcriptional regulation of
EMT transcription factors in cancer. Semin Cancer Biol. 97:21–29.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Farghadani R and Naidu R: The anticancer
mechanism of action of selected polyphenols in triple-negative
breast cancer (TNBC). Biomed Pharmacother. 165:1151702023.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Robson M, Im SA, Senkus E, Xu B, Domchek
SM, Masuda N, Delaloge S, Li W, Tung N, Armstrong A, et al:
Olaparib for metastatic breast cancer in patients with a germline
BRCA mutation. N Engl J Med. 377:523–533. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Levine AJ: p53: 800 million years of
evolution and 40 years of discovery. Nat Rev Cancer. 20:471–480.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Emens LA, Asquith JM, Leatherman JM,
Kobrin BJ, Petrik S, Laiko M, Levi J, Daphtary MM, Biedrzycki B,
Wolff AC, et al: Timed sequential treatment with cyclophosphamide,
doxorubicin, and an allogeneic granulocyte-macrophage
colony-stimulating factor-secreting breast tumor vaccine: A
chemotherapy dose-ranging factorial study of safety and immune
activation. J Clin Oncol. 27:5911–5918. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hwang JP, Fisch MJ, Lok AS, Zhang H,
Vierling JM and Suarez-Almazor ME: Trends in hepatitis B virus
screening at the onset of chemotherapy in a large US cancer center.
BMC Cancer. 13:5342013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
De Mattos-Arruda L, Weigelt B, Cortes J,
Won HH, Ng CKY, Nuciforo P, Bidard FC, Aura C, Saura C, Peg V, et
al: Capturing intra-tumor genetic heterogeneity by de novo mutation
profiling of circulating Cell-free tumor DNA: A proof-of-principle.
Ann Oncol. 25:1729–1735. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Adrada BE, Moseley TW, Kapoor MM, Scoggins
ME, Patel MM, Perez F, Nia ES, Khazai L, Arribas E, Rauch GM and
Guirguis MS: Triple-negative breast cancer: Histopathologic
features, genomics, and treatment. Radiographics. 43:e2300342023.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zou S, Tong Q, Liu B, Huang W, Tian Y and
Fu X: Targeting STAT3 in cancer immunotherapy. Mol Cancer.
19:1452020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Poli V and Camporeale A: STAT3-mediated
metabolic reprograming in cellular transformation and implications
for drug resistance. Front Oncol. 5:1212015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ouyang S, Li H, Lou L, Huang Q, Zhang Z,
Mo J, Li M, Lu J, Zhu K, Chu Y, et al: Inhibition of
STAT3-ferroptosis negative regulatory axis suppresses tumor growth
and alleviates chemoresistance in gastric cancer. Redox Biol.
52:1023172022. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sadrkhanloo M, Entezari M, Orouei S,
Ghollasi M, Fathi N, Rezaei S, Hejazi ES, Kakavand A, Saebfar H,
Hashemi M, et al: STAT3-EMT axis in tumors: Modulation of cancer
metastasis, stemness and therapy response. Pharmacol Res.
182:1063112022. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
El-Tanani M, Al Khatib AO, Aladwan SM,
Abuelhana A, McCarron PA and Tambuwala MM: Importance of STAT3
signalling in cancer, metastasis and therapeutic interventions.
Cell Signal. 92:1102752022. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sadrkhanloo M, Paskeh MDA, Hashemi M,
Raesi R, Bahonar A, Nakhaee Z, Entezari M, Beig Goharrizi MAS,
Salimimoghadam S, Ren J, et al: STAT3 signaling in prostate cancer
progression and therapy resistance: An oncogenic pathway with
diverse functions. Biomed Pharmacother. 158:1141682023. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ma JH, Qin L and Li X: Role of STAT3
signaling pathway in breast cancer. Cell Commun Signal. 18:332020.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jin W: Role of JAK/STAT3 signaling in the
regulation of metastasis, the transition of cancer stem cells, and
chemoresistance of cancer by Epithelial-mesenchymal transition.
