Introduction
Esophageal squamous cell carcinoma (ESCC) is the
ninth most frequent cancer and the sixth most frequent cause of
death from malignant tumors in Japan. The number of deaths due to
esophageal cancer has increased steadily. ESCC is often diagnosed
at an advanced stage, although even in early stage a number of
patients develop local tumor recurrence or distant metastasis
within a short period after curative surgery. Although
pre-operative chemotherapy and chemoradiotherapy are currently used
for patients with advanced-stage ESCC, the effects of such
modalities are unsatisfactory, prompting a search for new treatment
strategies.
MicroRNAs (miRNAs) are a species of small non-coding
single-stranded RNA of approximately 21–23 nucleotides that
post-transcriptionally modulate gene expression by negatively
regulating the stability or translational efficiency of their
target mRNAs. miRNAs are a class of gene products that were
recently found to play a role in several types of cancer (1–4).
miRNAs may function as potent regulators of gene expression and
altered miRNA levels result in aberrant expression of gene products
that may contribute to cancer biology (5). Furthermore, certain miRNAs may
function as either oncogenes or tumor-suppressor genes (6).
A previous study investigated the expression of 73
mature miRNAs in 30 ESCC patients using Taq Man quantitative PCR
(7). The expression of miR-34b was
high in all of the ESCC tissues. This study investigated miR-34b
expression in 88 patients with ESCC and evaluated its correlation
with clinicopathological features and postoperative survival. The
miR-34b expression in ESCC cell lines was also examined and the
effects of suppressing or overexpressing miR-34b on the
proliferation of ESCC cells were analyzed.
Materials and methods
Patients and tumor samples
Esophageal cancer samples were obtained from 88
patients who had undergone surgery at the Nagoya City University
Hospital, Japan, between January 1996 and December 2002. The study
design was approved by the Institutional Review Board of our
university hospital, and a written consent was obtained from all of
the patients. Tumors were classified according to the Guidelines
for the Clinical and Pathological Studies on Carcinoma of the
Esophagus. The tumor and corresponding normal tissue were also
obtained. Normal esophageal mucosa was removed from the apparently
non-cancerous mucosa as far as possible from the tumor. The samples
were frozen immediately in liquid nitrogen and stored at −80°C
until use. The characteristics of the 88 patients with ESCC are
shown in Table I.
 | Table ICorrelation of miR-34b expression with
clinicopathological factors in esophageal cancer. |
Table I
Correlation of miR-34b expression with
clinicopathological factors in esophageal cancer.
| Characteristics | No. of patients
(n=88) | miR-34b
expressiona | P-value |
|---|
| Normal | 88 | 1.00 | |
| Tumor | 88 | 34.18±12.55 | 0.004 |
| Age at surgery |
| ≤65 years | 48 | 20.89±9.61 | |
| >65 years | 40 | 50.12±25.04 | 0.250 |
| Gender |
| Male | 72 | 39.96±15.26 | |
| Female | 16 | 8.14±2.85 | 0.330 |
| Tumor status |
| T1 | 21 | 7.41±3.08 | |
| T2 | 12 | 32.44±21.09 | |
| T3 | 32 | 59.68±29.71 | |
| T4 | 23 | 24.04±21.26 | |
| T1 vs. T2, -3,
-4 | | | 0.200 |
| T1, -2 vs. T3,
-4 | | | 0.630 |
| T1, -2, -3 vs.
T4 | | | 0.540 |
| Lymph node
status |
| N0 | 24 | 44.63±33.69 | |
| N1 | 14 | 33.81±29.25 | |
| N2 | 27 | 10.07±3.84 | |
| N3 | 12 | 4.43±2.07 | |
| N4 | 9 | 85.04±58.27 | |
| Unknown | 2 | 186.29±185.93 | |
| N0 vs. N1, -2,
-3 | | | 0.730 |
| Pathological
stage |
| 0 | 5 | 7.51±4.43 | |
| I | 10 | 1.64±0.73 | |
| II | 17 | 66.67±47.07 | |
| III | 26 | 23.59±15.96 | |
| IV | 30 | 40.23±21.46 | |
| 0, I vs. II, III,
IV | | | 0.039 |
| Histological
differentiation |
| Well | 31 | 7.94±4.26 | |
| Moderate | 47 | 55.92±22.94 | |
| Poor | 10 | 13.30±5.30 | |
| Well vs. moderate,
poor | | | 0.120 |
| Well, moderate vs.
