Introduction
Chronic myeloid leukemia (CML) is a clonal malignant
disorder of pluripotent hematopoetic stem cells progressing from a
chronic to an accelerated to a blast phase (1). The cytogenetic hallmark of CML is the
Philadelphia (Ph) chromosome, resulting from t(9;22)(q34;q11),
which reflects the rearrangement of the ABL and BCR genes (2). The Ph chromosome is present in more
than 90% of CML cases (3). In
Ph-positive CML, expression of the BCR/ABL chimeric protein p210
with an increased tyrosine kinase activity is essential for
multiple signaling pathways to confer the leukemia phenotype
(4).
Complex chromosomal rearrangements involving one or
more additional chromosomes have been described in more than 600
CML cases (5). Using conventional
cytogenetic analysis, two variant subgroups have traditionally been
identified: complex t(9;22;V) where V represents a third
translocation partner chromosome, and simple t(9;V) or t(22;V)
(6). Only a few cases exhibit a
chromosomal fragment from the third chromosome translocated to the
der(22)t(9;22) producing a ‘masked Ph’ (7). In the majority of Ph-variant cases,
the segment 22q11-qter shifts to a third chromosome, while a part
of the third chromosome is located on 9q34. Deletions on the
derivative chromosome 9 occur with a much higher frequency in
patients with variant Ph translocations (45%) compared to those
with classic Ph (17%) (8).
We present a CML case with a translocated BCR to
der(2), involving four different chromosomal breakpoints
characterized by molecular cytogenetics.
Materials and methods
Case report
A 47-year-old female patient was admitted to our
Human Genetics Division initially presented with a WBC of
9.66×109/l and splenomegaly. Chromosome analysis using
banding cytogenetics revealed a karyotype in accordance with the
clinical diagnosis of CML in the chronic phase. She was treated
with hydroxyurea (1000 mg/day) for four years and three months. At
her initial admission, her hematological parameters were: 85.4%
neutrophils, 7.7% lymphocytes and 6.9% immature cells. The platelet
count was 372×109/l and the hemoglobin level was 11.8
g/dl. She was initially treated with hydroxyurea for 18 months.
Then, 33 months later, following hydroxyurea treatment, her WBC was
130.91×109/l (79.8% neutrophils, 8.5% lymphocytes and
11.7% immature cells). The platelet count was 340×109/l,
and the hemoglobin level was 11.9 g/dl.
Banding cytogenetics
Chromosome analyses were performed by the
GTG-banding technique according to standard procedures (9). Twenty metaphases, obtained from the
unstimulated bone marrow of the patient, were analyzed. Karyotypes
were described according to the International System for Human
Cytogenetic Nomenclature (10).
Fluorescence in situ hybridization
(FISH)
FISH was conducted using commercially available
probes. LSI BCR/ABL dual-color dual-fusion translocation probe
(Abbott molecular/Vysis, USA), whole chromosome painting (WCP)
probe for chromosomes 1, 2 and 22 (MetaSystems, Germany) and α
satellite probe (CEP) for chromosome 9 (Abbott molecular/Vysis)
were applied according to the standard method (11). Twenty metaphase spreads were
analyzed, using a fluorescence microscope (Axio Imager Z1 mot;
Zeiss, Germany) equipped with appropriate filter sets to
discriminate between a maximum of five fluorochromes and the
counterstain 4′6-diamino-2-phenylindole. Image capturing and
processing were carried out using an Isis image analysis system
(MetaSystems).