Cells. 9:2172020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Qin J, Shen X, Zhang J and Jia D:
Allosteric inhibitors of the STAT3 signaling pathway. Eur J Med
Chem. 190:1121222020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yu M, Peng X, Li H, Xu Y, Sun X and Chen
J: Gankyrin has a potential role in embryo implantation via
activation of STAT3. Reproduction. 163:157–165. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang Q, Zhang R, He Y, Mao G and Kong Z:
Taraxasterol enhanced bladder cancer cells radiosensitivity via
inhibiting the COX-2/PGE2/JAK2/STAT3/MMP pathway. Int J Radiat
Biol. 100:791–801. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Aigner P, Just V and Stoiber D: STAT3
isoforms: Alternative fates in cancer? Cytokine. 118:27–34. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Taifour T, Attalla SS, Zuo D, Gu Y,
Sanguin-Gendreau V, Proud H, Solymoss E, Bui T, Kuasne H,
Papavasiliou V, et al: The tumor-derived cytokine Chi3l1 induces
neutrophil extracellular traps that promote T cell exclusion in
triple-negative breast cancer. Immunity. 56:2755–2772.e8. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Balachandran K, Ramli R, Karsani SA and
Abdul Rahman M: Identification of potential biomarkers and small
molecule drugs for Bisphosphonate-related osteonecrosis of the jaw
(BRONJ): An integrated bioinformatics study using big data. Int J
Mol Sci. 24:86352023. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Roy ME, Veilleux C and Annabi B: In vitro
biomaterial priming of human mesenchymal stromal/stem cells:
Implication of the Src/JAK/STAT3 pathway in vasculogenic mimicry.
Sci Rep. 14:214442024. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Huang X, Liu Y, Qian C, Shen Q, Wu M, Zhu
B and Feng Y: CHSY3 promotes proliferation and migration in gastric
cancer and is associated with immune infiltration. J Transl Med.
21:4742023. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pan S, Zhu J, Liu P, Wei Q, Zhang S, An W,
Tong Y, Cheng Z and Liu F: FN1 mRNA 3′-UTR supersedes traditional
fibronectin 1 in facilitating the invasion and metastasis of
gastric cancer through the FN1 3′-UTR-let-7i-5p-THBS1 axis.
Theranostics. 13:5130–5150. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liu X, Meng L, Li X, Li D, Liu Q, Chen Y,
Li X, Bu W and Sun H: Regulation of FN1 degradation by the
p62/SQSTM1-dependent autophagy-lysosome pathway in HNSCC. Int J
Oral Sci. 12:342020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li XF, Selli C, Zhou HL, Cao J, Wu S, Ma
RY, Lu Y, Zhang CB, Xun B, Lam AD, et al: Macrophages promote
anti-androgen resistance in prostate cancer bone disease. J Exp
Med. 220:e202210072023. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang Y, Wang Y, Guo Y, Liao Z, Xu R and
Ruan Z: miR-135b promotes the invasion and metastasis of
hepatocellular carcinoma cells. Xi Bao Yu Fen Zi Mian Yi Xue Za
Zhi. 31:1316–1321. 2015.(In Chinese). PubMed/NCBI
|
|
36
|
Gang D, Qing O, Yang Y, Masood M, Wang YH,
Linhui J, Haotao S, Li G, Liu C, Nasser MI and Zhu P: Cyanidin
prevents cardiomyocyte apoptosis in mice after myocardial
infarction. Naunyn Schmiedebergs Arch Pharmacol. 397:5883–5898.
2024. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang XX, Luo JH and Wu LQ: FN1
overexpression is correlated with unfavorable prognosis and immune
infiltrates in breast cancer. Front Genet. 13:9136592022.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Matsuoka T and Yashiro M: Bioinformatics
analysis and validation of potential markers associated with
prediction and prognosis of gastric cancer. Int J Mol Sci.
