Introduction
Lung cancer is the leading cause of cancer-related
death in worldwide, and accounts for one third of all deaths
(1). Current treatment of lung
cancer is still based on traditional surgery. Unfortunately, most
lung cancer patients, especially non-small cell lung cancer (NSCLC)
patients present advanced disease that is surgically unresectable.
It is current practice to treat these patients with a combination
of chemotherapy and external beam irradiation. But treatment
outcomes for NSCLC remain unsatisfactory, with low long-term
survival rates (<15%). Emerging TARGETED therapy with epidermal
growth factor receptor tyrosine kinase inhibitors (EGFR-TKIs) are
currently being evaluated in NSCLC.
EGFR is a receptor tyrosine kinase (TK) which is
widely distributed in mammalian epithelial cells, glial cells,
fibroblasts, keratinocytes and other cell surface. EGFR family
includes four structurally-related tyrosine kinase receptors: EGFR
(ErbB-1), HER2/c-neu (ErbB-2), Her3 (ErbB-4). These receptors are
related to 70% of cancer (2).
Abnormal activation of EGFR can promote tumor cell proliferation,
differentiation, migration. EGFR has been found to be overexpressed
in a variety of human malignancies (2,3).
Activation of EGFR results in the initiation of a diverse range of
cellular signaling pathways, including cell proliferation and
protection of the cell from apoptosis (4). In 2004, Lynch et al (5) and Paez et al (6) proposed the EGFR gene mutations in lung
cancer can be predicted, which is considered a milestone for NSCLC
individualized targeted molecular therapies. These mutations are
located in EGFR exons 18–21 (7) and
~90% of the sensitizing mutations are in-frame deletions in exon19,
and the point mutation L858R in exon 21 (8). These mutations cluster around the ATP
binding pocket of the tyrosine kinase domain of EGFR, leading to
ligand-independent activation of the receptor and prolonged
activation time compared to wild-type EGFR. Although exon 20
mutations were less, it is associated with resistance to anti-EGFR
therapies (9–11).
The treatment of NSCLC using EGFR-TKIs, such as
gefitinib and erlotinib was confirmed, showing promising outcomes
for some patients with NSCLC, especially those with mutated EGFR
(5,6,12).
EGFR mutations increase sensitivity to TK inhibitors, most likely
through induction of critical structural modifications of the
ATP-binding site in the TK domain (13). EGFR mutation testing may be
prognostically important to guide identify potential responders (or
non-responders) to therapy. Therefore, a rapid, sensitive and
reliable method for EGFR mutations testing is required.
The classic method for detecting EGFR mutations is
direct sequencing. However, sequencing for routine testing is
impeded by relatively high cost, requirement of sophisticated
equipment and technical expertise, which are often only available
in specialized molecular pathology laboratories (14). HRM is a recently developed technique
that shows great potential for somatic mutations (15), but rarely uses serum samples. In
1989, studies have shown that the DNA in the plasma of cancer
patients had tumor characteristics (16). In 1994, Sorenson et al and
Vasioukhin et al detected the same gene mutation in tumor
and plasma, this discovery confirmed that a part of free DNA from
tumor existed in plasma (17,18).
However, the clinical development of EGFR mutation detection was
impeded by the low sensitivity of conventional methods, because the
tumor DNA levels in peripheral blood is extremely low. In this
study, we aimed to establish a sensitive and reliable HRM method
for routine EGFR mutation screening using serum, which will benefit
many NSCLC patients, especially those surgically unresectable.
Materials and methods
Samples
A total of 126 FFPE specimens (71 adenocarcinomas,
53 squamous cell carcinomas, one large cell carcinoma, and one
adenosquamous carcinoma) between December 2008 and September 2010,
47 fresh frozen surgically resected tumor tissues (28
adenocarcinomas, 19 squamous cell carcinomas) between October 2010
and June 2011 were obtained from tumor tissue bank of Department of
Pathology at Jinan General Hospital of PLA. All tissues were
finally diagnosed as NSCLC by Pathology. We also successfully
collected 47 matched pre-operation serum specimens for the 47 NSCLC
patients between October 2010 and June 2011.
