Introduction
Immunohistochemical staining has traditionally been
performed on whole tissue sections from paraffin blocks to evaluate
the value of proteins as prognostic markers. However, this
technique, which requires the processing and staining of hundreds
or even thousands of slides, is extremely time-consuming when a
large number of tumors or various markers are investigated. In
1998, Kononen et al (1)
introduced tissue microarray (TMA) technology which facilitated the
retrospective study of a large number of archival formalin-fixed,
paraffin-embedded samples. Using this relatively new technology,
hundreds of specimens can be inserted into a recipient TMA block
that can be tested with different techniques, including
immunohistochemical methods. TMA analysis has the added advantage
that hundreds of specimens are processed simultaneously using
identical conditions. Furthermore, TMA analysis markedly conserves
reagents, saves time and greatly decreases the amount of archival
tissue required for a particular study, thus preserving tissue for
other research or diagnostic needs (2,3). In
addition, analysis of serial TMA sections facilitates the
identification of associations between multiple markers (4,5).
However, the main criticism of TMA is that the amount of tissue
analysed using this technique is limited and may not be
representative of the whole specimen (6). This may pose a significant problem in
malignant epithelial tumors where intratumoral heterogeneity exists
(7,8).
Numerous studies have concentrated upon the
validation of TMA for different tumors by comparing the
immunohistochemical staining results of various core biopsies on
TMA with the results of whole section analysis to consider the
number of cores required to adequately represent the expression of
the antigen (3,6,9–11). Few
studies, however, have correlated survival and clinical data with
both TMA and the results obtained from whole tissue sections
(12–14).
Previously, we identified Ki-67 and p16 (15), using whole tissue sections and
immunohistochemistry, in a large series of stage III ovarian cancer
and compared the results with clinical information. Using
univariate analysis a high expression of Ki-67 (P=0.0001) and p16
(P=0.005) was found to be associated with poor survival. However,
in multivariate analysis only a high expression of Ki-67 was
significantly associated with shorter survival (P=0.025) (15). Therefore, TMAs were generated from
the 171 paraffin-embedded ovarian carcinomas. Each tumor is
represented by 3 punches with a diameter of 0.6 mm, and obtained
from different sites within the active and representative tumor
regions. This study aimed to compare the immunohistochemical
expression of Ki-67 and p16 in whole tissue sections and TMAs and
establish whether data derived from TMAs can be reliably used for
survival analysis in ovarian cancer patients.
Materials and methods
Patients and samples
Tumor tissue samples from 171 patients with stage
III epithelial ovarian cancer, treated in the period from January
1988 to May 1993 at The Norwegian Radium Hospital, were collected
during primary surgery. The median age at diagnosis was 54 years
(range 21–70). The patients were followed-up until they succumbed
to the disease or until December 31st, 2003. Detailed patient
information was reported in our previous study (15). The Regional Committee for Medical
Research Ethics South of Norway (S-06277a), The Social and Health
Directorate (06/3280) and The Data Inspectorate (06/5345) approved
the study.
TMA construction
A total of 171 ovarian carcinomas were used for TMA
construction. Prior to insertion into a TMA block, hematoxylin and
eosin (H&E)-stained sections were created from each tissue
donor block to identify the most appropriate (absence of necrosis,
poorly differentiated) tumor areas. A total of 3 tissue cylinders
with a diameter of 0.6 mm were removed from selected areas of each
tissue donor block using Beecher Instruments (Beecher Instruments,
Silver Spring, MD, USA) and placed into a TMA paraffin block.
H&E-stained sections from the TMA blocks were generated for
histological evaluation.
