Introduction
Chronic myeloid leukemia (CML) is a clonal
myeloproliferative disorder of pluripotent hematopoieitic stem
cells characterized by the BCR and ABL somatic gene rearrangement.
In 90–95% of cases with CML, the BCR/ABL fusion gene is the result
of the reciprocal translocation between chromosomes 9 and 22, with
cytogenetically observable small derivative chromosome 22, which is
known as the Philadelphia (Ph) chromosome (1,2).
Expression of the BCR/ABL chimeric protein with an increased
tyrosine kinase activity plays an essential role in the
pathogenesis of CML (3). However,
little is known about the corresponding reciprocal translocation
product ABL/BCR. The fusion gene ABL/BCR is transcriptionally
active in only one-third of CML patients (4). A deletion involving the
arginosuccinate synthase (ASS) and ABL genes, located on the
derivative chromosome 9, is found in 10–15% of CML patients and is
associated with a shortened chronic phase and a decrease in
survival in the absence of imatinib treatment (5). Even when deletion of these genes
directly leads to loss of ABL/BCR, ABL-BCR expression does not
correlate with poor prognosis (6).
The progression of CML from the chronic phase (CP) to blast crisis
(BC) is frequently associated with non-random secondary chromosomal
aberrations, such as +8, i(17q), +19 and an extra Ph (7). At the molecular level, mutation of the
tumor suppressor gene TP53 located at 17p13 (8) is detected in 25–30% of CML-myeloid BC.
However, no mutation of the remaining TP53 allele in CML cases with
i(17q) has been noted (9).
This study examined a rare case of CML with inverted
chromosome 9 and unbalanced translocation between chromosomes 10
and 17.
Materials and methods
Case report
A 39-year-old woman was referred to our laboratory
in December 2005, due to an occasional blood test showing moderate
leukocytosis [the white blood count (WBC) was
113.5×109/l (74% neutrophils, 25% lymphocytes and 1%
eosinophils)]. The platelet count was 144.5×109/l and
the hemoglobin level was 11.9 g/dl. A physical examination showed
splenomegaly. The WBC differential, as well as the bone marrow
aspirate, were consistent with CP of CML. The patient had not
received any treatment prior to the date of the test. Additionally,
patient consent was obtained for this study.
Chromosome analysis
Chromosome analysis using GTG- banding was conducted
according to standard procedures (10). A total of 20 metaphases, derived
from unstimulated bone marrow of the patient, were analyzed.
Karyotypes were described according to the International System for
Human Cytogenetic Nomenclature (11).
Fluorescence in situ hybridization
analysis
Fluorescence in situ hybridization (FISH)
using the LSI BCR/ABL+9q34 Tricolor dual fusion translocation probe
(Abbott Molecular/Vysis, USA) and 17p13(p53)/alpha-satellite 17,
dual color (Q-Biogene, USA) were applied according to the
manufacturer’s instructions. A multicolor banding probe (MCB),
based on microdissection-derived region-specific libraries for
chromosome 9, was applied as previously described (12,13). A
total of 20 metaphase spreads were analyzed, each using a
fluorescence microscope (Axio Imager.Z1 mot; Zeiss) equipped with
the appropriate filter sets to discriminate between a maximum of
five fluorochromes and the counter-stain DAPI
(4′,6-diamino-2-phenylindole). Image capturing and processing were
carried out using an ISIS imaging system (MetaSystems, Altlussheim,
Germany) for the MCB evaluation.