DNA extraction
The paraffin rolls were cut from each block (5
sections of 5-μm thickness) for DNA extraction. About 300 mg fresh
frozen tissue was used for DNA extraction. QIAamp DNA FFPE tissue
kit (Qiagen) and QIAamp DNA mini kit (Qiagen) was used to extract
FFPE and fresh frozen tissue DNA, respectively. DNA was extracted
from 200 μl serum using QIAamp DNA blood mini kit (Qiagen). All DNA
solutions were quantitated with the Lambda Bio Spectrophotometer
system (PerkinElmer Company, USA), adjusted to the same
concentration of 30 ng/μl, and then stored at −20°C until use.
Design of HRM primers
Primers specific for EGFR exons 18 to 21 were
designed using Primer Designer Software (primer premier 5.0).
Primers that flanked the exons as closely as possible were chosen.
All primers were analyzed for specificity and to ensure similar
melting temperatures using Primer-BLAST software. The sequences of
primers for the EGFR exon 18 to 21 were listed in Table I. All amplicons were less than 250
bp and covered the most common EGFR mutations.
 | Table IHRM primer sequences. |
Table I
HRM primer sequences.
| Exon | Primer | T (°C) | G/C (%) | Amplicon size
(bp) |
|---|
| 18 | F:
CTGAGGTGACCCTTGTCTCTGTGTTC | 66.63 | 53.85 | 183 |
| R:
AGGCCTGTGCCAGGGACCTTA | 67.36 | 61.90 | |
| 19 | F:
GCATGTGGCACCATCTCACAA | 64.42 | 52.38 | 204 |
| R:
CCTGAGGTTCAGAGCCATGGA | 64.88 | 57.14 | |
| 20 | F:
CATTCATGCGTCTTCACCTG | 60.30 | 50.00 | 228 |
| R:
TCTTTGTGTTCCCGGACATAG | 60.00 | 47.60 | |
| 21 | F:
GCAGAGCTTCTTCCCATGATGA | 63.98 | 50.00 | 236 |
| R:
GCTGACCTAAAGCCACCTCCT | 62.01 | 57.14 | |
Real-time PCR and HRM assay
Real-time PCR and HRM assays were performed with
LightCycler® 480 Real-time system (Roche Diagnostics,
Switzerland). Each 20 μl reaction system contained ~30 ng DNA, 1X
LightCycler HRM Master reaction mix (Roche Diagnostics), 2.5 mM
MgCl2, and 4 μM forward and reverse primer (HPLC
purified). The same PCR program was used for all amplicons: 95°C
for 10 min; 50 cycles of 95°C for 10 sec, 60°C for 15 sec. After
amplification, a post amplification melting curve program was
initiated by heating to 95°C for 1 min, cooling to 60°C for 1 min,
and increasing the temperature to 95°C while continuously measuring
fluorescence at 25 acquisitions per degree. Each PCR run contained
a negative (no template) control. Data were acquired and analyzed
using LC480 Gene Scanning software V1.5 (Roche Diagnostics). All
curves were analyzed following normalization, temperature shifting,
and the inspection of difference plots.
Specificity and sensitivity of HRM
assays
All amplified products were sent to sequence, and
then were analyzed for specificity to ensure the homology of EGFR
gene using BLAST (19). Sequence
analysis was performed with Chromas 2.31 software. To test the
sensitivity, we mixed the genomic DNA of the EGFR-mutated sample
with wild-type DNA sample in dilutions of 50, 25, 12.5, 10, 5 and
2.5%, and then HRM were performed, PCR products were also
sequenced.
EGFR mutation rate screening by HRM
All 126 FFPE specimens, 47 fresh frozen tissue
samples and 47 serum specimens were tested by HRM for the detection
of mutations in EGFR exons 18–21. Samples were amplified in 96-well
plates.
Statistical analysis
SPSS statistical software (version 17) was used for
statistical analysis. Difference of EGFR mutation between FFPE and
fresh frozen tissue samples were analyzed with Fisher’s exact test.
The correlation between the presence of EGFR mutation status and
the clinicopathological categorical characteristics was assessed by
Logistic Regression. A two-tailed p-value of <0.05 was
statistically significant.