Immunohistochemistry
Whole tissue sections and TMA slides were
immunostained using the Dako EnVisionTM + System,
Peroxidase (DAB) (K4007, Dako Corporation, CA, USA) and Dako
Autostainer, as previously described (15). Briefly, after deparaffinization, the
sections were incubated with monoclonal antibodies against p16
(clone 16P04, diluted 1:200, 1 μg IgG1/ml) from NeoMarkers, Inc.,
CA, USA and Ki-67 (clone Ki-S5, 1:50, 0.9 μg IgG1/ml) from Dako
A/S, Glostrup, Denmark, for 30 min at room temperature. The
sections were then incubated with peroxidase-labeled polymer
conjugated to goat anti-mouse for 30 min and 3′3-diaminobenzidine
tetrahydrochloride (DAB) for 10 min. Positive controls (cervical
carcinomas for p16 and tonsils for Ki-67) were included in the
series. Negative controls included substitution of the monoclonal
antibody with mouse myeloma protein of the same subclass and
concentration as the monoclonal antibody. The controls yielded
satisfactory results. Only distinct nuclear staining was considered
to be positive. A total of 4 semiquantitative classes were used to
describe the number of positively-stained tumor cells: none,
<10% positive; 10–50% positive and >50% positive. Two
independent investigators (M.H.K. and R.H.) scored the whole tissue
sections and TMA slides with no knowledge of the clinical data.
Conflicting results were reviewed until a final agreement was
achieved. Protein levels were classified as high when ≥10% of the
cells were positive for Ki-67 and >50% of the cells were
positive for p16. The levels were classified based on our previous
publication (15). This study
showed the results for each of the three tissue cores (1, 2 and 3),
TMAs (a tumor was considered to be a high expression of Ki-67 or
p16 if at least one of the three tissue cores was scored as a high
expression) and the whole tissue sections.
Statistical analysis
Differences in proportions were evaluated by the
Chi-square or Fisher’s exact test as required. The Kaplan-Meier
method was used to calculate the disease-free and corrected
survival rates (16). The long-rank
test was used for univariate analysis and a Cox proportional
hazards regression model was used for multivariate evaluation of
the survival rates (17). A
backward stepwise selection procedure was used for the multivariate
analysis. The hazard proportionality was verified by computing the
log - log against time. Statistical analysis was performed using
the SPSS 15.0 software package (SPSS, Chicago, IL, USA). P<0.05