Results
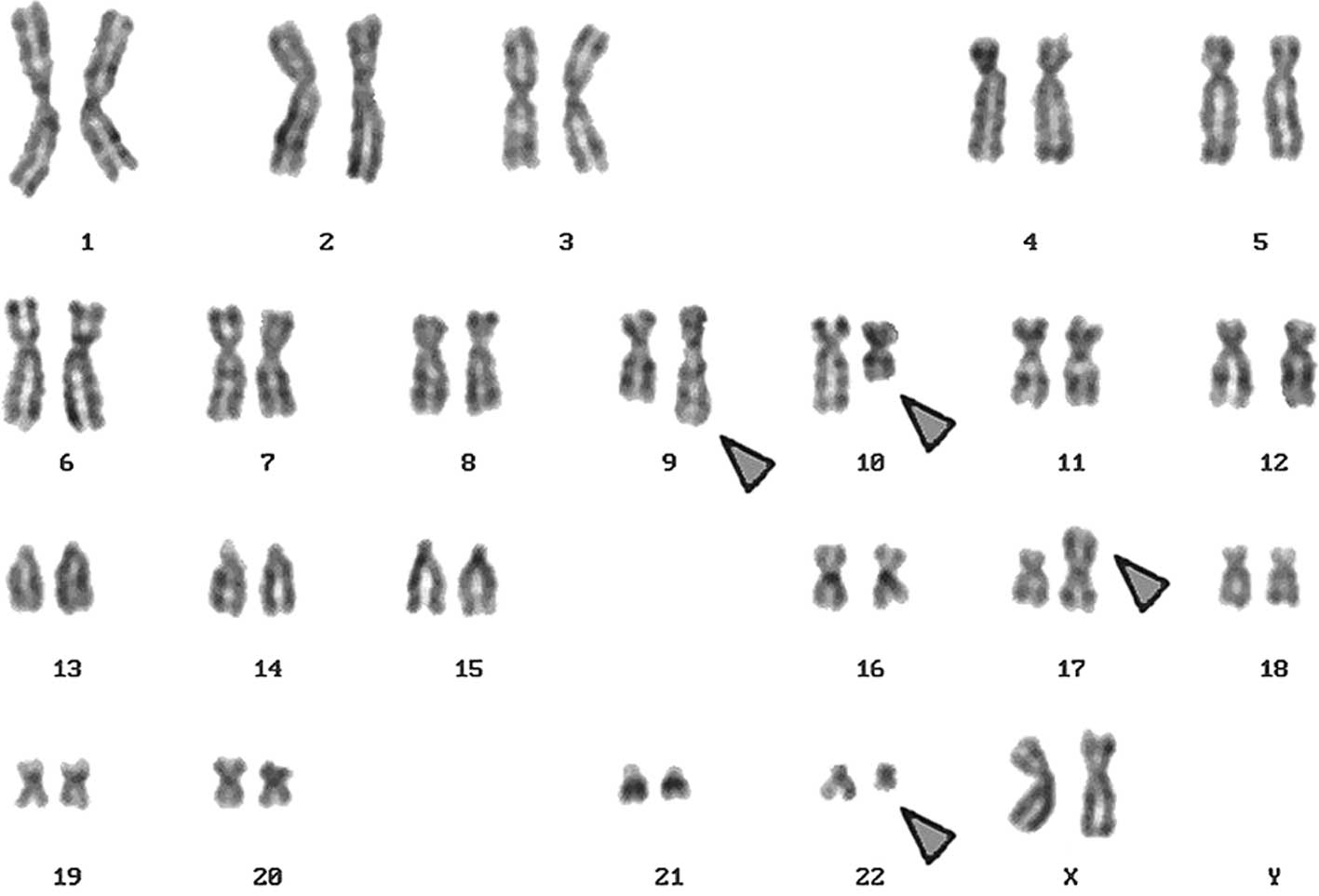
A complex karyotype, 46,XX,t(9;22),t(10;17), was
determined in the GTG-banding (Fig.
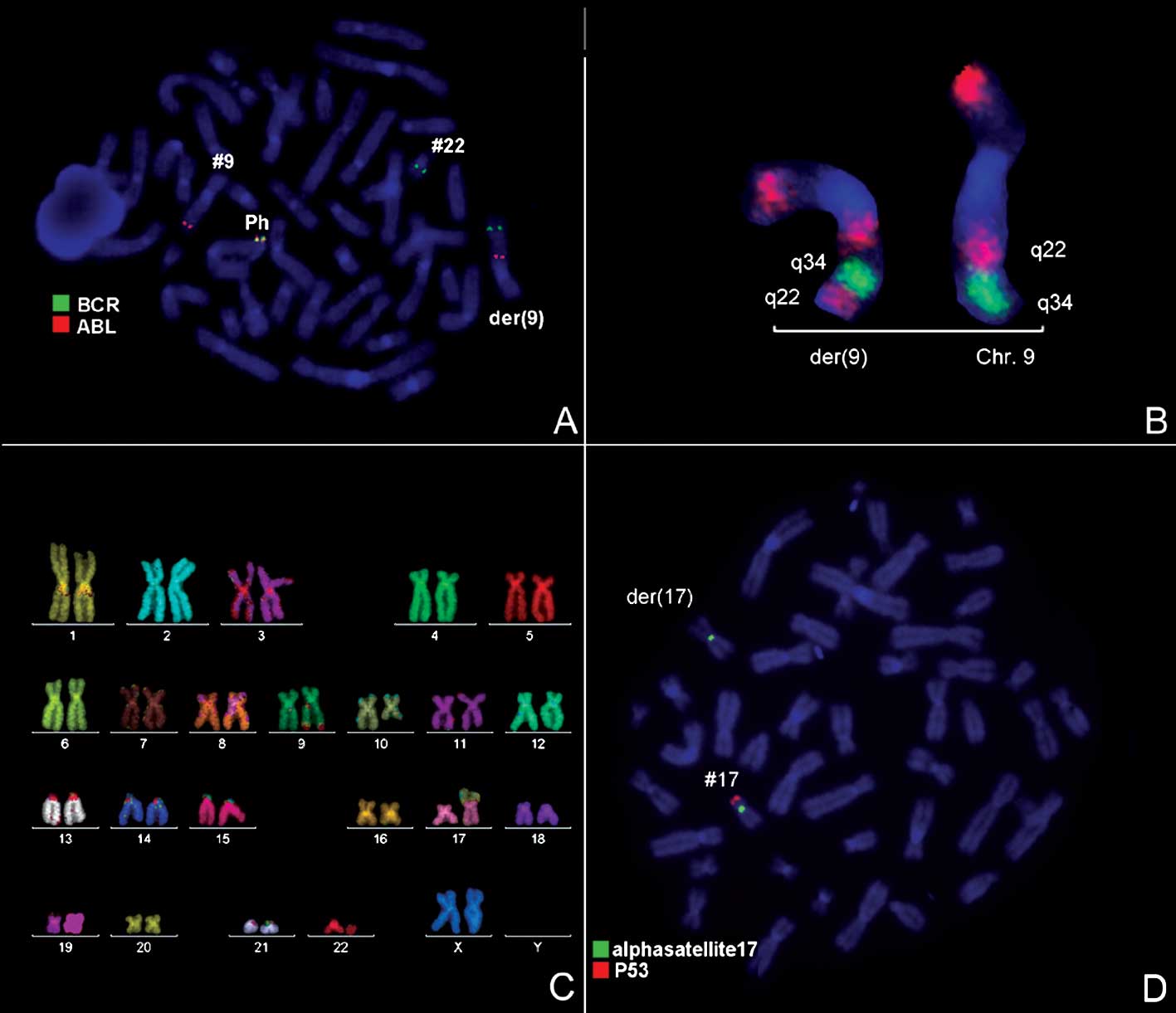
1) and was further studied by molecular cytogenetics (Fig. 2). Tri-color FISH using a probe
specific for BCR, ABL and the ASS gene revealed a typical Ph with
the BCR/ABL fusion gene. However, the ASS gene was localized more
proximal to the centromere, as 3′BCR was in the terminal part of
the derivative chromosome 9 (Fig.
2A). Inversion of 9q22q34 was detected using MCB 9 (Fig. 2B). Thus, M-FISH was applied to
confirm the GTG-banding results (Fig.
2C). The locus-specific probe 17p13(p53)/alpha-satellite 17
demonstrated the absence of 17p13 (Fig.
2D). The result obtained was: 46,XX,der(9)t(9;22)(q34;q11)inv(9)(q22q34),der(10)t(10;17)(q22.3;p13.1),der(17)t(10;17)(q22.3;p13.1)del(17)(p13.1),der(22)t(9;22)(q34;q11).
Discussion
The present study identified two additional
chromosomal alterations, inv(9)(q22q34) and
t(10;17)(q22.3;p13.1)del(17)(p13.1), in Ph+ CML-CP. To
our knowledge, the translocation between 10q22.3 and 17p13.1 was
previously described in clear-cell sarcoma (2 cases) and
endometrial stroma sarcoma (3 cases). Thus, this translocation has
never been observed in CML, and the breakpoint 10q22 has been shown
to be a partner in the variant translocation t(9;22;V) (14). Although 3′BCR was localized on
derivative chromosome 9, the ABL/BCR fusion gene was absent due to
inv(9)(q22q34). Therefore, we
suggest that the derivative chromosome 9 was formed in a two-event
rearrangement. The first step was the reciprocal translocation
between chromosome 9 and 22, resulting in subsequent inversion.
Reid et al described two CML cases with variant t(9;22;V),
where 5′ABL and 3′BCR signals remained present on the long arm of
the derivative chromosome 9, but were separated by additional
material of a third partner (15).
The breakpoint 9q22 was previously shown to be involved in the
formation of variant translocation t(9;22;V) in 2 cases with CML
(14).
The unbalanced translocation on
t(10;17)(q22.3;p13.1) resulted in the loss of a region of the
chromosome 17p13.1, including the TP53 gene. During CML
progression, isochromosome (17)(q10) is one of the non-random changes.
This aberration is associated with loss of the tumor suppressor
gene TP53 and mostly with poor prognosis (16). Several relevant genes are affected
due to the deletion of 17p and simultaneous formation i(17q). Point
mutation and/or deletion of the TP53 gene are regarded as
potentially important steps in the development of various
hematological malignances (17).