Results
Specificity and sensitivity of EGFR HRM
assay
Amplicons sequences were verified using BLAST to
search the NCBI GenBank. The sensitivity was evaluated by detection
of serial dilutions of EGFR-mutated sample. HRM assays for EGFR
exons 18, 19, 20, and 21 mutations were detected in dilution of 5,
2.5, 5, and 5%, respectively (Fig.
1). While EGFR mutations were detected by direct sequencing in
dilutions of 50 and 25, 12.5, but not <10%.
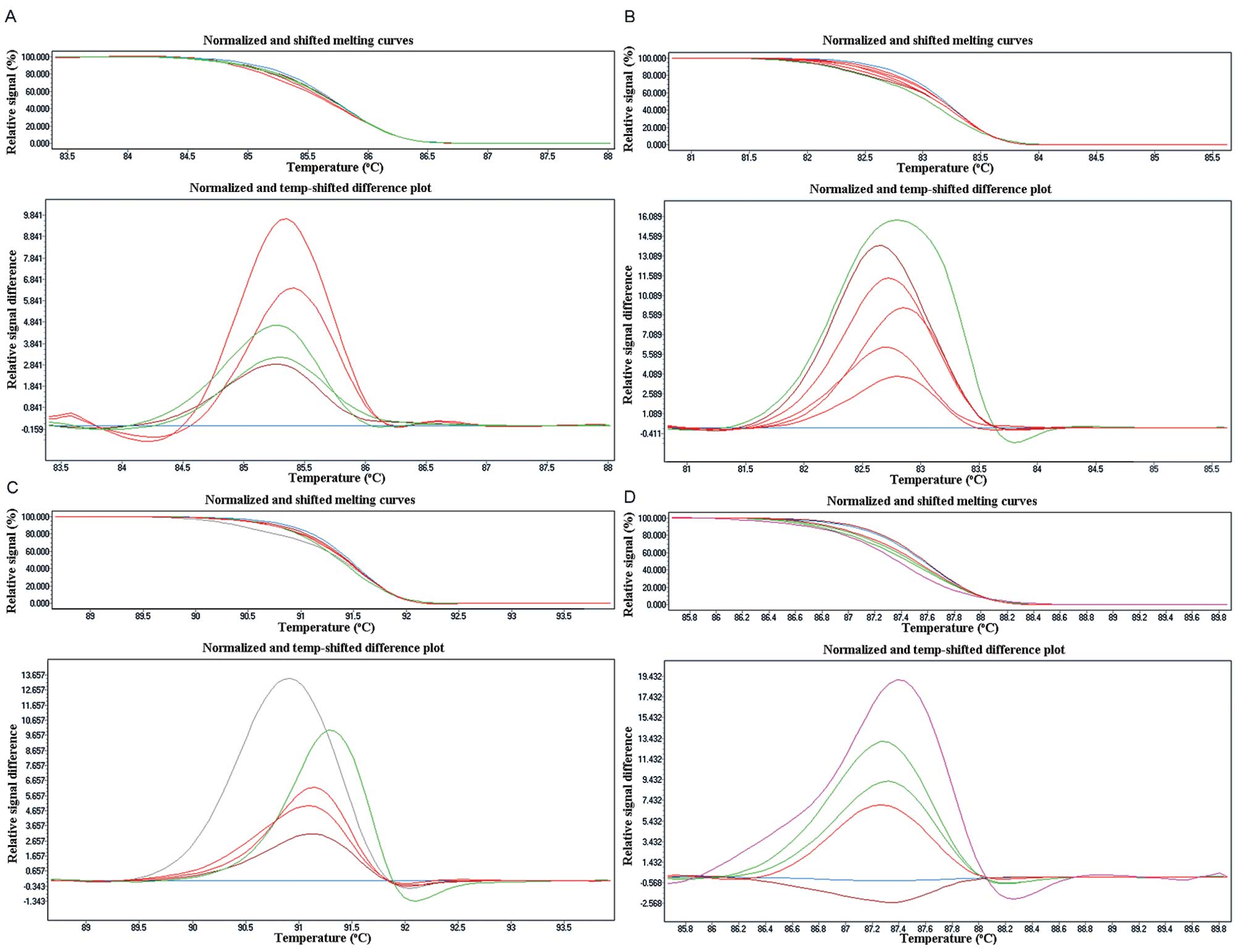 | Figure 1Sensitivity of the EGFR HRM assay.