was regarded as statistically significant.
Results
Table I shows a
summary of the immunohistochemical results for Ki-67 and p16 in the
three cores, TMAs and whole tissue sections. Only 88/1026 cores
(8.6%) were missing, including 39/513 (7.6%) for Ki-67 and 49/512
(9.6%) for p16, resulting in 2/171 (1.2%) cases for Ki-67 and 7/171
(4.1%) cases for p16 that were not noted in the TMAs. Excluding the
missing tissue cores, a high expression of Ki-67 was identified in
85.0% of core 1, 85.5% of core 2, 85.8% of core 3, 90.5% of TMAs
and 84% of whole tissue sections. For p16, a high expression was
found in 36.5% of core 1, 31.4% of core 2, 30.3% of core 3, 46.3%
of TMAs and 31.0% of whole tissue sections. The high expression of
Ki-67 and p16 in whole tissue sections significantly correlated
with a high expression of Ki-67 and p16 in core 1 (P<0.0001 and
P<0.0001, respectively), core 2 (P<0.0001 and P<0.0001,
respectively), core 3 (P<0.0001 and P<0.0001, respectively)
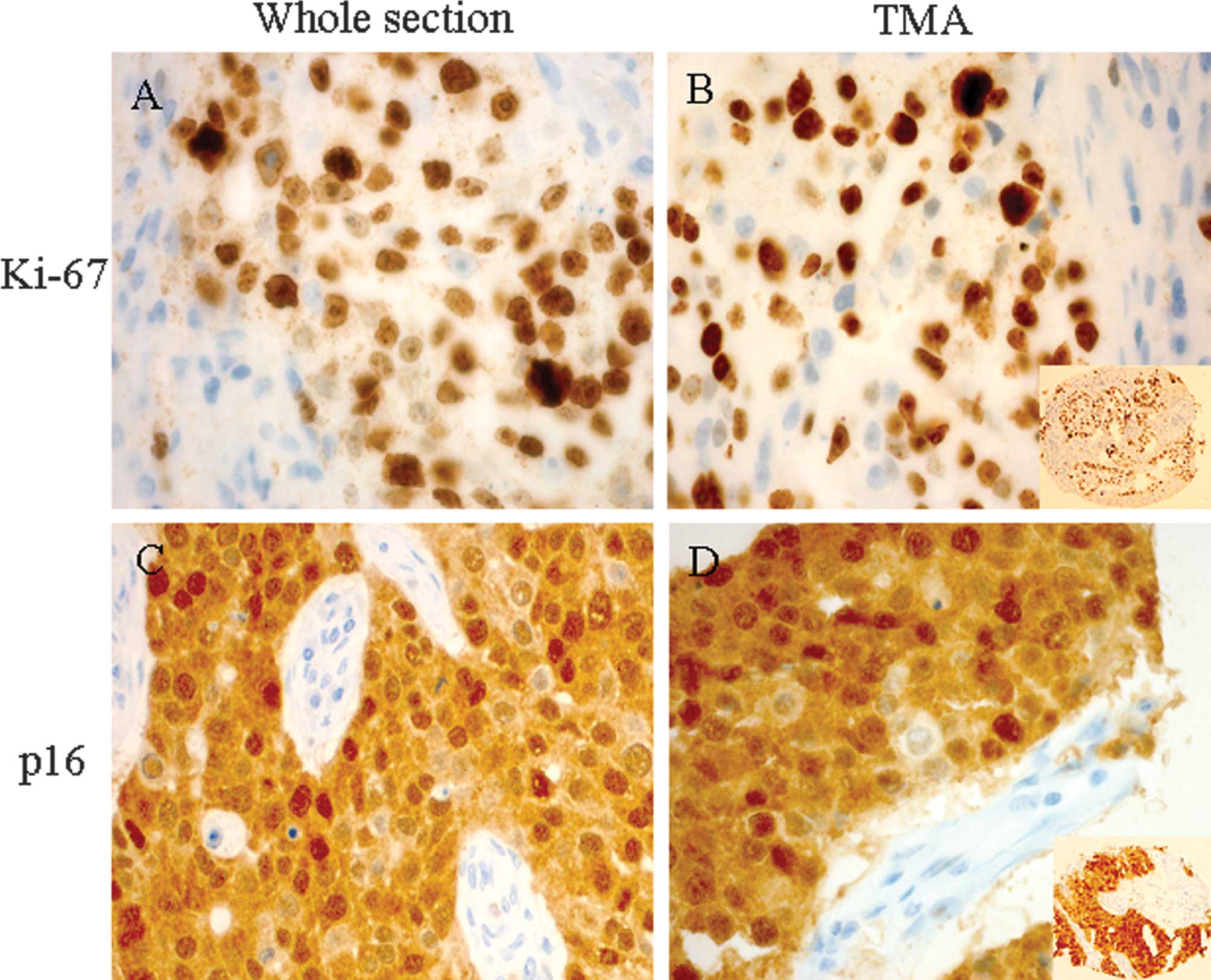
and TMAs (P<0.0001 and P<0.0001, respectively). Examples of
the high expression for Ki-67 and p16 are shown for a core in TMA
and whole sections (Fig. 1).
 | Table ISummary of immunohistochemical
analysis. |
Table I
Summary of immunohistochemical
analysis.
| Ki67 | p16 |
|---|
|
|
|
|---|
| C1 | C2 | C3 | TMA | Whole | C1 | C2 | C3 | TMA | Whole |
|---|
| Low | 24 (14.0)a | 23 (13.5) | 22 (12.9) | 16 (9.4) | 27 (16.0) | 99 (57.9) | 107 (62.6) | 106 (61.9) | 88 (51.5) | 118 (69.0) |
| High | 136 (79.5) | 136 (79.5) | 133 (77.8) | 153 (89.5) | 144 (84.0) | 57 (33.3) | 49 (28.6) | 46 (26.9) | 76 (44.4) | 53 (31.0) |
| Missing | 11 (6.4) | 12 (7.0) | 16 (9.4) | 2 (1.2) | 0 | 15 (8.8) | 15 (8.8) | 19 (11.1) | 7 (4.1) | 0 |
| Total | 171 | 171 | 171 | 171 | 171 | 171 | 171 | 171 | 171 | 171 |
Previously, we found, in the same patient
population, that in the univariate analysis, FIGO substage
(P=0.0002), presence of ascites (P=0.0012), presence of residual
disease after surgery (P<0.0001), degree of differentiation
(P=0.0001), Silverberg histopathological grade (grade 1 vs. grade
2/3) (P=0.0019) and age (P=0.045) were correlated to
disease-related survival (15). In
univariate analysis, high expression levels for Ki-67 on core 2
(P=0.008), core 3 (P=0.012), TMA (P=0.012) and whole tissue
sections (P=0.0001) were significantly correlated to
disease-related survival, whereas, Ki-67 expression on core 1 was
not significantly correlated to disease-related survival (P=0.054).
