Introduction
Gastric cancer is one of the most frequently
occurring aggressive malignancies, and is the second most common
cause of cancer-related mortality worldwide (1). The majority of gastric cancer cases
are already in the advanced stages when diagnosed and the prognosis
is generally poor, with a 5-year survival rate <30% (2). Preoperative chemotherapy has been
successfully used in the treatment of locally advanced gastric
cancer, as it is capable of shrinking the tumor, thereby increasing
the possibility of complete resection. Previous studies indicated
that this treatment strategy may significantly improve overall
survival in patients with resectable gastric cancer (3). However, only ~20% of patients
experienced complete or subtotal tumor regression (4), with chemotherapy resistance
representing the major obstacle for successful treatment.
Leucine-rich repeat-containing G protein-coupled
receptor 5 (Lgr5), also known as GPR49, is considered a target of
Wnt signaling (5–8). Lgr5 is a potential marker of adult
stem cells of the small intestine, colon, stomach and hair follicle
bulge (9–11). Increased expression of Lgr5 has been
investigated in several types of human cancer, including
hepatocellular carcinoma (7),
gastric (12), colorectal (13–15)
and ovarian cancer (16), basal
cell carcinoma (17), esophageal
adenocarcinoma (18) and brain
cancer (19). In colorectal cancer,
increased Lgr5 expression was identified in the spheroid cells
(20), making it an ideal marker of
colorectal cancer stem cells (CSCs) (13,14).
It is well documented that CSCs can survive radiation therapy and
chemotherapy. Becker et al suggested that Lgr5 may represent
a better marker for CSCs in colorectal cancer (21). Previous studies indicated that Lgr5
expression level in rectal cancer specimens was elevated after
preoperative chemoradiotherapy (CRT). Moreover, elevated Lgr5 gene
expression was associated with poor pathological response and poor
survival (22,23) suggesting that Lgr5 may contribute to
CRT resistance.
Previous studies identified elevated Lgr5 expression
in gastric cancer (21).
Furthermore, Lgr5-positive cells were correlated with gastric
cancer carcinogenesis and progression (12). Previously, gastric cancer was
considered to be a chemosensitive tumor; however, responses to
chemotherapy were partial and short lived (24). We propose that Lgr5 plays a
potential role in chemotherapy resistance in gastric cancer. To our
knowledge, no studies investigating the value of Lgr5 expression in
the prediction of preoperative chemotherapy efficacy in gastric
cancer patients have been reported. In this study, we investigated
the potential of Lgr5 as a specific biomarker in predicting tumor
response and overall survival in advanced gastric cancer patients
treated with preoperative chemotherapy. Furthermore, the possible
association between Lgr5 expression and chemotherapy resistance was
also investigated.
Materials and methods
Patients and specimens
A total of 68 patients with gastric cancer were
included in this study. All patients had undergone preoperative
(oxaliplatin-based) chemotherapy followed by gastrectomy between
2007 and 2009 in the Chinese People’s Liberation Army (PLA) General
Hospital (Beijing, China). All patients received three cycles of
oxaliplatin-based adjuvant chemotherapy, including 56 patients
treated with oxaliplatin/leucovorin/5-fluorouracil (5-FU) [FOLFOX4,
oxaliplatin (85 mg/m2) and leucovorin (400
mg/m2) followed on days 1 and 2 by 5-FU (400
mg/m2) administered by intravenous (i.v.) bolus, then
subsequently (600 mg/m2) over a 22-h continuous
infusion] repeated every 2 weeks, and 12 patients treated with
S-1/oxaliplatin [SOX, oxaliplatin (130 mg/m2)
administered by i.v. injection on day 1, with S-1 administered
orally (80 mg/m2/day) for 14 days] repeated every 3
weeks. Patient eligibility was based on fulfillment of the
following institute criteria: i) patients had received no previous
radiotherapy or immunotherapy, ii) performance status was 0–2
(Eastern Cooperative Oncology Group scale) (25), iii) aged between 18 and 79 years,
and iv) clinical tumor was stage II or III according to clinical
TNM stage revised by the International Union Against Cancer (UICC)
in 2009, with no evidence of distant metastases. Pre-treatment
endoscopic biopsy specimens and surgically resected tumors were
routinely fixed in 10% formalin and embedded in paraffin wax.
Tissue sections were stained with hematoxylin and eosin (HE).
Histopathological features and responsiveness to neoadjuvant
chemotherapy were evaluated by light microscopy. All patients were
followed up for survival analysis. The follow-up period was
calculated from the date of surgery until June 30, 2012.
Histological assessment of
chemotherapeutic effects
Histological slides were independently reviewed by
two pathologists blinded to all clinical pathology data. In the
event of discordant observations, the slides were reassessed on a
double-headed microscope to establish a final result.
Post-chemotherapy, histological tumor regression grading (TRG) was
evaluated according to the Becker et al score (26), based on an estimation of the
percentage of vital tumor tissue in relation to the macroscopically
identifiable tumor bed that was evaluated. Tumor regression was
classified into three categories: TRG 1, complete or subtotal
regression (<10% residual tumor/tumor bed); TRG 2, partial tumor
regression (10–50% residual tumor/tumor bed) and TRG 3, minimal or
no tumor regression (>50% residual tumor/tumor bed). All
patients with grade 1 and 2 regression were classified as
responders, while patients with grade 3 regression were defined as
pathologic non-responders. Specimens with no residual cancer cells
and only a fibrotic mass were excluded, as our immunohistochemical
staining targeted residual cancer tissue.