Using HRM, the mutation was readily detectable at 5% mutation
frequency in exon 18 (A), exon 20 (C) and exon 21 (D). Adjusted
melting curves (top) and differential plots (bottom) showing the
presence of 50, 25, 12.5, 10, 5 and 0% mutant. The 2.5% dilution
was not distinct from the normal DNA. (B) The sensitivity of exon
19, it can detect 2.5% dilution. Adjusted melting curves (top) and
differential plots (bottom) showing the presence of 50, 25, 12.5,
10, 5, 2.5 and 0% mutant. |
EGFR mutation status in FFPE and fresh
frozen tissues of NSCLC
Difference plots and sequence traces of
representative mutations for each amplicon are depicted in Fig. 2, with Table II showing the results obtained.
EGFR mutations (78) were detected in 70 NSCLC FFPE samples by HRM,
with a total mutation rate of 55.56% (70/126). Among all the
mutations, there were 5 in exon 18 (3.97%), 23 in exon 19 (18.25%),
27 in exon 20 (21.42%), and 23 in exon 21 (18.25%). 74 EGFR
mutation identified by HRM were confirmed by direct sequencing.
There were 5 in exon 18 (G719S), 22 in exon 19 (2236-225Del), 25 in
exon 20 (3 insertion mutation, 22 Q787Q nonsense mutation), and 22
in exon 21 (20 L858R, 2 L861Q). In addition, 8 patients were found
to have double EGFR mutations, 4 in exon 19 and exon 20, 3 in exon
20 and exon 21, one in exon 18 and exon 21. We found 25 EGFR
mutations in 24 fresh frozen NSCLC tissues; the total mutation rate
was 51.06% (24/47). There was one in exon 18 (2.13%, G719S), 8 in
exon 19 (17.02%, 2236–2250 Del), 9 in exon 20 (19.15%, Q787Q
nonsense mutation), and 7 in exon 21 (14.89%, L858R), one in exon
19 and exon 20. There was no difference in the mutation status
between FFPE and fresh frozen tissues (P>0.05).
 | Table IIEGFR mutations detected by HRM and
sequencing. |
Table II
EGFR mutations detected by HRM and
sequencing.
| HRM (%) | Gene
sequencing | |
|---|
|
|
| |
|---|
| Mutation in | FFPE | Frozen | FFPE | Frozen | Genotype |
|---|
| Exon 18 | 5 (3.97) | 1 (2.13) | 5 | 1 | G719S |
| Exon 19 | 23 (18.25) | 8 (17.02) | 22 | 8 | 2236–2250Del |
| Exon 20 | 27 (21.42) | 9 (19.15) | 22 | 8 | Q787Q |
| | | 3 | 0 |
2310–2311insGGT |
| Exon 21 | 23 (18.25) | 7 (14.89) | 20 | 6 | L858R |
| | | 2 | 0 | L861Q |
| Exon 18 + exon
21 | 1 (1.28) | 0 | 1 | 0 | G719S and
L858R |
| Exon 19 + exon
20 | 4 (5.13) | 1 (4.17) | 4 | 1 | 2236–2250Del and
Q787Q |
| Exon 20 + exon
21 | 3 (3.85) | 0 | 3 | 0 | Q787Q and
L858R |
| Total
mutations | 78 | 25 | 74 | 23 | |
| Cases | 70 (55.56) | 24 (51.06) | 66 | 22 | |
In addition, there was an obvious Tm shift between
the insertion mutation (2310–2311 ins GGT) and Q787Q nonsense
mutation in exon 20, so we could easily differentiated the two
mutation types by setting positive mutation standard. However, the
two exon 21 mutation types could not be identified through HRM.