High expression levels for p16 on core 1 (P=0.0007), core 2
(P=0.0005), TMA (P=0.0008) and whole tissue sections (P=0.005) were
correlated to disease-related survival. No significant association
was observed between p16 expression on core 3 and disease-related
survival (P=0.055).
The variables that reached significance in the
univariate survival analysis were incorporated in a multivariate
analysis, with disease-related survival used as the end point. In
this study, only residual disease (P=0.0003), differentiation grade
(P=0.012) and Ki-67 on whole tissue sections (P=0.025) retained
independent prognostic significance, with a high expression of
Ki-67 indicating shorter disease-related survival (15). Prognostic significance was not
achieved in Ki-67 expression on the three individual cores and the
TMAs. Furthermore, prognostic significance was not achieved in the
multivariate analysis in p16 on whole tissue sections, the three
individual cores and the TMAs.
Discussion
TMA is proving to be a user-friendly technique in
the evaluation of immunohistochemical markers in tumors and may be
an alternative for whole sections especially in studies that
investigate a large number of cases. Numerous studies have used
this technology to examine the associations between molecular
changes and clinicopathological characteristics of tumors (4,15,18).
However, only a few studies have correlated survival and clinical
data with both TMA and results obtained from whole sections to
establish whether data from TMAs can be reliably used for
clinicopathological correlations and survival analysis (12,14).
The majority of studies investigated the ability of TMA to
represent whole sections in immunohistochemical studies of certain
antigens as well as the optimal number of TMA cores needed to
achieve acceptable representation of the specimens (6,10,19–21).
Our results showed that even one 0.6 mm TMA core represented whole
sections as significantly as the score of all three TMA cores. In
agreement with our results, a study on breast cancer (3) concluded that one or two TMA cores per
case result in outcomes that are 95% similar to those achieved
using conventional tissue sections. However, the majority of
validation studies have found that analysis of two to three 0.6 mm
cores produces higher concordance rates than the use of one core;
the percentage of missing cores is also reduced (3,6,22–24).
Jourdan et al (9) analyzed
eight (0.6 mm) cores per tumor in colorectal carcinomas and
concluded that compared with one or two cores, three cores
significantly increase the concordance rate with the whole
sections, and decrease the risk of missing cases. However, another
study on colorectal adenoma (19)
indicated that, due to the heterogeneous staining pattern, four
(0.6 mm) cores are required to reliably assess the expression
levels of Ki-67. Furthermore, other studies suggested that even two
cores, but with a larger diameter (1 mm), provide sufficient
information to achieve results similar to those obtained from whole
sections (11). The discrepancy in
the number of cores required to obtain acceptable representation of
the specimens may be due to variation of the heterogeneity of the
antigens in the tumors tested.
The missing cores in our study comprised 8.6% of the
entire number of cores. This result was acceptable considering the
large number of cores, and the comparison of missing core
percentages of other similar studies; these core percentages
deteriorated from 6.6 to 17% (11,19,20).
Considering the previous results, our study was also
concerned with the reliability of the TMA immunohistochemical
results of certain antibodies as prognostic markers. Our results
showed a significant correlation of high expression levels for
Ki-67 and p16 in TMA and whole tissue sections to disease-related
survival in univariate analyses. However, prognostic significance
was not achieved when the same variables were incorporated in a
multivariate analysis. In this case, only certain clinical
parameters retained independent prognostic significance, and a high
expression of Ki-67 and p16 was not significant on each of the
three individual cores and the TMAs. In contrast, in the
multivariate analysis whole tissue sections in Ki-67 expression
retained independent prognostic significance (15). A review of the literature showed
that various studies investigated the clinico-immunologic
correlations using TMAs (25).
However, only a few studies have correlated survival and clinical
data with both TMA and results obtained from whole sections to
establish whether data from TMAs can be reliably used for
clinicopathological correlations and survival analysis (12,14).