25:58802024. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Schneeweiss A, Denkert C, Fasching PA,
Fremd C, Gluz O, Kolberg-Liedtke C, Loibl S and Lück HJ: Diagnosis
and therapy of Triple-negative breast cancer (TNBC)-Recommendations
for daily routine practice. Geburtshilfe Frauenheilkd. 79:605–617.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Long L, Fei X, Chen L, Yao L and Lei X:
Potential therapeutic targets of the JAK2/STAT3 signaling pathway
in triple-negative breast cancer. Front Oncol. 14:13812512024.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Mou J, Xu X, Wang F, Kong W, Chen J and
Ren J: HMGN4 plays a key role in STAT3-mediated oncogenesis of
triple-negative breast cancer. Carcinogenesis. 43:874–884. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Novotny-Diermayr V, Sangthongpitag K, Hu
CY, Wu X, Sausgruber N, Yeo P, Greicius G, Pettersson S, Liang AL,
Loh YK, et al: SB939, a novel potent and orally active histone
deacetylase inhibitor with high tumor exposure and efficacy in
mouse models of colorectal cancer. Mol Cancer Ther. 9:642–652.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Novotny-Diermayr V, Hart S, Goh KC, Cheong
A, Ong LC, Hentze H, Pasha MK, Jayaraman R, Ethirajulu K and Wood
JM: The oral HDAC inhibitor pracinostat (SB939) is efficacious and
synergistic with the JAK2 inhibitor pacritinib (SB1518) in
preclinical models of AML. Blood Cancer J. 2:e692012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Chen J, Li N, Liu B, Ling J, Yang W, Pang
X and Li T: Pracinostat (SB939), a histone deacetylase inhibitor,
suppresses breast cancer metastasis and growth by inactivating the
IL-6/STAT3 signalling pathways. Life Sci. 248:1174692020.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Sumanadasa SD, Goodman CD, Lucke AJ,
Skinner-Adams T, Sahama I, Haque A, Do TA, McFadden GI, Fairlie DP
and Andrews KT: Antimalarial activity of the anticancer histone
deacetylase inhibitor SB939. Antimicrob Agents Chemother.
56:3849–3856. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Jenuwein T and Allis CD: Translating the
histone code. Science. 293:1074–1080. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Piekna-Przybylska D, Bambara RA and
Balakrishnan L: Acetylation regulates DNA repair mechanisms in
human cells. Cell Cycle. 15:1506–1517. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Banerjee K and Resat H: Constitutive
activation of STAT3 in breast cancer cells: A review. Int J Cancer.
138:2570–2578. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhou Y, Cao G, Cai H, Huang H and Zhu X:
The effect and clinical significance of FN1 expression on
biological functions of gastric cancer cells. Cell Mol Biol
(Noisy-le-grand). 66:191–198. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Costanzo L, Soto B, Meier R and Geraghty
P: The biology and function of tissue inhibitor of
metalloproteinase 2 in the lungs. Pulm Med. 2022:36327642022.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Li Z, Wei H, Li S, Wu P and Mao X: The
Role of progesterone receptors in breast cancer. Drug Des Devel
Ther. 16:305–314. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Burstein HJ, Curigliano G, Thurlimann B,
Weber WP, Poortmans P, Regan MM, Senn HJ, Winer EP and Gnant M;
Panelists of the St Gallen Consensus Conference, : Customizing
local and systemic therapies for women with early breast cancer:
The St. Gallen International Consensus Guidelines for treatment of
early breast cancer 2021. Ann Oncol. 32:1216–1235. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zagami P and Carey LA: Triple negative
breast cancer: Pitfalls and progress. NPJ Breast Cancer. 8:952022.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Yang Y, Li H, Yang W and Shi Y: Improving