Results
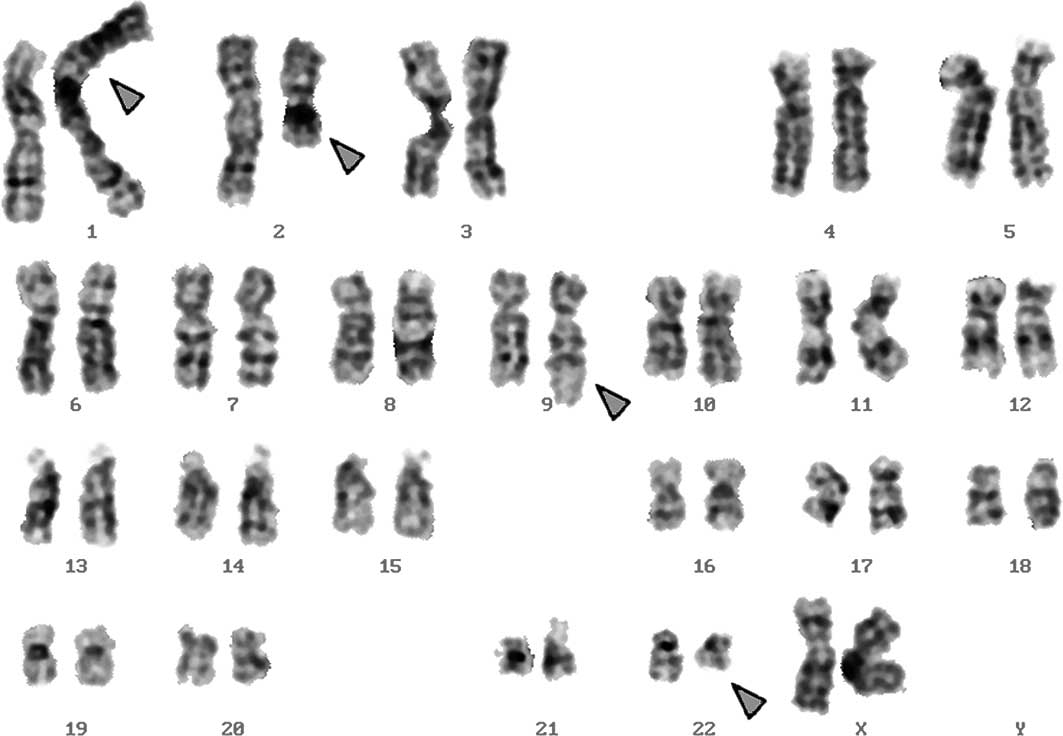
Karyotyping was performed at 3 and 10 months after
the initiation of hydroxyurea treatment. The same karyotypic
changes were noted. A complex karyotype 46,XX,t(1;2;9;22) was
determined in GTG-banding (Fig. 1),
and was further studied by molecular cytogenetics (Figs. 2–4).
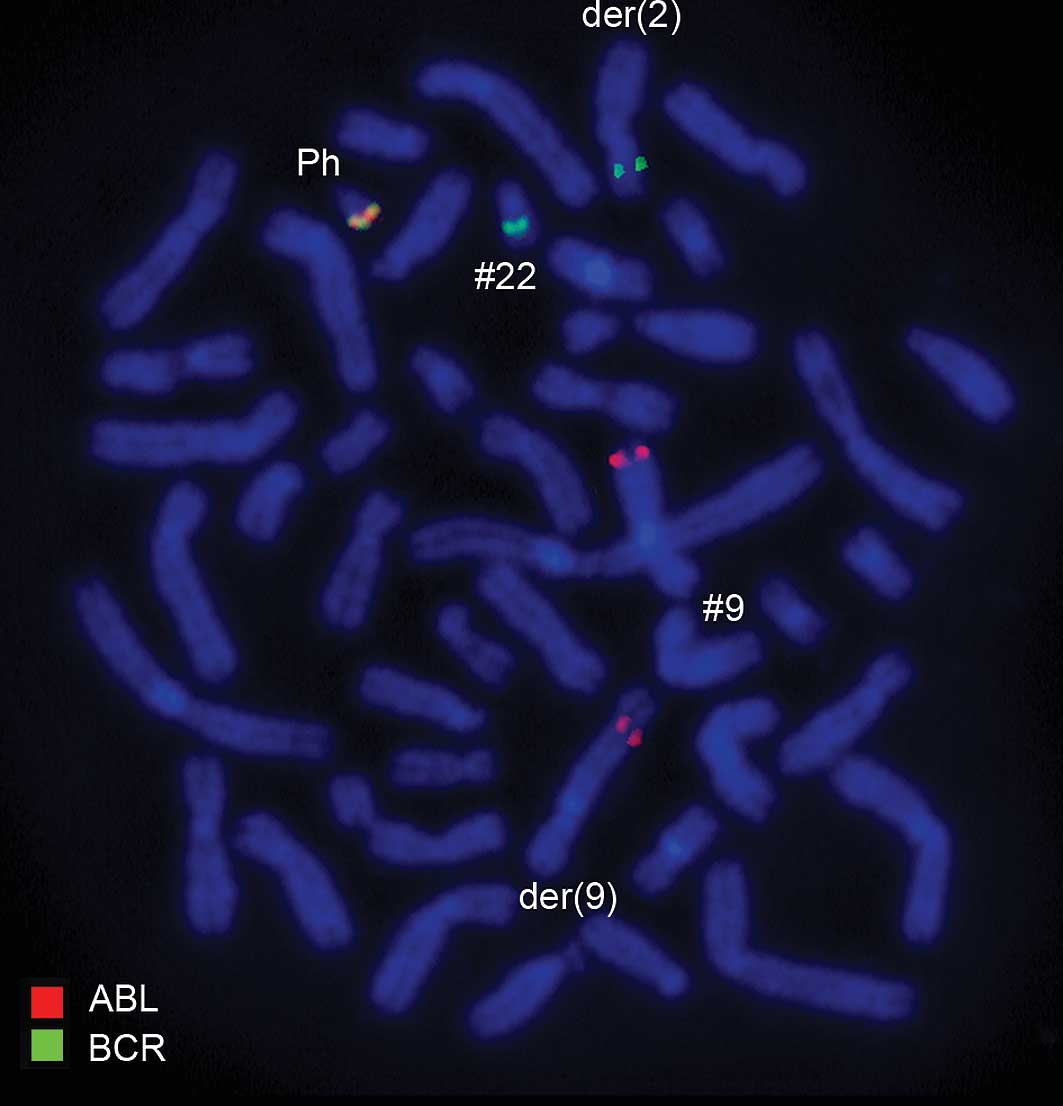
Using a commercially available probe specific for BCR/ABL,
dual-color FISH showed that the typical Ph chromosome with the
BCR/ABL translocation was present. However, BCR was translocated to
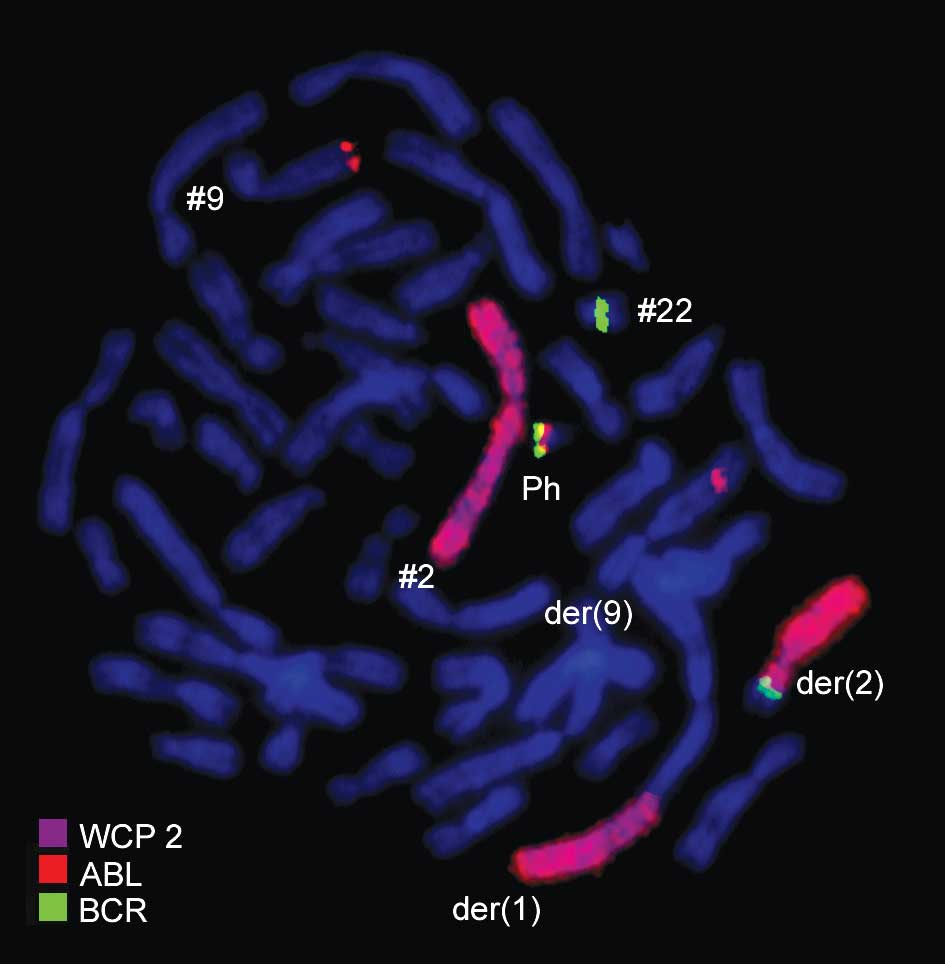
der(2) (Figs. 2 and 3). Another commercially available probe
specific for WCP1 + WCP2 + CEP9 confirmed the involvement of
chromosome 1 with chromosomes 2 and 9 (Fig. 4). Thus, FISH was performed using
probes for the involved chromosomes according to GTG-banding
(Figs. 2–4). The result obtained was:
46,XX,t(1;2;9;22)(p32;q21;q34;q11.2).
Discussion
The present study identified one additional