In concordance with our results, a study of 199 patients with
prostatic carcinoma (13) revealed
that the prognostic value of p53 and bcl-2 were not confirmed using
TMA technology in contrast to radical prostatectomy sections. In
this study, two cores of 0.6 mm from each representative area of
each case were selected. It was found that the value of TMAs in the
search for prognostic markers may be limited to biomarkers with
diffuse expression and not focally clustered biomarkers such as p53
and bcl-2. Another study on breast carcinomas (14) proved that an analysis of three
antibodies on four cores per tumor yielded more significant
associations with tumor-specific survival than large section
analyses. By contrast, a study on bladder carcinoma, using four
replica TMAs of 0.6 mm, showed a correlation between the Ki-67 LI
(labeling index) and tumor stage or prognoses that highly reproduce
whole section data (12). These
studies indicate that the prognostic value of TMA compared to whole
sections may depend on the type of cancers, number of cores and the
antigen investigated.
In conclusion, the question regarding to what extent
TMAs are able to represent whole sections is currently being
investigated by a growing number of studies that have proved the
reliability of this technique. However, a crucial point is whether
TMA is able to reproduce clinicopatholological correlations that
exist on whole section analyses. Additionally, the number of cores
required to achieve this particular purpose should be investigated.
Although TMA may have advantages, such as being cost-effective and
saving time and tissues, our results indicate that further studies,
with the use of a larger number of cores are necessary to determine
the efficacy of TMA to reflect the prognostic value of different
antibodies. Moreover, this method should be evaluated for each type
of tumor and each separate antigen. Confirmation of the clinical
associations on whole sections prior to investigating the same
parameters on TMAs should also be obtained.
Acknowledgements
We thank Ellen Hellesylt and Mette Førsund for their
excellent technical assistance. This study was supported in part by
grants from The Norwegian Cancer Society and Inger and John
Fredriksen Foundation for Ovarian Cancer Research.
References
|
1
|
Kononen J, Bubendorf L, Kallioniemi A,
Barlund M, Schraml P, Leighton S, Torhorst J, Mihatsch MJ, Sauter G
and Kallioniemi OP: Tissue microarrays for high-throughput
molecular profiling of tumor specimens. Nat Med. 4:844–847. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mills SE, Fechner RE, Frierson HF, Kempson
RL, Wick MR, Dehner LP, Swanson PE and Humphrey PA: Guardians of
the wax and the patient. Am J Clin Pathol. 104:365–367.
1995.PubMed/NCBI
|
|
3
|
Camp RL, Charette LA and Rimm DL:
Validation of tissue microarray technology in breast carcinoma. Lab
Invest. 80:1943–1949. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tolgay OI, Dolled-Filhart M, D’Aquila TG,
Camp RL and Rimm DL: Tissue microarray-based studies of patients
with lymph node negative breast carcinoma show that met expression
is associated with worse outcome but is not correlated with
epidermal growth factor family receptors. Cancer. 97:1841–1848.
2003. View Article : Google Scholar
|
|
5
|
Korsching E, Packeisen J, Agelopoulos K,
Eisenacher M, Voss R, Isola J, van Diest PJ, Brandt B, Boecker W
and Buerger H: Cytogenetic alterations and cytokeratin expression
patterns in breast cancer: integrating a new model of breast
differentiation into cytogenetic pathways of breast carcinogenesis.
Lab Invest. 82:1525–1533. 2002. View Article : Google Scholar
|
|
6
|
Griffin MC, Robinson RA and Trask DK:
Validation of tissue microarrays using p53 immunohistochemical
studies of squamous cell carcinoma of the larynx. Mod Pathol.
16:1181–1188. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kuwabara S, Ajioka Y, Watanabe H, Hitomi
J, Nishikura K and Hatakeyama K: Heterogeneity of p53 mutational
status in esophageal squamous cell carcinoma. Jpn J Cancer Res.