efficacy of TNBC immunotherapy: Based on analysis and subtyping of
immune microenvironment. Front Immunol. 15:14416672024. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Reddy Baddam S, Ganta S, Nalla S, Banoth
C, Vudari B, Akkiraju PC, Srinivas E and Tade RS: Polymeric
nanomaterials-based theranostic platforms for triple-negative
breast cancer (TNBC) treatment. Int J Pharm. 660:1243462024.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Soria JC, Ohe Y, Vansteenkiste J,
Reungwetwattana T, Chewaskulyong B, Lee KH, Dechaphunkul A, Imamura
F, Nogami N, Kurata T, et al: Osimertinib in untreated EGFR-mutated
advanced Non-Small-Cell lung cancer. N Engl J Med. 378:113–125.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Hou Y, Yang K, Wang L, Wang J, Huang X,
Piffkó A, Luo SZ, Yu X, Rao E, Martinez C, et al: Radiotherapy
enhances metastasis through immune suppression by inducing PD-L1
and MDSC in distal sites. Clin Cancer Res. 30:1945–1958. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Hu Z, Wei F, Su Y, Wang Y, Shen Y, Fang Y,
Ding J and Chen Y: Histone deacetylase inhibitors promote breast
cancer metastasis by elevating NEDD9 expression. Signal Transduct
Target Ther. 8:112023. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Yang T, Wang P, Yin X, Zhang J, Huo M, Gao
J, Li G, Teng X, Yu H, Huang W and Wang Y: The histone deacetylase
inhibitor PCI-24781 impairs calcium influx and inhibits
proliferation and metastasis in breast cancer. Theranostics.
11:2058–2076. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Eigl BJ, North S, Winquist E, Finch D,
Wood L, Sridhar SS, Powers J, Good J, Sharma M, Squire JA, et al: A
phase II study of the HDAC inhibitor SB939 in patients with
castration resistant prostate cancer: NCIC clinical trials group
study IND195. Invest New Drugs. 33:969–976. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Razak AR, Hotte SJ, Siu LL, Chen EX, Hirte
HW, Powers J, Walsh W, Stayner LA, Laughlin A, Novotny-Diermayr V,
et al: Phase I clinical, pharmacokinetic and pharmacodynamic study
of SB939, an oral histone deacetylase (HDAC) inhibitor, in patients
with advanced solid tumours. Br J Cancer. 104:756–762. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Sumanadasa SD, Goodman CD, Lucke AJ,
Skinner-Adams T, Sahama I, Haque A, Do TA, McFadden GI, Fairlie DP,
Andrews KT, et al: Antimalarial activity of the anticancer histone
deacetylase inhibitor SB939. Antimicrob Agents Chemother.
56:3849–3856. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Levy DE and Darnell JE Jr: Stats:
Transcriptional control and biological impact. Nat Rev Mol Cell
Biol. 3:651–662. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
64
|
Yu H, Pardoll D and Jove R: STATs in
cancer inflammation and immunity: A leading role for STAT3. Nat Rev
Cancer. 9:798–809. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Wendt MK, Balanis N, Carlin CR and
Schiemann WP: STAT3 and epithelial-mesenchymal transitions in
carcinomas. JAKSTAT. 3:e289752014.PubMed/NCBI
|
|
66
|
Johnson DE, O'Keefe RA and Grandis JR:
Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat Rev
Clin Oncol. 15:234–248. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Hrabia A: Matrix metalloproteinases (MMPs)
and inhibitors of MMPs in the avian reproductive system: An
Overview. Int J Mol Sci. 22:80562021. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Hashmi F, Mollapour M, Bratslavsky G and
Bourboulia D: MMPs, tyrosine kinase signaling and extracellular
matrix proteolysis in kidney cancer. Urol Oncol. 39:316–321. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Sohel M: Comprehensive exploration of
Biochanin A as an oncotherapeutics potential in the treatment of
multivarious cancers with molecular insights. Phytother Res.