Immunohistochemistry
Immunohistochemical staining of Lgr5 was performed
as previously described (27).
Sections (4 μm) were cut from paraffin-embedded tissue blocks,
deparaffinized in xylene and rehydrated. Antigen retrieval was
performed by heating in 0.01 mol/l citrate buffer (pH 6.0) in a
microwave oven for 2 min at 100°C. Sections were subsequently
incubated with 3% hydrogen peroxidase-methanol for 15 min to
inhibit endogenous peroxidase activity. After washing with
phosphate-buffered saline (PBS), and blocking with 10% goat serum,
sections were incubated with primary monoclonal rabbit antibody to
human Lgr5 (1:50 in blocking solution; Abcam, Cambridge, MA, USA)
or PBS (negative control) and incubated overnight at 4°C. Slides
were subsequently washed three times with PBS and incubated for 30
min with biotinylated secondary antibody
(polyperoxidase-anti-mouse/rabbit IgG; Zymed Laboratories, San
Francisco, CA, USA). Peroxidase reactivity was visualized using a
3,3′-diaminobenzidine (DAB) substrate kit (Zymed Laboratories) and
slides were counterstained with hematoxylin.
Evaluation of immunohistochemistry
Immunohistochemical sections were independently
reviewed by two experienced pathologists with no prior knowledge of
patient pathology data. In discrepant cases, a final score was
established by reassessment of section on a double-headed
microscope. Lgr5 expression was evaluated according to a score
incorporating both staining intensity and the percentage of cells
stained (28). Staining intensity
was scored as follows: 0, no staining; 1+, weak staining; 2+,
moderate staining; and 3+, intense staining. The percentage of
staining was scored as follows: 0, no staining in all cells; 1+,
positive staining in <10% of cells; 2+, positive staining in
10–50% cells; and 3+, >50% positive staining. The final score
was determined by the combined staining score. A score (extent +
intensity) ≤1 was considered negative and a score between 2 and 6
was considered positive (29,30).
Cell culture
Previously, we demonstrated that Lgr5 levels were
significantly higher in the gastric cancer cell lines (MGC803,
SGC7901, AGS, BGC823, MKN45) than in the gastric epithelial cell
line (GES-1). Lgr5 expression was the highest in AGS cells, which
were selected for subsequent RNA interference experiments. The AGS
human gastric carcinoma cell line was purchased from the American
Type Culture Collection (ATCC; Manassas, VA, USA). Cells were
cultured in RPMI-1640 medium supplemented with 10% fetal bovine
serum (both from Gibco-BRL, Gaithersburg, MD, USA), 100 U/ml
penicillin and 100 μg/ml streptomycin. Cells were cultured and
passaged at 37°C in culture flasks with a humidified atmosphere of
5% CO2.
Transient transfection of Lgr5 small
interfering RNAs (siRNAs)
siRNAs were synthesized by GenePharma Co. (Shanghai,
China). The sequences were as follows: 5′-GCAGA AUAAUCAGCUAAGATT-3′
(sense), and 5′-UCUUAGCUGA UUAUUCUGCTT-3′ (antisense);
Lgr5-homo-1555, 5′-GGAC GACCUUCAUAAGAAATT-3′ (sense), and
5′-UUUCUUAU GAAGGUCGUCCTT-3′ (antisense); Lgr5-homo-2664, 5′-GC
UCCAGCAUCACUUAUGATT-3′ (sense), and 5′-UCAUAAG UGAUGCUGGAGCTT-3′
(antisense); negative control, 5′-UUCUCCGAACGUGUCACGUTT-3′ (sense),
and 5′-ACG UGACACGUUCGGAGAATT-3′ (antisense); GAPDH positive
control, 5′-GUAUGACAACAGCCUCAAGTT-3′ (sense), and
5′-CUUGAGGCUGUUGUCAUACTT-3′ (antisense).
AGS cells were cultured in 6-well plates and were
then transiently transfected with 4 μl of siRNA using 2 μl of
Lipofectamine 2000 (Invitrogen, USA), according to the
manufacturer’s instructions.
Cell viability assay
Transfected AGS cells were seeded in 96-well plates
(1×104 cells/well). After 24 h, cells were exposed to
increasing concentrations of oxaliplatin, 0.1–10 μg/ml, or to 5-FU
(0.25–20 μg/ml; both from Sigma, St. Louis, MO, USA) for 72 h, and
cell viability was assessed by MTT assay. MTT reagent (20 μl of a 5
mg/ml stock) was added into each well and incubated at 37°C, in a
humidified atmosphere of 5% CO2. After 4 h, this mixture
was carefully removed and 150 μl of dimethyl sulfoxide was added to
each well and plates were incubated on a rocking shaker for 10 min.
Absorbance was measured at a wavelength of 490 nm with a microplate
reader and the background absorbance of the medium in the absence
of cells was subtracted. The cell survival rate (%) was calculated
as follows: Survival rate (%) = (mean A value of drug-treated group
− mean A value of blank control group)/(mean A value of the
negative control group − mean A value of blank control group) ×
100%. Each assay was performed in triplicate, and the results are
presented as the means ± standard deviation (SD).