89:405–410. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Baisse B, Bouzourene H, Saraga EP, Bosman
FT and Benhattar J: Intratumor genetic heterogeneity in advanced
human colorectal adenocarcinoma. Int J Cancer. 93:346–352. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jourdan F, Sebbagh N, Comperat E, Mourra
N, Flahault A, Olschwang S, Duval A, Hamelin R and Flejou JF:
Tissue microarray technology: validation in colorectal carcinoma
and analysis of p53, hMLH1, and hMSH2 immunohistochemical
expression. Virchows Arch. 443:115–121. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gomaa W, Ke Y, Fujii H and Helliwell T:
Tissue microarray of head and neck squamous carcinoma: validation
of the methodology for the study of cutaneous fatty acid-binding
protein, vascular endothelial growth factor, involucrin and Ki-67.
Virchows Arch. 447:701–709. 2005. View Article : Google Scholar
|
|
11
|
Rosen DG, Huang X, Deavers MT, Malpica A,
Silva EG and Liu J: Validation of tissue microarray technology in
ovarian carcinoma. Mod Pathol. 17:790–797. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nocito A, Bubendorf L, Tinner EM, et al:
Microarrays of bladder cancer tissue are highly representative of
proliferation index and histological grade. J Pathol. 194:349–357.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Merseburger AS, Kuczyk MA, Serth J, et al:
Limitations of tissue microarrays in the evaluation of focal
alterations of bcl-2 and p53 in whole mount derived prostate
tissues. Oncol Rep. 10:223–228. 2003.
|
|
14
|
Torhorst J, Bucher C, Kononen J, Haas P,
Zuber M, Kochli OR, Mross F, Dieterich H, Moch H, Mihatsch M,
Kallioniemi OP and Sauter G: Tissue microarrays for rapid linking
of molecular changes to clinical endpoints. Am J Pathol.
159:2249–2256. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Khouja MH, Baekelandt M, Nesland JM and
Holm R: The clinical importance of Ki-67, p16, p14, and p57
expression in patients with advanced ovarian carcinoma. Int J
Gynecol Pathol. 26:418–425. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kaplan EL and Meier P: Nonparametric
estimation from incomplete observation. J Am Stat Assoc.
53:457–481. 1958. View Article : Google Scholar
|
|
17
|
Cox DR: Regression models and life tables.
J R Stat Soc B. 34:187–220. 1972.
|
|
18
|
Sapino A, Marchio C, Senetta R, Castellano
I, Macri L, Cassoni P, Ghisolfi G, Cerrato M, D’Ambrosio E and
Bussolati G: Routine assessment of prognostic factors in breast
cancer using a multicore tissue microarray procedure. Virchows
Arch. 449:288–296. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Su Y, Shrubsole MJ, Ness RM, Cai Q,
Kataoka N, Washington K and Zheng W: Immunohistochemical
expressions of Ki-67, cyclin D1, beta-catenin, cyclooxygenase-2,
and epidermal growth factor receptor in human colorectal adenoma: a
validation study of tissue microarrays. Cancer Epidemiol Biomarkers
Prev. 15:1719–1726. 2006. View Article : Google Scholar
|
|
20
|
Leversha MA, Fielding P, Watson S, Gosney
JR and Field JK: Expression of p53, pRB, and p16 in lung tumours: a
validation study on tissue microarrays. J Pathol. 200:610–619.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tawfik El-Mansi M and Williams AR:
Validation of tissue microarray technology using cervical
adenocarcinoma and its precursors as a model system. Int J Gynecol
Cancer. 16:1225–1233. 2006.PubMed/NCBI
|
|
22
|
Kallioniemi OP, Wagner U, Kononen J and
Sauter G: Tissue microarray technology for high-throughput
molecular profiling of cancer. Hum Mol Genet. 10:657–662. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fernebro E, Dictor M, Bendahl PO, Ferno M
and Nilbert M: Evaluation of the tissue microarray technique for
immunohistochemical analysis in rectal cancer. Arch Pathol Lab Med.
126:702–705. 2002.PubMed/NCBI
|
|
24
|
Fons G, Hasibuan SM, van der Velden J and
ten Kate FJ: Validation of tissue microarray technology in
endometrioid cancer of the endometrium. J Clin Pathol. 60:500–503.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zerkowski MP, Camp RL, Burtness BA, Rimm
DL and Chung GG: Quantitative analysis of breast cancer tissue
microarrays shows high cox-2 expression is associated with poor
outcome. Cancer Invest. 25:19–26. 2007. View Article : Google Scholar : PubMed/NCBI
|