poor | | | 0.550 |
| Lymphatic
invasion |
| Negative | 17 | 4.77±2.02 | |
| Positive | 57 | 35.92±13.43 | 0.850 |
| Unknown | 14 | 62.80±57.49 | |
| Blood vessel
invasion |
| Negative | 29 | 23.57±16.80 | |
| Positive | 45 | 32.11±13.47 | 0.640 |
| Unknown | 14 | 62.80±57.49 | |
| p53 IHC |
| Negative | 23 | 21.05±12.31 | |
| Positive | 36 | 23.43±13.70 | 0.950 |
| Unknown | 29 | 57.93±32.68 | |
Cell lines and cell culture
ESCC cell lines (TE1-15) were obtained from the
Japanese Collection of Research Bioresources. Cultures were
maintained in RPMI-1640 (Sigma) supplemented with 10% fetal bovine
serum (FBS) (Gibco) at 37°C in a humidified 5% CO2
incubator.
A human esophageal squamous epithelial cell line
(Het-1A) was obtained from the American Type Culture Collection.
Het-1A was maintained in serum-free medium (LHC-9; BioSource) at
37°C in a humidified 5% CO2 incubator.
RNA extraction
Total RNA was extracted from ESCC tissue, as well as
its corresponding non-cancerous mucosa, using the Absolutely
RNA™ RT-PCR Miniprep kit (Stratagene, La Jolla, CA, USA)
according to the manufacturer’s instructions. The concentration of
total RNA was adjusted to 2 ng/μl using a spectrophotometer.
Quantitative reverse
transcription-polymerase chain reaction
Taq Man miRNA assays (ABI PRISM, Forest City, CA,
USA) employed the stemloop method to detect the expression level of
mature miR-34b. For reverse transcription (RT) reactions, 10 ng of
total RNA was used in each reaction (5 μl) and mixed with the RT
primer (3 μl). The RT reaction was carried out at 16°C for 30 min;
42°C for 30 min; 85°C for 5 min; and then maintained at 4°C. After
the RT reaction, 1.33 μl of the cDNA was used for the polymerase
chain reaction (PCR) along with Taq Man primers (2 μl). The PCR
reaction was conducted at 95°C for 10 min followed by 40 cycles of
95°C for 15 sec and 60°C for 60 sec in the ABI 7500 real-time PCR
system. The real-time PCR results were analyzed and expressed as a
relative miRNA expression of threshold cycle (CT) value. RT and PCR
primers for miR-34b were purchased from ABI PRISM. The expression
of miR-34b was calculated using the 2−ΔΔCt analysis
method (8). The expression of
miR-34b was normalized to that of the U6B small nuclear RNA gene
(RNU6B).
p53 evaluation by immunohistochemistry
(IHC)
Immunohistochemical staining of p53 was performed on
4-mm sections from the paraffin-embedded tumors. Antigen retrieval
was carried out by microwaving the tissue sections in
phosphate-buffered saline (PBS) (0.1 M, pH 7.2) for 15 min followed
by incubation with a monoclonal anti-p53 antibody (p-53 protein
PAb240; Dako, Glostrup, Denmark) diluted 1:75 in PBS overnight at
4°C. Primary antibody was detected by EnVision™/HRP
Mouse code K4001 (Dako) and visualized by Dako Liquid DAB. Counter
staining was performed with Mayers hematoxylin. Nuclear p53
staining intensity was defined as high or low. Immunoreactivity for
p53 was evaluated semiquantitatively by two observers and all
tumors showing p53 immunoreactivity were assumed to have p53
mutation.
Transfection
Nuclear transfection was performed by using the
Nucleofector system (Amaxa Biosystems, Koln, Germany). Cells
(1×105) were suspended in 150 μl Nucleofector solution
(Amaxa Biosystems) containing 100 nM miR-34b precursor, antisense
miR-34b inhibitor or respective controls at room temperature. Each
assay was performed in triplicate.
MTT assay
Transfected cells were seeded in 96-well plates at a
density of 1×105 cells/100 μl. Cell proliferation was
measured using the MTT method. After 72 h, 20 μl of 5 mg/ml MTT
solution was added to each well and plates were incubated for 3 h
at 37°C. Absorbance at 490 nm was determined using a SPECTRAmax 340
(Molecular Devices Corporation). Six wells were assayed for each
set of conditions and stadard deviations (SDs) were determined.