Quantitative real-time reverse
transcription-polymerase chain reaction (qRT-PCR)
Total RNA was extracted from cells using an RNeasy
Mini kit (Qiagen, Tokyo, Japan) and reverse transcribed using a
cDNA reverse transcription kit (Applied Biosystems, Foster City,
CA, USA). qRT-PCR analysis was performed on an ABI PRISM 7700
Sequence Detection System using SYBR-Green PCR Master Mix (both
from Applied Biosystems) and the following cycling conditions: 95°C
for 10 min, followed by 50 cycles of 95°C for 15 sec and 60°C for 1
min. Amplification was performed using the following primer sets:
Lgr5 (NM_003667.2) (161 bp), forward primer
5′-TTTGGACAAGGGAGACCTGGAGAAT-3′, and reverse primer
5′-GAAAGCCACAGGGCAGTTTAGGAT-3′; MMP2 (NM_001127891) (160 bp),
forward primer 5′-AGCA TGTCCCTACCGAGTCT-3′, and reverse primer,
5′-AAACAG ATGGCAAACACGGC-3′; GAPDH (266 bp), forward primer
5′-AGAAGGCTGGGGCTCATTTG-3′, and reverse primer,
5′-AGGGGCCATCCACAGTCTTC-3′. Relative transcript levels were
calculated using the 2−ΔΔCT method (31). Lgr5 mRNA expression levels
were normalized to those of GAPDH. All experiments were performed
in biological triplicate.
Western blot analysis
BGC-823 and AGS cells were lysed in RIPA lysis
buffer, and cell lysates were clarified by centrifugation (13,000
rpm, 4°C, 20 min). The concentration of protein lysates was
assessed using the Bradford method (Bio-Rad, Hercules, CA, USA).
Equal quantities of protein (50 μg/lane) were then separated by 12%
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
(SDS-PAGE), and transferred to nitrocellulose membranes (Amersham
Biosciences, Piscataway, NJ, USA). After blocking with 5% non-fat
milk in TBS-T [50 mmol/L Tris-HCl (pH 7.6), 150 mmol/l NaCl, 0.1%
Tween-20] at room temperature for 1 h, membranes were incubated
with primary antibodies (anti-Lgr5, 1:100; anti-GAPDH, 1:1,500;
Abcam, Cambridge, MA, USA) in blocking buffer overnight at 4°C.
After washing three times with TBS-T, membranes were incubated with
a horseradish peroxidase-coupled goat anti-rabbit secondary
antibody (1:2,000; Santa Cruz Biotechnology) for 2 h at room
temperature. Bands were visualized by chemiluminesence. The protein
quantity using Quantity One v4.4 software (Bio-Rad). Target protein
expression was evaluated by the relative intensity ratio of target
protein-to-β-actin loading control.
Apoptosis analysis with Annexin
V-FITC/propidium iodide (PI) dual staining
Apoptosis was quantified by dual staining with
Annexin V and PI using the Annexin V-FITC apoptosis detection kit
(Biosea Biotechnology, Beijing, China). All gastric cancer cell
lines (2×105 cells/well) were cultured in 6-well plates
to 70–80% confluence. Cells were then treated with the indicated
concentrations of oxaliplatin or 5-FU for 24 h. Cells were
harvested and washed with ice-cold PBS (x3) and resuspended in 200
μl 1× binding buffer. Annexin V-FITC (10 μl) and PI (5 μl) were
then added and cells were incubated for 15 min at room temperature
in the dark. Cells were analyzed by flow cytometry using a FACSAria
cytometer (Becton-Dickinson, San Jose, CA, USA). All experiments
were performed in biological triplicate.
Statistical analysis
Statistical analyses were performed using SPSS
V.13.0 (SPSS, Inc., Chicago, IL, USA). The Pearson χ2
test was used to examine the various clinicopathological
characteristics and TRG with the expression of Lgr5. Cumulative
survival curves were drawn by the Kaplan-Meier method. The
difference between the curves was analyzed by the log-rank test.
Multivariate survival analysis was based on the Cox proportional
hazard model. The data of cell survival rate are presented as the
means ± SD of at least three independent experiments. Differences
of the variables between groups were analyzed by the Student’s
t-test. A value of P<0.05 was considered statistically
significant.
Results
Patients and tumor characteristics
A total of 68 patients with gastric cancer, aged
between 31 and 79 years (median, 62.5 years; mean, 58.5 years) were
enrolled in this study. These included 36 cases of well or
moderately differentiated adenocarcinoma and 32 cases of poorly
differentiated, signet ring cell, or mucinous adenocarcinoma. The
post-chemotherapy, pathological T stages were ypT0–2
(n=32) and ypT3+4 (n=36). Forty-five patients exhibited
lymph node metastases. Histological TRG was as follows: TRG 1, 28
patients (41.2%); TRG 2, 15 patients (22.0%) and TRG 3, 25 patients
(36.8%) (Table I). During the
follow-up period, 37 patients died while 31 patients remained alive
(median survival time, 48 months; mean survival time, 45.8±6.2
months). Representative HE staining patterns in the pre-treatment
endoscopic biopsy specimens and surgically resected tumors are
shown in Fig. 1A.