translocation, 46,XX,t(1;2;9;22)(p32;q21;q34;q11.2), in CML-CP. To
the best of our knowledge, this translocation has never been
described in the literature (12).
In 5–8% of CML cases, the fusion gene BCR/ABL is the
result of a complex translocation (13). At present, it appears that variant
translocations can affect any chromosome. However, it has been
suggested that the distribution of the breakpoints is non-random,
with the chromosomal bands 1p36, 3p21, 5q31, 6p21, 9q22, 10q22,
11q13, 12p13, 17p13, 17q21, 17q25, 19q13, 21q22, 22q12 and 22q13
being the most susceptible to breakage (5). None of the above-mentioned breakpoints
were noted in our study. However, the fusion gene is located on
chromosome 22.
Two possible mechanisms for variant translocation
formation have been suggested. The first is a single-event
rearrangement via the simultaneous breakage of several chromosomes
followed by mismatched joining (14). Nacheva et al proposed a
classic Ph translocation followed by a further translocation event
between chromosomes 9 and 22 plus a third chromosome (15). The mechanism of the formation of a
variant Ph translocation may have prognostic importance in that a
two-event mechanism represents clonal evolution, whereas a variant
translocation occurring via a single genomic rearrangement may
confer a similar prognosis to the classic Ph translocation
(16).
Acknowledgements
We thank Dr I. Othman, the Director General of the
Atomic Energy Commission of Syria (AECS) and Dr N. Mirali, Head of
the Molecular Biology and Biotechnology Department for the support.
This study was supported by the Syrian Atomic Energy
Commission.
References
|
1
|
Gale RP and Canaani E: An 8-kilobase abl
RNA transcript in chronic myelogenous leukemia. Proc Natl Acad Sci
USA. 81:5648–5652. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
De Klein A, van Kessel AG, Grosveld G,
Bartram CR, Hagemeijer A, Bootsma D, Spurr NK, Heisterkamp A,
Groffen J and Stephenson JR: A cellular oncogene is translocated to
the Philadelphia chromosome in chronic myelogenous leukaemia.