Serum EGFR mutations testing
EGFR mutations were detected in 22 serum samples
from 24 fresh frozen tissue EGFR mutations positive NSCLC patients
(Table III). The serum positive
rate was 100% (13/13) for those tissue EGFR mutations positive
patients in stages II-IV, 81.8% (9/11) for stage I. No false
negative was found in stage II-IV patients, while there was 18.2%
false negatives for stage I patients. Therefore, the sensitivity of
serum EGFR screening by HRM assay was 91.67%, specificity 100%.
 | Table IIISummary of data of 24 serum
samples. |
Table III
Summary of data of 24 serum
samples.
| Case | Gender | Age | Histologic
type | Staging | Smoking status | Mutation |
|---|
| 1 | M | 55 | Ad | IIIA | C | Exon 20 |
| 2 | M | 58 | Ad | IIA | N | Exon 20 |
| 3 | F | 54 | SCC | IV | N | Exon 19 |
| 4 | M | 65 | SCC | IA | C | Exon 20 |
| 5 | M | 57 | SCC | IB | C | Exon 20 |
| 6 | M | 70 | Ad | IB | C | Exon 21 |
| 7 | F | 75 | Ad | IIA | N | Exon 21 |
| 8 | M | 59 | Ad | IIB | N | Exon 18 |
| 9 | M | 64 | SCC | IB | C | Exon 20 |
| 10 | F | 51 | Ad | IIIA | N | Exon 19 |
| 11 | F | 51 | Ad | IIA | N | Exon 21 |
| 12 | M | 74 | SCC | IIA | C | Exon 19 |
| 13 | F | 62 | Ad | IIIA | N | Exon 20 |
| 14 | M | 76 | SCC | IIA | C | Exon 19 |
| 15 | F | 59 | Ad | IIIA | N | Exon 19 |
| 16 | F | 70 | Ad | IA | N | Exon 21 |
| 17 | F | 63 | Ad | IA | N | Exon 21 |
| 18 | M | 53 | SCC | IB | C | Exon 20 |
| 19 | F | 63 | SCC | IB | N | Exon 19 |
| 20 | M | 62 | SCC | IIA | C | Exon 20 |
| 21 | M | 46 | SCC | IIA | C | Exon 19 |
| 22 | F | 68 | Ad | IA | N | Exon 19 |
| 23 | F | 76 | Ad | IA | N | Not detected |
| 24 | F | 74 | Ad | IB | C | Not detected |
Relationship between EGFR mutations and
clinicopathological factors
The correlation between clinicopathological
characteristics of the patients and EGFR mutations is summarized in
Table IV. EGFR mutations were
detected more frequently in never-smokers than smokers (81.03%
versus 33.82%; P<0.01), females than males (90.48% versus
38.10%; P<0.05), adenocarcinoma histology compared to squamous
cell carcinoma (76.06% versus 28.30%; P<0.01). EGFR mutation
status did not show any significant association with clinical
staging (P=0.197).
 | Table IVRelationship between EGFR mutations
and clinicopathological factors. |
Table IV
Relationship between EGFR mutations
and clinicopathological factors.
| EGFR mutation | | |
|---|
|
| | |
|---|
| No. patient | P-value | EGFR wild-type | Total |
|---|
| Total no.
patient | 70 | - | 56 | 126 |
| Gender |
| Male | 32 | | 52 | 84 |
| Female | 38 | 0.028 | 4 | 42 |
| Smoking
history |
| Never | 47 | | 11 | 58 |
|
Former/current | 23 | 0.004 | 45 | 68 |
| Histological
diagnosis |
|
Adenocarcinoma | 54 | | 7 | 71 |
| Squamous cell
carcinoma | 15 | 0.003 | 38 | 53 |
| Large cell
carcinoma | - | | 1 | 1 |
| Adenosquamous
carcinoma | 1 | | - | 1 |
| Staging |
| I-II | 49 | | 40 | 89 |
| III–IV | 21 | 0.197 | 14 | 35 |
Discussion
Personalized medicine for cancer has raised new hope
of cancer treatment, and become widely used in clinical settings
over the last decade. Based on specific genetic information of
various cancers, it had become possible to determine the genetic
type of cancer, select the appropriate treatment, then enhance drug
efficacy and reduce toxicity. However, the effectiveness of these
new introduced molecular targeted drugs was closely dependent on
the presence of specific genetic mutations in the tumor context
(20–22). Gene mutation testing is the premise
to using molecular-targeting drugs. EGFR gene mutations have
recently been identified as a predictor for first-line EGFR-TKIs
sensitivity (23). It was shown
that 70% of EGFR mutation-positive NSCLC patients were effective to
EGFR-TKIs, while only 10% of EGFR mutation- negative NSCLC patients
(24). In addition, the responses
of patients with different EGFR mutation types to EGFR-TKIs
treatment were also different (25–27).