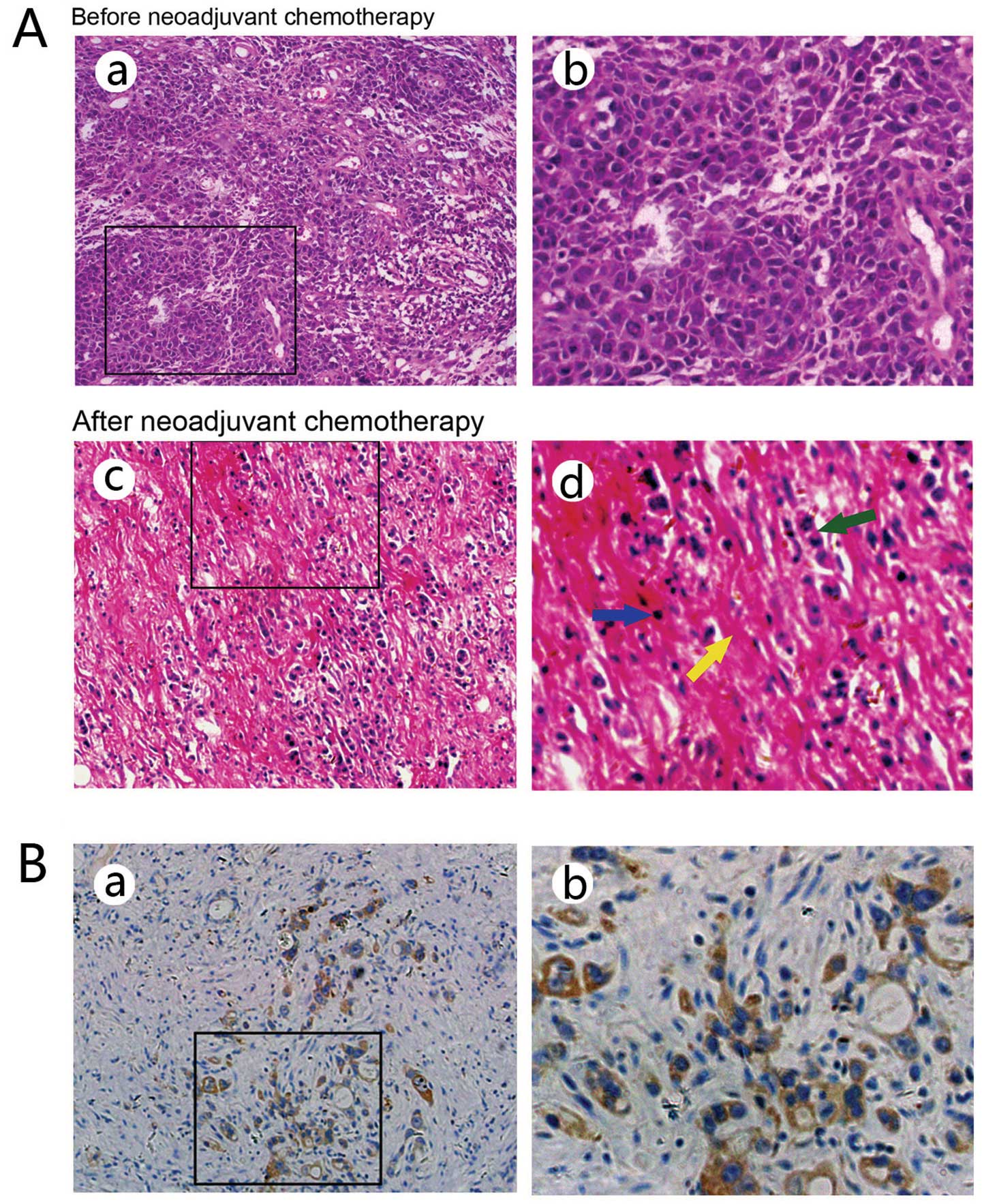 | Figure 1(A) Hematoxylin and eosin (HE)
staining patterns in a pre-treatment endoscopic biopsy specimen (a
and b) and the resected tumor (c and d) from a representative
gastric cancer patient who responded to neoadjuvant SOX
chemotherapy. Chemotherapy-induced histological changes including
the degeneration of cancer cells with marked inflammatory cell
infiltration (blue arrow), fibrosis (yellow arrow), and
significantly reduced cancer cells were noted in the resected tumor
compared with the pre-treatment biopsy specimen. The green arrow
indicates residual cancer cells (a and c, magnification, ×100; b
and d, magnification, ×200). (B) Immunohistochemical analysis of
Lgr5 expression in residual cancer cells. Immunoreactive Lgr5
protein was located in the cytoplasm (a, magnification, ×100; b,
magnification, ×200). Lgr5, leucine-rich repeat-containing G
protein-coupled receptor 5. |
 | Table ICorrelation between Lgr5 expression
and clinicopathological features in gastric carcinoma. |
Table I
Correlation between Lgr5 expression
and clinicopathological features in gastric carcinoma.
| Lgr5 | |
|---|
|
| |
|---|
| Variables | Positive (%) | Negative (%) | P-value |
|---|
| Gender | | | 0.867 |
| Male | 36 (66.7) | 18 (33.3) | |
| Female | 9 (64.3) | 5 (35.7) | |
| Age (years) | | | 0.582 |
| <45 | 5 (83.3) | 1 (16.7) | |
| ≥45 and
<60 | 26 (66.7) | 13 (33.3) | |
| ≥60 | 14 (60.9) | 9 (39.1) | |
| Tumor size
(cm) | | | 0.011 |
| d <4 | 7 (41.2) | 10 (58.8) | |
| d ≥4 and
<8 | 30 (69.8) | 13 (30.2) | |
| d ≥8 | 8 (100) | 0 (0.0) | |
| Histology | | | 0.001 |
| Well/moderate | 17 (47.2) | 19 (52.8) | |
|
Poor/signet/mucinous | 28 (87.5) | 4 (12.5) | |
| ypT category | | | 0.008 |
|
ypT0–2 | 16 (50.0) | 16 (50.0) | |
|
ypT3+4 | 29 (80.6) | 7(19.4) | |
| ypN category | | | 0.001 |
|
ypN0 | 7 (30.4) | 16 (69.6) | |
|
ypN1–3 | 38 (84.4) | 7 (15.6) | |
| Postoperative TNM
stage | | | 0.001 |
| I/II | 16 (42.1) | 22 (57.94) | |
| III | 29 (96.7) | 1 (3.3) | |
| Tumor regression
grading | | | 0.001 |
| TRG 1 | 8 (28.6) | 20 (71.4) | |
| TRG 2 | 13 (86.7) | 2 (13.3) | |
| TRG 3 | 24 (96.0) | 1 (4) | |
Correlation of Lgr5 expression in
residual cancer cells with clinicopathological variables
Lgr5 expression was observed in the cytoplasm of
residual cancer cells (Fig. 1B). A
significant correlation was observed between Lgr5 immunoreactivity
and histological differentiation (P=0.001). Lgr5 expression was
more frequently observed in advanced ypT-stage cancer (P=0.008).