38:489–506. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Verdugo E, Puerto I and Medina MA: An
update on the molecular biology of glioblastoma, with clinical
implications and progress in its treatment. Cancer Commun (Lond).
42:1083–1111. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Tan X, Liu Z, Wang Y, Wu Z, Zou Y, Luo S,
Tang Y, Chen D, Yuan G and Yao K: miR-138-5p-mediated HOXD11
promotes cell invasion and metastasis by activating the
FN1/MMP2/MMP9 pathway and predicts poor prognosis in penile
squamous cell carcinoma. Cell Death Dis. 13:8162022. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
He X, Huang Z, Liu P, Li Q, Wang M, Qiu M,
Xiong Z and Yang S: Apatinib inhibits the invasion and metastasis
of liver cancer cells by downregulating MMP-Related proteins via
regulation of the NF-κB signaling pathway. Biomed Res Int.
2020:31261822020. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
El-Ashmawy NE, Khedr EG, Abo-Saif MA and
Hamouda SM: Long noncoding RNAs as regulators of epithelial
mesenchymal transition in breast cancer: A recent review. Life Sci.
336:1223392024. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Zhang K, Liu P, Tang H, Xie X, Kong Y,
Song C, Qiu X and Xiao X: Corrigendum: AFAP1-AS1 promotes
Epithelial-Mesenchymal transition and tumorigenesis through
Wnt/β-Catenin signaling pathway in Triple-negative breast cancer.
Front Pharmacol. 11:11072020. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Masuda T, Fukuda A, Yamakawa G, Omatsu M,
Namikawa M, Sono M, Fukunaga Y, Nagao M, Araki O, Yoshikawa T, et
al: Pancreatic RECK inactivation promotes cancer formation,
Epithelial-mesenchymal transition, and metastasis. J Clin Invest.
133:e1618472023. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Tan X, Liu Z, Wang Y, Wu Z, Zou Y, Luo S,
Tang Y, Chen D, Yuan G and Yao K: miR-138-5p-mediated HOXD11
promotes cell invasion and metastasis by activating the
FN1/MMP2/MMP9 pathway and predicts poor prognosis in penile
squamous cell carcinoma. Cell Death Dis. 13:8162022. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Fan S, Guo C, Yang G, Hong L, Li H, Ma J,
Zhou Y, Fan S, Xue Y and Zeng F: GPR160 regulates the self-renewal
and pluripotency of mouse embryonic stem cells via JAK1/STAT3
signaling pathway. J Genet Genomics. 51:1055–1065. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Jiang H, Yang J, Li T, Wang X, Fan Z, Ye Q
and Du Y: JAK/STAT3 signaling in cardiac fibrosis: A promising
therapeutic target. Front Pharmacol. 15:13361022024. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Leszczynska KB, Freitas-Huhtamäki A,
Jayaprakash C, Dzwigonska M, Vitorino FNL, Horth C, Wojnicki K,
Gielniewski B, Szadkowska P, Kaza B, et al: H2A.Z histone variants
facilitate HDACi-dependent removal of H3.3K27M mutant protein in
pediatric high-grade glioma cells. Cell Rep. 43:1137072024.
View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Kuramoto K, Liang H, Hong JH and He C:
Exercise-activated hepatic autophagy via the FN1-α5β1 integrin
pathway drives metabolic benefits of exercise. Cell Metab.
35:620–632.e5. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Islam P, Rizzieri D, Lin C, de Castro C,
Diehl L, Li Z, Moore J, Morris T and Beaven A: Phase II study of
Single-agent and combination everolimus and panobinostat in
relapsed or refractory diffuse large B-cell lymphoma. Cancer
Invest. 39:871–879. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Arrowsmith CH, Bountra C, Fish PV, Lee K
and Schapira M: Epigenetic protein families: A new frontier for
drug discovery. Nat Rev Drug Discov. 11:384–400. 2012. View Article : Google Scholar : PubMed/NCBI
|