Statistical analysis
Data are expressed as means ± SD. Statistical
analyses were performed using the software package StatView (Abacus
Concepts, Berkeley, CA, USA). The Wilcoxon signed-rank,
Mann-Whitney U and Kruskal-Wallis tests were used to evaluate the
significance of differences in the expression levels of miR-34b. In
all analyses, P<0.05 was considered to be statistically
significant.
Results
Expression of miR-34b in esophageal
cancer tissue
Quantitative RT-PCR was used to evaluate the miRNA
expression in 88 tumors and paired normal esophageal tissues. The
expression level of miR-34b was significantly higher in the tumor
tissue than in the corresponding non-cancerous mucosa (Table I). The expression of miR-34b in the
normal esophageal mucosa was very low (Fig. 1).
The relationship between miR-34b expression, in 88
ESCC samples, and patient clinicopathological factors was examined
(Table I). The miR-34b expression
levels in patients with advanced stage 2, 3 or 4 were higher than
those in patients with stage 0 or 1 tumors (P=0.039, Mann-Whitney U
test) (Fig. 2). No significant
differences were noted in miR-34b expression with respect to age,
gender, the depth of invasion (T-factor), lymph node status,
histological differentiation, lymphatic invasion, blood vessel
invasion and p53 IHC. Additionally, no significant relationship was
found between miR-34b expression and patient survival (data not
shown).
Expression of miR-34b in esophageal
cancer cell lines
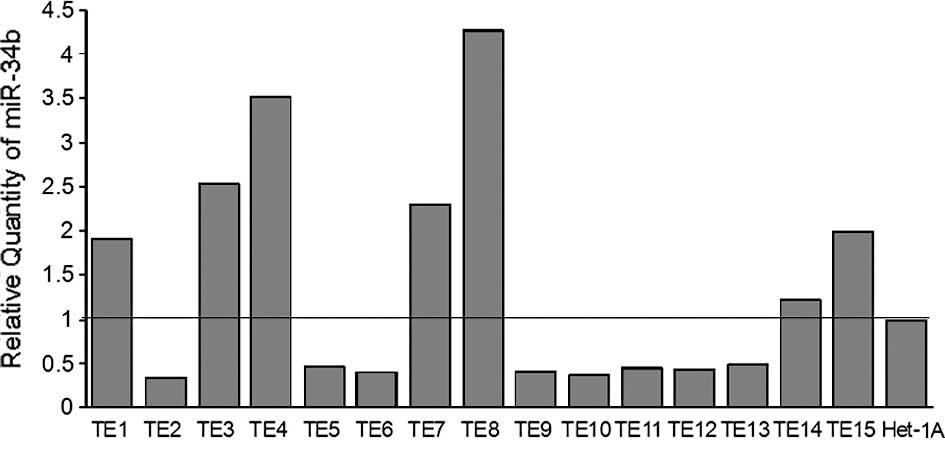
The expression of miR-34b was investigated in 15
esophageal cancer cell lines (TE1-15) and 1 non-cancerous
esophageal squamous epithelial cell line (Het-1A) using quantative
RT-PCR. The expression of miR-34b and U6B was detectable in all of
the cell lines studied. The level of expression of miR-34b varied