Furthermore, Lgr5 expression positively correlated with metastasis
in the regional lymph nodes (P=0.001) and with progression of the
ypTNM stage (P=0.001). Lgr5 expression was also significantly
associated with TRG after preoperative chemotherapy. Finally,
patients with poor tumor regression exhibited a significantly
higher rate of positive Lgr5 expression than patients with
regressed tumors (P=0.001) (Table
I).
Survival analysis
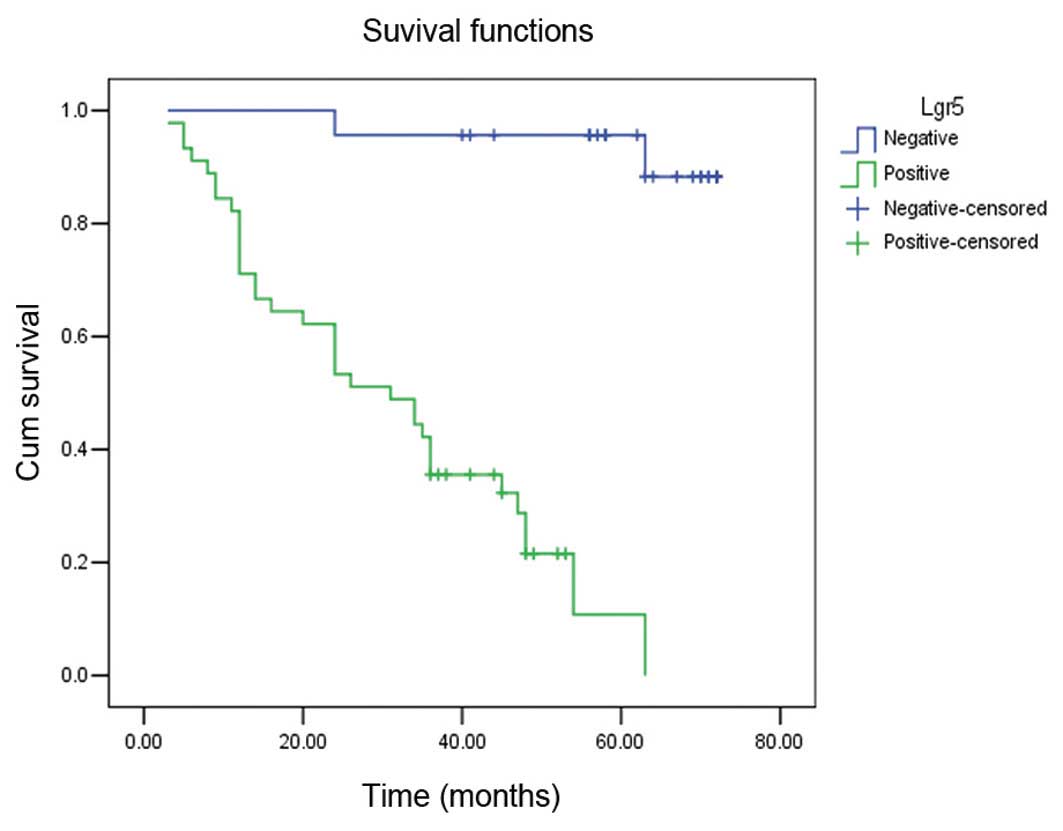
Survival analysis by log-rank test revealed that
Lgr5-positive patients had a significantly shorter survival time
(median, 31 months; mean, 31.29±5.92 months) compared with
Lgr5-negative patients (median, 63 months; mean, 69.25±4.13 months)
(log-rank=33.12, P=0.001) (Fig. 2).
Multivariate analysis using the Cox regression model revealed that
Lgr5 expression significantly affected the outcome of gastric
cancer after preoperative chemotherapy and appeared to be an
independent prognostic factor (P=0.039) (hazard ratio=6.270, 95%
confidence interval=1.097–35.829) (Table II).
 | Table IICox regression analysis of prognostic
factors in gastric carcinoma after preoperative chemotherapy. |
Table II
Cox regression analysis of prognostic
factors in gastric carcinoma after preoperative chemotherapy.