Thus, it is of great importance to detect EGFR mutation status and
type for NSCLC patients, which can effectively predict the benefit
of taking an EGFR-TKI, and allow the physician to prescribe the
most suitable therapy.
There are several methods for EGFR mutation testing,
such as PCR-SSCP, gene chips and gene sequencing, with the gene
sequencing being the current gold standard (28). HRM is a technique recently developed
on the basis of real-time PCR amplification, which shows great
potential for somatic mutations. A double stranded DNA binding dye
is utilized in melting analysis in order to characterize
primer-related non-specific amplification (or primer dimer) for
detection of a specific target (29). Compared with traditional real-time
PCR, HRM which adopt saturated dye have higher resolution, even
single-base change can make the melting temperature of PCR products
different (30). All the processes
of PCR amplification followed by HRM take place in the same tube
during a real-time run in <2 h (31). Therefore, HRM is the method of
choice for rapid EGFR mutation screening.
In our study, we confirmed that both FFPE and fresh
frozen tissue can be used for the HRM method. Mutation rates of
FFPE and fresh frozen tissue are compatible; fresh frozen tissues
showed no better than FFPE as others have reported (29). In addition, we found that HRM method
could be used to differentiate 2310–2311insGGT and Q787Q mutation
in exon 20 if known positive mutations control were properly set.
However, HRM failed to identify the two mutation types in exon 21;
sequencing had to be done to discriminate them.
Owing to the fact that tissue samples cannot be
obtained from most of the lung cancer patients, the use of EGFR
mutation screening in tissue samples is limited in practice. Plasma
or serum which has free DNA from tumor provides as good
alternative. However, the tumor DNA levels in peripheral blood are
extremely low, which needs a more sensitive method for detection.
The HRM assays described herein successfully identified EGFR
mutations not only in tissue specimens but also in serum samples
from NSCLC patients. The concordance rate between serum and fresh
frozen tissues was better than in a previous report (32). The HRM assays acted perfectly in
serum from NSCLC patients in stage II-IV, and no false negative was
found. In addition, for NSCLC patients in stage I, most mutations
can also be detected despite the 18.2% false negative rate. The
sensitivity and specificity for serum EGFR screening by HRM was
91.67 and 100%, respectively. Therefore, HRM assays using serum
samples for EGFR mutation screening can act as a good alternative
for tissue screening, provide convincing and valuable results for
the physician, and benefit surgically unresectable NSCLC
patients.
The reported EGFR mutation rate in NSCLC patients
differs and varies from 5 to 48.3% (30,32,33).
It was shown that EGFR mutations were found in ~10% of cases of
NSCLC in North America and 30–50% of NSCLC patients of East Asian
descent (7). The frequency of the
activating mutations is higher in Asian populations (34). In our research, the total EGFR
mutation rate was 55.56%, which is the highest so far. This result
may suggest the EGFR mutation rate of people in eastern China must
be higher than other regions. In addition, many reports suggest
major activating EGFR mutations are primarily in exons 19 and 21,
with the inframe deletion in exon 19 and L858R accounting for 90%
of reported mutations, additional mutations do occur in exons 18
and 20 that account for 8% of mutants (35). However, we found in this region of
china, EGFR mutation occurs in exon 20 more often, especially
Q787Q, and then in exons 19 and 21. Therefore, we should not only
detect EGFR mutation in exons 19 and 21, but must attach great
importance in EGFR exon 20 mutation detection in this region. We
recommend that EGFR mutations in exons 18–21 should be detected
before prescribing an EGFR-TKI for NSCLC patients from eastern
China.
There have been debates on the relationships between
EGFR mutation state and clinical characteristics (14,33,36).
Our data showed EGFR mutations were more frequently observed in
never-smokers, females, and those with adenocarcinoma histology,
and there was no correlation between different clinical stages. Our
results are comparable to those of Sriram et al (14) and Takano et al (33).