among the cell lines: TE2 had the lowest expression and TE8 the
highest (Fig. 3).
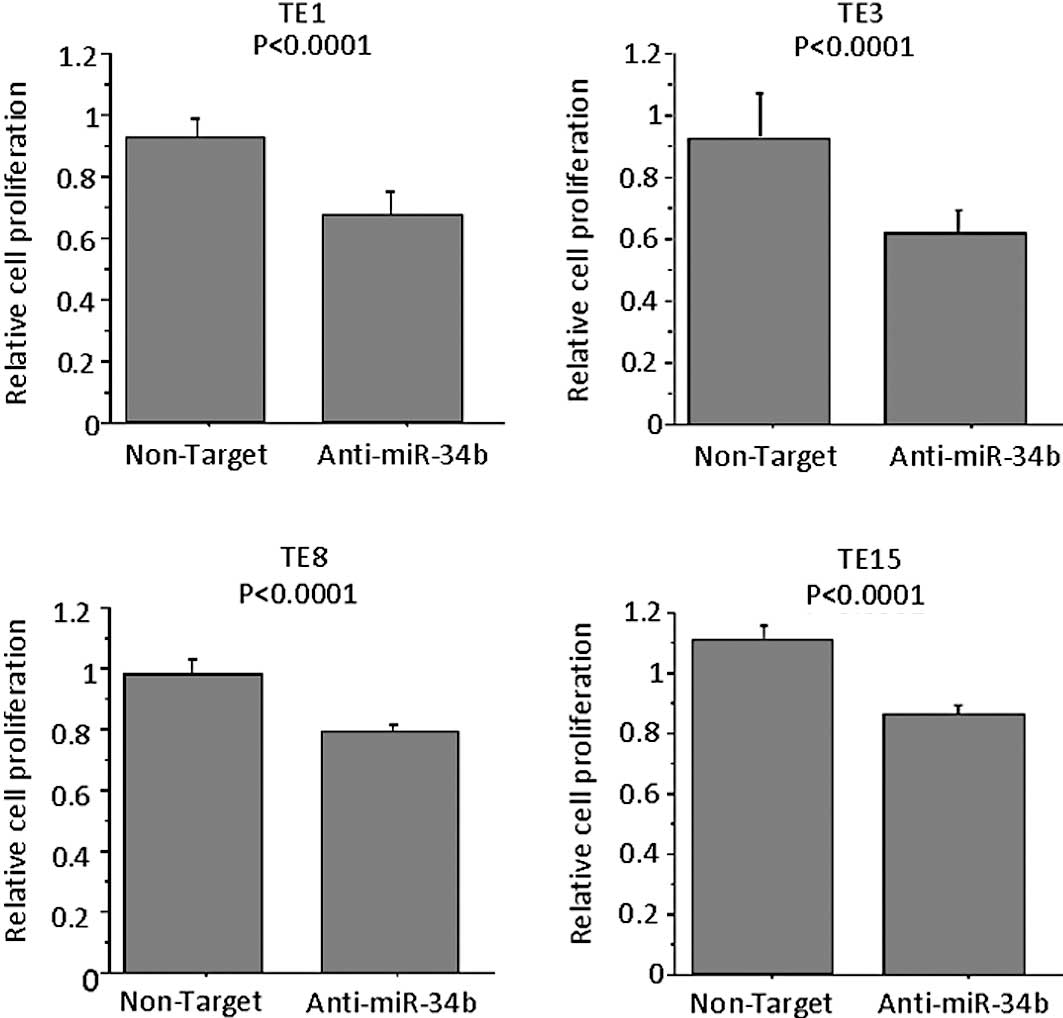
Effects of suppression of miR-34b in ESCC
cell lines
To examine the role of miR-34b on the proliferation
of ESCC cells, antisense miRNA inhibitor or respective controls
were transfected into TE8 cells (the highest expressor of miR-34b
among the TE series cell lines). MTT assay confirmed that the
proliferation of anti-miR-34b-transfected TE1, TE3, TE8 and TE15
cells was significantly lower as compared to control
inhibitor-transfected cells on Day 3 (Fig. 4). When TE6, a miR-34b-low cell line,
was used no difference was noted in the proliferation following the
transfection of antisense miR-34b (data not shown). Anti-miR miRNA
inhibitors (patent pending) are designed to bind to and inhibit not
the expression of miR-34b, but the activity of endogenous miRNAs
when introduced into cells.
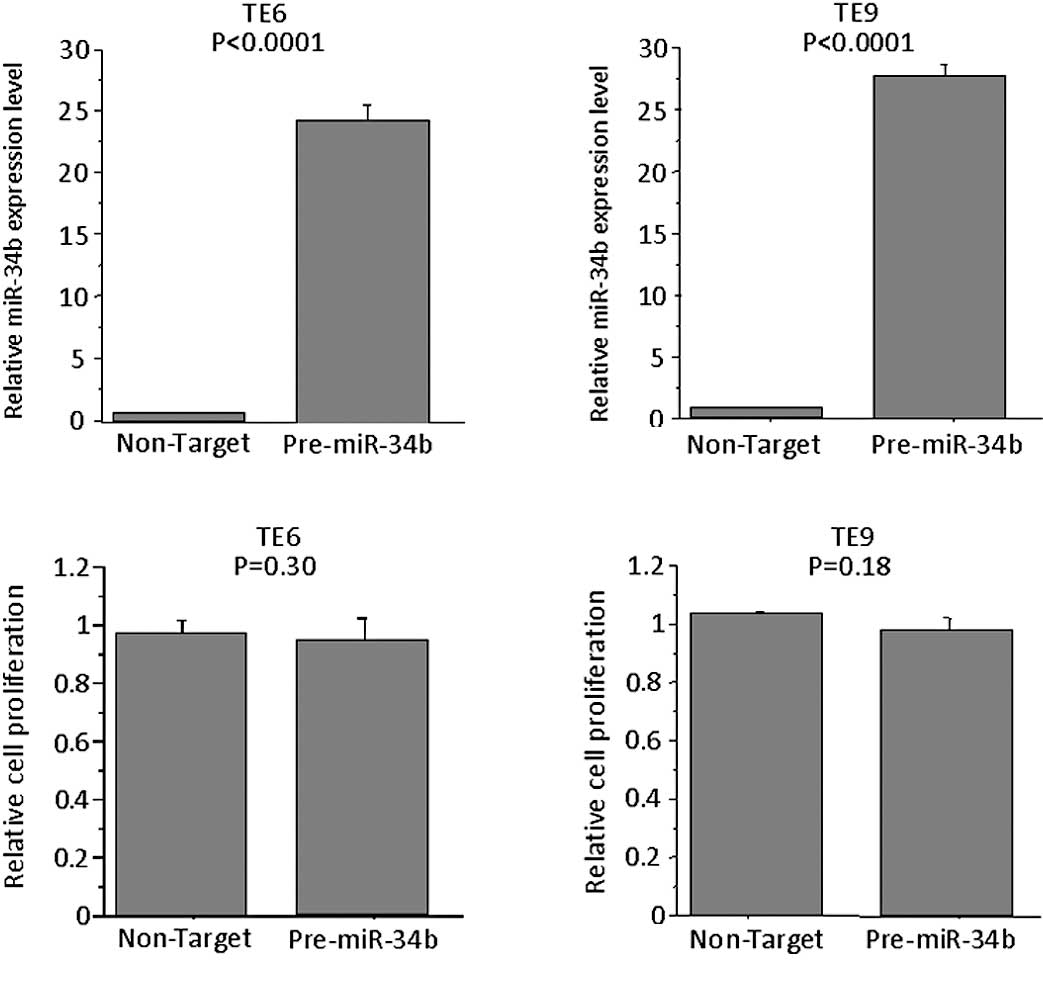
Effects of overexpression of miR-34b in
TE6 and TE9 cells
To determine whether the overexpression of miR-34b
affects the proliferation of TE6 and -9 cells, the miRNA precursor
was transfected into these cells, which express miR-34b at low
levels. The expression levels of miR-34b in pre-miR-34b-
transfected cells were higher than control precursor
miR-transfected cells. However, no significant difference was
observed in the proliferation of these cells as compared to cells
transfected with control precursor miR on Day 3 (Fig. 5).
Discussion
We analyzed the expression of miR-34b in 88 ESCC
samples by RT-PCR. The expression levels of miR-34b in ESCC were
significantly higher than those in normal esophageal tissues.
Moreover, miR-34b was more highly expressed in tumors with stages