| Variables | B | SE | Wald value | P-value | HR | 95.0% CI for
HR |
|---|
|
|---|
| Lower | Upper |
|---|
| Gender | −0.419 | 0.453 | 0.853 | 0.356 | 0.658 | 0.271 | 1.600 |
| Age | 0.251 | 0.365 | 0.472 | 0.492 | 1.285 | 0.629 | 2.625 |
| Histology | 1.533 | 0.470 | 10.635 | 0.001 | 4.630 | 1.843 | 11.630 |
| Size | 0.092 | 0.344 | 0.071 | 0.790 | 1.096 | 0.558 | 2.150 |
| ypT category | 0.423 | 0.441 | 0.920 | 0.337 | 1.526 | 0.643 | 3.620 |
| ypN category | 0.886 | 0.805 | 1.211 | 0.271 | 2.425 | 0.500 | 11.751 |
| ypTNM stage | 1.513 | 0.598 | 6.408 | 0.011 | 4.540 | 1.407 | 14.645 |
| TRG | 0.623 | 0.640 | 0.947 | 0.330 | 1.864 | 0.532 | 6.532 |
| Lgr5 | 1.836 | 0.889 | 4.261 | 0.039 | 6.270 | 1.097 | 35.829 |
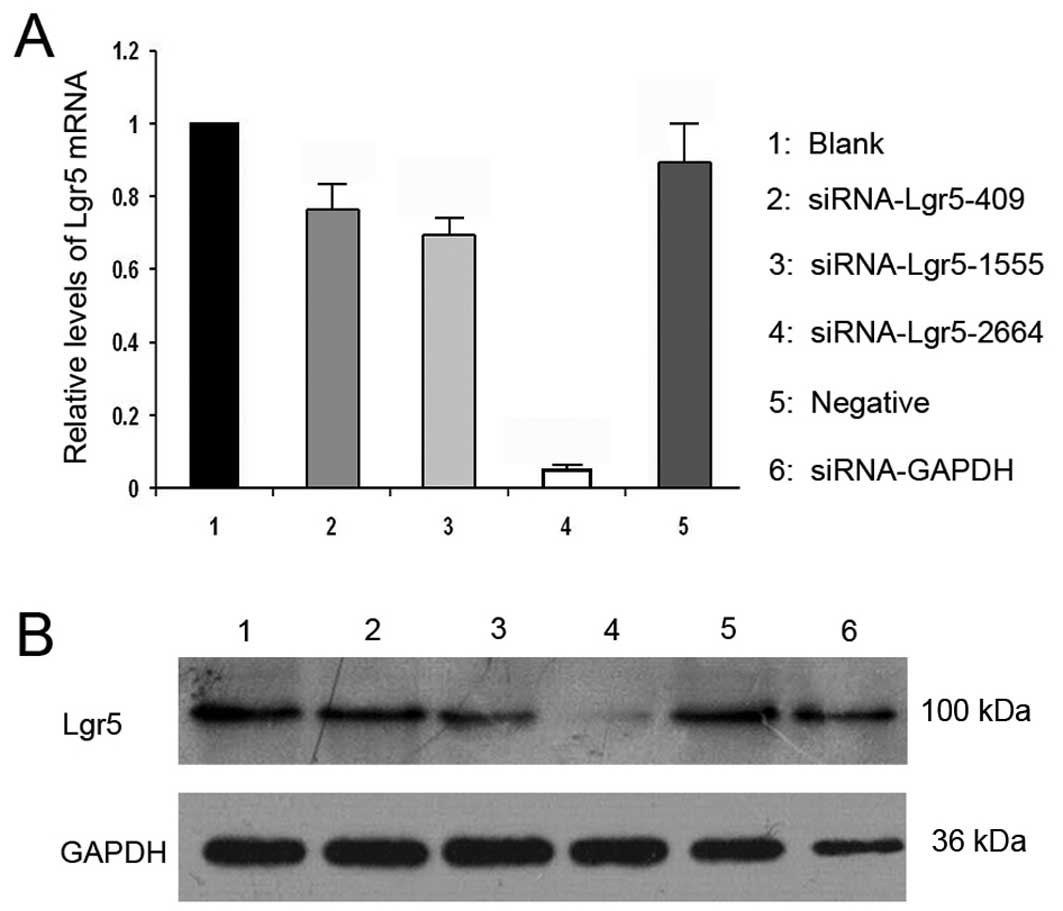
Silencing of Lgr5 expression in gastric
cancer cells
To specifically silence Lgr5 expression in gastric
cancer cells, three siRNA duplexes targeting different coding
regions of the Lgr5 mRNA (Lgr5-homo-409, Lgr5-homo-1555 and
Lgr5-homo-2664) were designed and synthesized. siRNAs were
transiently transfected into AGS gastric cancer cell lines, and the
expression of lgr5mRNA and protein was examined by qRT-PCR and
western blot analysis, respectively. The Lgr5-homo-2664 siRNA
exhibited the highest Lgr5 silencing efficiency (Fig. 3). The siRNA (Lgr5-homo-2664) was
transfected into AGS cells, and the transfectants were then
selected for further experiments.