Nature. 300:765–777. 1982.PubMed/NCBI
|
|
3
|
Rowley JD: A new consistent chromosomal
abnormality in chronic myelogenous leukemia identified by
quinacrine fluorescence and Giemsa staining. Nature. 243:290–291.
1973. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lugo T, Pendergast A, Müller A and Witte
O: Tyrosine kinase activity and transformation potency of bcr-abl
oncogene products. Science. 247:1079–1082. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Johansson B, Fioretos T and Mitelman F:
Cytogenetic and molecular genetic evolution of chronic myeloid
leukemia. Acta Haematol. 107:76–94. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Huret JL: Complex translocations, simple
variant translocations and Ph-negative cases in chronic myelogenous
leukaemia. Hum Genet. 85:565–568. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hagemeijer A, de Klein A, Godde-Salz E,
Turc-Carel C, Smit EME, van Aghtoven AJ and Grosveld GC:
Translocation of c-abl to ‘masked’ Ph in chronic myeloid leukemia.
Cancer Genet Cytogenet. 18:95–104. 1985.
|
|
8
|
Reid A, Gribble SM, Huntly BJ, Andrews KM,
Campbell L, Grace CD, Wood ME, Green AR and Nacheva EP: Variant
Philadelphia translocations in chronic myeloid leukaemia can mimic
typical blast crisis chromosome abnormalities or classic t(9;22): a
report of two cases. Br J Haematol. 113:439–442. 2001. View Article : Google Scholar
|
|
9
|
Claussen U, Michel S, Mühlig P, Westermann
M, Grummt UW, Kromeyer-Hauschild K and Liehr T: Demystifying
chromosome preparation and the implications for the concept of
chromosome condensation during mitosis. Cytogenet Genome Res.
98:136–146. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mitelman F: ISCN An International System
for Human Cytogenetic Nomenclature. Karger; Basel: 1995
|
|
11
|
Al-Achkar W, Wafa A and Nweder MS: A
complex translocation t(5;9;22) in Philadelphia cells involving the
short arm of chromosome 5 in a case of chronic myelogenous
leukemia. J Exp Clin Cancer Res. 26:411–415. 2007.PubMed/NCBI
|
|
12
|
Mitelman F, Johansson B and Mertens F:
http://cgap.nci.nih.gov/chromosomes/Mitelman.
Mitelman database on chromosome abberrations in cancer. Accessed
Oct. 25, 2007
|
|
13
|
La Starza R, Testoni N, Lafage-Pochitaloff
M, Ruggeri D, Ottaviani E, Perla G, Martelli MF, Marynen P and
Mecucci C: Complex variant Philadelphia translocations involving
the short arm of chromosome 6 in chronic myeloid leukemia.
Haematologica. 87:143–147. 2002.
|
|
14
|
Fitzgerald PH and Morris CM: Complex
chromosomal translocations in the Philadelphia chromosome
leukemias. Serial translocations or a concerted genomic
rearrangement? Cancer Genet Cytogenet. 57:143–151. 1991. View Article : Google Scholar
|
|
15
|
Nacheva E, Holloway T, Brown K, Bloxham D
and Green AR: Philadelphia-negative chronic myeloid leukaemia:
detection by FISH of BCR-ABL fusion gene localized either to
chromosome 9 or chromosome 22. Br J Haematol. 87:409–412. 1994.
View Article : Google Scholar
|
|
16
|
Reid AG, Huntly BJP, Grace C, Green AR and
Nacheva EP: Survival implications of molecular heterogeneity in
variant Philadelphia-positive chronic myeloid leukaemia. Br J
Haematol. 121:419–427. 2003. View Article : Google Scholar : PubMed/NCBI
|