2–4 than in those with stages 0 or 1. These results suggest that
the expression of miR-34b is oncogenic in ESCC. miR-34b is located
at chromosome 11q23.1. Recently, 11q23 amplification was described
as a new cytogenetic entity in myeloid malignancies (9). However, no study is currently
available on the amplification of 11q23.1 in ESCC.
It was reported that the expression of miR-34b is
high in squamous cell carcinoma of the tongue (10) and undifferentiated gastric cancer
(11). To study the functional
roles of miR-34b in ESCC, we knocked down miR-34b using the
specific inhibitor in the ESCC cell lines. We found that the
proliferation of ESCC cell lines with a high expression of miR-34b
was inhibited by antisense miR-34b. The results indicate a close
association between miR-34b expression and the proliferation of
ESCC cells. However, not all of the ESCC cell lines expressed
miR-34b at higher levels than the normal esophageal cell line
(Het-1A) (Fig. 3). Additionally,
the overexpression of miR-34b in the miR-34b-low TE6 or -9 cell
lines had no effect on proliferation. These results indicate that
the role of miR-34b in cell proliferation is limited to subsets of
ESCC patients.
miR-34b was recently identified as a p53 target and
a potential tumor suppressor (12–15).
Over 50% of human cancers have mutant p53 and the expression of
miR-34a, b and c appears to be correlated with p53 (16,17).
In human tumors, the selective pressure to lose miR-34s may be
relieved by the frequent mutation of p53. Thus, genetic alterations
in miR-34s are more likely to occur in tumor types that
characteristically contain wild-type p53 (18). The mutation of p53 was found in
approximately 50% of cell lines and the primary tumor of the
esophagus (19). In our study, p53
immunohistochemistry was positive in 59.3% of the ESCC patients.