RNAi-mediated downregulation of Lgr5
decreases resistance to oxaliplatin or 5-FU in gastric cancer
cells
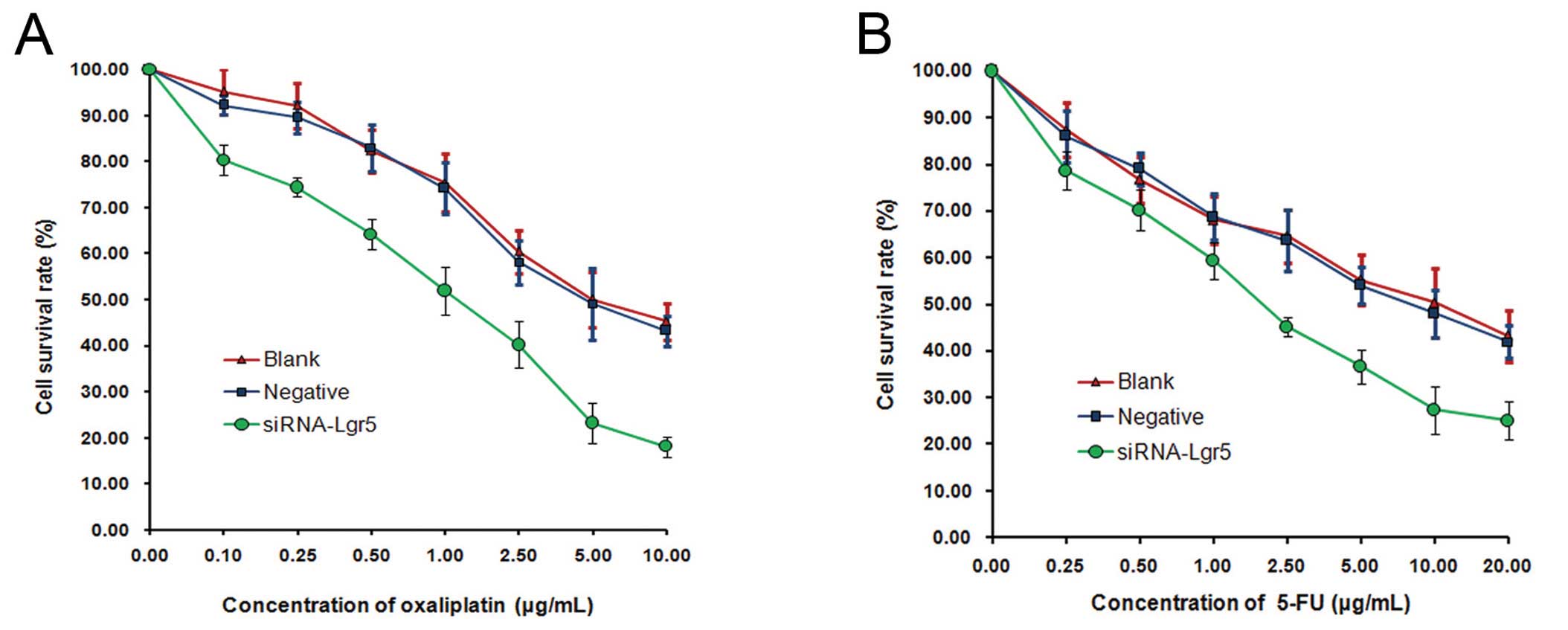
To investigate whether Lgr5 is associated with
chemosensitivity in gastric cancer cells, the effects of silencing
Lgr5 expression on oxaliplatin or 5-FU sensitivity in AGS cells was
examined by an MTT assay. RNAi-mediated suppression of Lgr5 in AGS
cells led to a significant decrease in cell survival rate following
treatment with oxaliplatin (0.1–10 μg/ml) compared with the blank
and negative control groups (all P=0.001) (Fig. 4A). Furthermore, we observed a
significant decrease in the survival rate of Lgr5-silenced AGS
cells following treatment with 5-FU (0.25–20 μg/ml) (P=0.001)
(Fig. 4B). These results suggest
that downregulation of Lgr5 in gastric cancer cells may increase
sensitivity to the cytotoxic effects of oxaliplatin or 5-FU.
Effect of Lgr5 interference on AGS cell
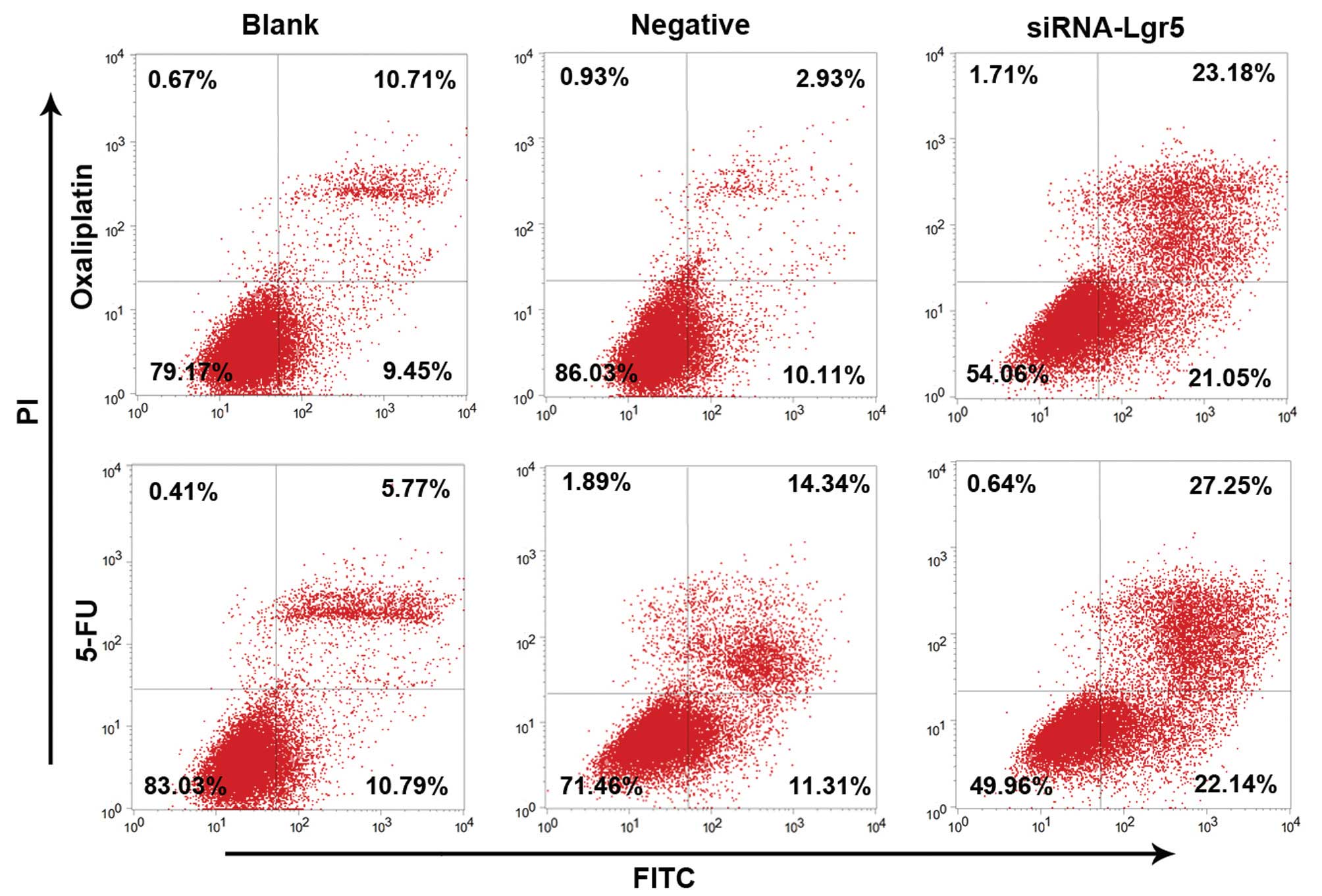
apoptosis induced by oxaliplatin or 5-FU
The effect of modulating Lgr5 expression on
oxaliplatin or 5-FU-induced cell apoptosis was next examined by
Annexin V-FITC/PI dual staining analysis (Fig. 5). The rate of apoptosis was
significantly higher in AGS cells transfected with Lgr5 siRNA
following treatment with (22.08%±1.95) compared with the blank
group (9.83%±0.72) and the negative control group (10.02%±1.22)
(all P=0.001). No significant difference was observed between the
blank and the negative control groups. Similar trends in apoptosis
were observed following treatment with 5-FU. The AGS transfected
with Lgr5 siRNA cell group exhibited significantly higher rates of
apoptosis (24.15%±2.56) compared with those in the blank
(10.87%±1.24) and negative control (11.56%±1.43) groups (all
P=0.001). No significant difference was observed between the blank
and the negative control group. Taken together, these results
indicate that the inhibition of Lgr5 expression in gastric cancer
cells using RNA interference technology may promote sensitivity to
chemotherapy in gastric cancer cells.
Discussion
Gastric cancer remains a major health problem
worldwide (4). To date, a number of
strategies exist for the treatment of gastric cancer, including
preoperative chemotherapy followed by surgery, which is now
increasingly used. Patients who respond to preoperative
chemotherapy have a significantly longer survival time than
non-responders (2). However,
response rates are very low (~20%) (32), and this is mostly attributed to