In summary, we have confirmed the feasibility of HRM
for screening EGFR mutations in serum. Serum EGFR gene mutation
screening by HRM assays are fast, relatively cheap, and robust, and
can benefit surgically unresectable NSCLC patients; provide rapid
scientific reference to the physician, and guide them to prescribe
the most suitable therapy.
References
|
1
|
Sasaki H, Endo K, Konishi A, et al: EGFR
mutation status in Japanese lung cancer patients: genotyping
analysis using LightCycler. Clin Cancer Res. 11:2924–2929. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Nicholson RI, Gee JM and Harper ME: EGFR
and cancer prognosis. Eur J Cancer. 37(Suppl 4): S9–S15. 2001.
View Article : Google Scholar
|
|
3
|
Ohsaki Y, Tanno S, Fujita Y, et al:
Epidermal growth factor receptor expression correlates with poor
prognosis in non-small cell lung cancer patients with p53
overexpression. Oncol Rep. 7:603–607. 2000.PubMed/NCBI
|
|
4
|
Scaltriti M and Baselga J: The epidermal
growth factor receptor pathway: a model for targeted therapy. Clin
Cancer Res. 12:5268–5272. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lynch TJ, Bell DW, Sordella R, et al:
Activating mutations in the epidermal growth factor receptor
underlying responsiveness of non-small-cell lung cancer to
gefitinib. N Engl J Med. 350:2129–2139. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Paez JG, Janne PA, Lee JC, et al: EGFR
mutations in lung cancer: correlation with clinical response to
gefitinib therapy. Science. 304:1497–1500. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shigematsu H, Lin L, Takahashi T, et al:
Clinical and biological features associated with epidermal growth
factor receptor gene mutations in lung cancers. J Natl Cancer Inst.
97:339–346. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Borras E, Jurado I, Hernan I, et al:
Clinical pharmacogenomic testing of KRAS, BRAF and EGFR mutations
by high resolution melting analysis and ultra-deep pyrosequencing.
BMC Cancer. 11:406–415. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yasuda H, Kobayashi S and Costa DB: EGFR
exon 20 insertion mutations in non-small-cell lung cancer:
preclinical data and clinical implications. Lancet Oncol.
13:e23–e31. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Murray S, Dahabreh IJ, Linardou H,
Manoloukos M, Bafaloukos D and Kosmidis P: Somatic mutations of the
tyrosine kinase domain of epidermal growth factor receptor and
tyrosine kinase inhibitor response to TKIs in non-small cell lung
cancer: an analytical database. J Thorac Oncol. 3:832–839. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Linardou H, Dahabreh IJ, Bafaloukos D,
Kosmidis P and Murray S: Somatic EGFR mutations and efficacy of
tyrosine kinase inhibitors in NSCLC. Nat Rev Clin Oncol. 6:352–366.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Han SW, Kim YT, Hwang PG, et al:
Predictive and prognostic impact of epidermal growth factor
receptor mutation in non-small-cell lung cancer patients treated
with gefitinib. J Clin Oncol. 23:2493–2501. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Carey KD, Garton AJ, Romero MS, et al:
Kinetic analysis of epidermal growth factor receptor somatic mutant
proteins shows increased sensitivity to the epidermal growth factor
receptor tyrosine kinase inhibitor, erlotinib. Cancer Res.
66:8163–8171. 2006. View Article : Google Scholar
|
|
14
|
Sriram KB, Tan ME, Savarimuthu SM, et al:
Screening for activating EGFR mutations in surgically resected
nonsmall cell lung cancer. Eur Respir J. 10:455–465.
2011.PubMed/NCBI
|
|
15
|
Taylor CF: Mutation scanning using
high-resolution melting. Biochem Soc Trans. 37:433–437. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Stroun M, Anker P, Maurice P, Lyautey J,
Lederrey C and Beljanski M: Neoplastic characteristics of the DNA
found in the plasma of cancer patients. Oncology. 46:318–322. 1989.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sorenson GD, Pribish DM, Valone FH, Memoli
VA, Bzik DJ and Yao SL: Soluble normal and mutated DNA sequences
from single copy genes in human blood. Cancer Epidemiol Biomarkers
Prev. 3:67–71. 1994.PubMed/NCBI
|
|
18
|
Vasioukhin V, Anker P, Maurice P, Lyautey
J, Lederrey C and Stroun M: Point mutations of the N-ras gene in
the blood plasma DNA of patients with myelodysplastic syndrome or
acute myelogenous leukaemia. Br J Haematol. 86:774–779. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
NCBI/BLAST. http://blast.ncbi.nlm.nih.gov/Blast.cgi.
|
|
20
|
Horn L and Pao W: EML4-ALK: Honing in on a
new target in non-small-cell lung cancer. J Clin Oncol.