However, no difference was found in the expression of miR-34b
between the p53-positive and -negative patients. Our study suggests
that miR-34b functions as an oncogene as opposed to a tumor
suppressor. It is possible that miR-34b functions in a
context-dependent manner or that other factors may regulate
miR-34b.
In esophageal cancer, it has been reported that a
high expression of miR-103/miR-107 is associated with a poor
prognosis (20). miRNA processing
enzyme (RNASEN) was elevated in a proportion of ESCC and a high
RNASEN expression correlates with poor prognosis in ESCC (21). Thus, it appears that miRs play a
role in ESCC.
In conclusion, further studies on the role of
miR-34b on ESCC progression including identification of its target
genes are warranted.
Acknowledgements
The authors would like to thank Ms. Shinobu Makino
for the excellent technical assistance.
References
|
1
|
McManus MT: MicroRNAs and cancer. Semin
Cancer Biol. 13:253–258. 2003. View Article : Google Scholar
|
|
2
|
Michael MZ, O’ Connor SM, van Holst
Pellekaan NG, Young GP and James RJ: Reduced accumulation of
specific microRNAs in colorectal neoplasia. Mol Cancer Res.
12:882–891. 2003.PubMed/NCBI
|
|
3
|
Calin GA and Croce CM: MicroRNA-cancer
connection: the beginning of a new tale. Cancer Res. 66:7390–7394.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Esquela-Kerscher A and Slack FJ: Oncomirs
– microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269.
2006.
|
|
5
|
Meng F, Henson R, Wehbe-Janek H, Ghoshal
K, Jacob ST and Patel T: MicroRNA-21 regulates expression of the
PTEN tumor suppressor gene in human hepatocellular cancer.
Gastroenterology. 133:647–658. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lu J, Getz G, Miska EA, et al: MicroRNA
expression profiles classify human cancers. Nature. 435:834–838.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ogawa R, Ishiguro H, Kuwabara Y, et al:
Expression profiling of microRNAs in human esophageal squamous cell
carcinoma using RT-PCR. Med Mol Morphol. 42:102–109. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Poppe B, Vandesompele J, Schoch C, et al:
Expression analyses identify MLL as a prominent target of 11q23
amplification and support an etiologic role for MLL gain function
in myeloid malignancies. Blood. 103:229–235. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP
and Wei WI: Mature miR-184 as potential oncogenic microRNA of
squamous cell carcinoma of tongue. Clin Cancer Res. 14:2588–2592.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Katada T, Ishiguro H, Kuwabara Y, et al:
microRNA expression profile in undifferentiated gastric cancer. Int
J Oncol. 34:537–542. 2009.PubMed/NCBI
|
|
12
|
Chang TC, Wentzel EA, Kent OA, et al:
Transactivation of miR-34a by p53 broadly influences gene
expression and promotes apoptosis. Mol Cell. 26:745–752. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
He L, He X, Lim LP, et al: A microRNA
component of the p53 tumour suppressor network. Nature.
447:1130–1134. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hermeking H: p53 enters the microRNA
world. Cancer Cell. 12:414–418. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Raver-Shapira N, Marciano E, Meiri E, et
al: Transcriptional activation of miR-34a contributes to
p53-mediated apoptosis. Mol Cell. 26:731–743. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bommer GT, Gerin I, Feng Y, et al:
p53-mediated activationof miRNA34 candidate tumor-suppressor genes.
Curr Biol. 17:1298–1307. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
He X, He L and Hannon GJ: The guardian’s
little helper: microRNAs in the p53 tumor suppressor network.
Cancer Res. 67:11099–11101. 2007.
|
|
18
|
Tarasov V, Jung P, Verdoodt B, et al:
Differential regulation of microRNAs by p53 revealed by massively
parallel sequencing: miR-34a is a p53 target that induces apoptosis
and G1-arrest. Cell Cycle. 6:1586–1593. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Nishihira T, Hashimoto Y, Katayama M, Mori
S and Kuroki T: Molecular and cellular features of esophageal
cancer cells. J Cancer Res Clin Oncol. 119:441–449. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Guo Y, Chen Z, Zhang L, et al: Distinctive
microRNA profiles relating to patient survival in esophageal
squamous cell carcinoma. Cancer Res. 68:26–33. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sugito N, Ishiguro H, Kuwabara Y, et al:
RNASEN regulates cell proliferation and affects survival in
esophageal cancer patients. Clin Cancer Res. 12:7322–7328. 2006.
View Article : Google Scholar : PubMed/NCBI
|