We reported on a novel, unique case of
Ph+ CML with a paracentric inversion of the derivative
chromosome 9 resulted in the absence of the ABL/BCR fusion gene and
the unbalanced translocation of chromosomes 10 and 17 with deletion
of the TP53 gene.
Acknowledgements
We thank Dr I. Othman, Director General of the
Atomic Energy Commission of Syria (AECS) and Dr N. Mirali, Head of
the Molecular Biology and Biotechnology Department for their
support. This study was supported by AECS, in part, by the
Stefan-Morsch-Stiftung, Monika-Kutzner-Stiftung and the DAAD
(D/07/09624).
References
|
1
|
Rooney DE: Human Cytogenetics: Malignancy
and Acquired Abnormalities. 3rd edition. Oxford University Press;
New York: pp. 372001
|
|
2
|
Sessions J: Chronic myeloid leukemia in
2007. Am J Health Syst Pharm. 64:S4–S9. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lugo T, Pendergast A, Müller A and Witte
O: Tyrosine kinase activity and transformation potency of bcr-abl
oncogene products. Science. 247:1079–1082. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Melo JV, Gordon DE, Cross NC and Goldman
JM: The ABL-BCR fusion gene is expressed in chronic myeloid
leukaemia. Blood. 81:158–165. 1993.PubMed/NCBI
|
|
5
|
Quintas-Cardama A, Kantarjian H, Talpaz M,
O’Brien S, Garcia-Manero G, Verstovsek S, Rios M, Hayes K, Glassman
A, Bekele B, Zhou X and Cortes J: Imatinib mesylate therapy may
overcome the poor prognostic significance of deletions of
derivative chromosome 9 in patients with chronic myelogenous
leukemia. Blood. 105:2281–2286. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Melo J, Hochhaus A, Yun X-H and Goldman J:
Lack of correlation of ABL/BCR expression and response to
interferon-α in chronic myeloid leukaemia. Br J Haematol.
92:684–686. 1996.PubMed/NCBI
|
|
7
|
Sandberg AA: The Chromosomes in Human
Cancer and Leukemia. 2nd edition. Elsevier Science; New York: pp.
151–172. 1990
|
|
8
|
Rege-Cambrim G, Caidano G, Serra A,
Scaravaglio P, Guglielmelli T, Guerrasio A, Giovinazzo B and Saglio
G: Analysis of the p53 in myeloid malignancies associated with
chromosomal abnormalities involving the short arm of chromosome 17.
Leukemia. 8:23–26. 1994.PubMed/NCBI
|
|
9
|
Calabretta B and Perrotti D: The biology
of CML blast crisis. Blood. 103:4010–4022. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Claussen U, Michel S, Mühlig P, Westermann
M, Grummt UW, Kromeyer-Hauschild K and Liehr T: Demystifying
chromosome preparation and the implications for the concept of
chromosome condensation during mitosis. Cytogenet Genome Res.
98:136–146. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shaffer L, Slovak M and Cambell L: ISCN
2009: An International System for Human Cytogenetic Nomenclature.
S. Karger; Basel: 2009
|
|
12
|
Weise A, Mrasek K, Fickelscher I, Claussen
U, Cheung SW, Cai WW, Liehr T and Kosyakova N: Molecular definition
of high-resolution multicolor banding probes: first within the
human DNA sequence anchored FISH banding probe set. J Histochem
Cytochem. 56:487–493. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liehr T, Heller A, Starke H, Rubtsov N,
Trifonov V, Mrasek K, Weise A, Kuechler A and Claussen U:
Microdissection based high resolution multicolor banding for all 24
human chromosomes. Int J Mol Med. 9:335–339. 2002.PubMed/NCBI
|
|
14
|
Mitelman F, Johansson B and Mertens F:
Mitelman Database of Chromosome Aberrations in Cancer. 2009,
http://cgap.nci.nih.gov/chromosomes/Mitelman.
|
|
15
|
Reid AG, Huntly BJ, Grace C, Green AR and
Nacheva EP: Survival implications of molecular heterogeneity in
variant Philadelphia-positive chronic myeloid leukaemia. Br J
Haematol. 121:419–427. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Johansson B, Fioretos T and Mitelman F:
Cytogenetic and molecular genetic evolution of chronic myeloid
leukemia. Acta Haematol. 107:76–94. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Imamura J, Miyoshi I and Koeffler HP: p53
in hematologic malignancies. Blood. 84:24121994.PubMed/NCBI
|