27:4232–4235. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Choi YL, Soda M, Yamashita Y, et al:
EML4-ALK mutations in lung cancer that confer resistance to ALK
inhibitors. N Engl J Med. 363:1734–1739. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pao W and Girard N: New driver mutations
in non-small-cell lung cancer. Lancet Oncol. 12:175–180. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sequist LV, Bell DW, Lynch TJ and Haber
DA: Molecular predictors of response to epidermal growth factor
receptor antagonists in non-small-cell lung cancer. J Clin Oncol.
25:587–595. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Uramoto H and Mitsudomi T: Which biomarker
predicts benefit from EGFR-TKI treatment for patients with lung
cancer? Br J Cancer. 96:857–863. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jackman DM, Yeap BY, Sequist LV, et al:
Exon 19 deletion mutations of epidermal growth treated with
gefitinib or erlotinib. Clin Cancer Res. 12:3908–3914. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Riely GJ, Pao W, Pham D, et al: Clinical
course of patients with non-small cell lung cancer and epidermal
growth factor receptor exon 19 and exon 21 mutations treated with
gefitinib or erlotinib. Clin Cancer Res. 12:839–844. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mitsudomi T, Kosaka T, Endoh H, et al:
Mutations of the epidermal growth factor receptor gene predict
prolonged survival after gefitinib treatment in patients with
non-small cell lung cancer with postoperative recurrence. J Clin
Oncol. 23:2513–2520. 2005. View Article : Google Scholar
|
|
28
|
Pao W and Ladanyi M: Epidermal growth
factor receptor mutation testing in lung cancer: searching for the
ideal method. Clin Cancer Res. 13:4954–4955. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bosquet JG, Calcei J, Wei JS, et al:
Detection of somatic mutations by high-resolution DNA melting (HRM)
analysis in multiple cancers. PLoS One. 6:e145222011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Nomoto K, Tsuta K, Takano T, et al:
Detection of EGFR mutations in archived cytologic specimens of
non-small cell lung cancer using high resolution melting analysis.
Am J Clin Pathol. 126:608–615. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Camila GC and Mauricio RL: The use of
high-resolution melting analysis for genotyping Symbiodinium
strains: a sensitive and fast approach. Mol Ecol Resour.
11:394–399. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sriram KB, Tan ME, Savarimuthu SM, et al:
Screening for activating EGFR mutations in surgically resected
nonsmall cell lung cancer. Eur Respir J. 38:903–910. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Takano T, Ohe Y, Tsuta K, et al: Epidermal
growth factor receptor mutation detection using high-resolution
melting analysis predicts outcomes in patients with advanced non
small cell lung cancer treated with gefitinib. Clin Cancer Res.
18:5385–5390. 2007. View Article : Google Scholar
|
|
34
|
Bell DW, Lynch TJ, Haserlat SM, et al:
Epidermal growth factor receptor mutations and gene amplification
in non-small-cell lung cancer: molecular analysis of the
IDEAL/INTACT gefitinib trials. J Clin Oncol. 23:8081–8092. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sharma SV, Bell DW, Settleman J and Haber
DA: Epidermal growth factor receptor mutations in lung cancer. Nat
Rev Cancer. 7:169–181. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
36
|
Smith GD, Chadwick BE, Willmore-Payne C
and Bentz JS: Detection of epidermal growth factor receptor gene
mutations in cytology specimens from patients with non-small cell
lung cancer utilising high-resolution melting amplicon analysis. J
Clin Pathol. 61:487–493. 2008. View Article : Google Scholar : PubMed/NCBI
|
