chemotherapy resistance. CSCs, capable of surviving chemotherapy,
may play an important role in this process (33–35).
Lgr5, a member of the G-protein-coupled receptor
(GPCR) superfamily, is a known downstream target gene activated by
Wnt signaling and a stem cell marker in the hair-follicle,
intestine, colon and stomach (9–11).
Lgr5 has also been reported as a marker of colorectal CSCs
(13,14). Recent studies reported that rectal
cancer specimens from patients with a poor pathological response
had significantly higher Lgr5 expression levels than those
exhibiting a positive response after CRT. This suggests that Lgr5
expression may be implicated in resistance to CRT in rectal cancer
(22,23).
Previously, it was suggested that Lgr5+
pyloric stem cells may represent a potential cell of origin in
Wnt-driven gastric cancer (11).
Moreover, Lgr5 expression levels were much higher in gastric cancer
than in normal mucosa (12).
However, to date, there has been no investigation into the
relationship between Lgr5 expression and chemotherapy
resistance.
In the present study, we observed that elevated Lgr5
expression in gastric cancer after preoperative chemotherapy was
significantly associated with histological differentiation,
ypT-stage and postoperative TNM stage. Positive expression of Lgr5
in gastric cancer specimens was associated with poor pathological
response to chemotherapy. Additionally, elevated expression of Lgr5
in gastric cancer was significantly correlated with poor survival.
These results suggest that Lgr5 may be a useful prognostic factor
for gastric cancer after preoperative chemotherapy and may also
predict patient response to preoperative chemotherapy. We also
examined the biological role of Lgr5 in oxaliplatin resistance of
gastric cancer cells following either overexpression or targeted
silencing of Lgr5. Analysis of cell growth by an MTT assay revealed
a significant decrease in AGS cell survival following suppression
of Lgr5. This suggests that Lgr5 increases chemotherapy resistance
in gastric cancer cells. There is a relative increase in the
frequency of CSCs after chemotherapy as, similar to normal stem
cells, CSCs are more resistant to chemotherapy compared with other
more differentiated cancer cells. Therefore, Lgr5, a cancer stem
cell marker in post-chemotherapy specimens, likely plays important
roles in predicting the clinical prognosis of gastric cancer
patients.
Furthermore, many signaling pathways have been
associated with chemotherapy resistance, including the Wnt
signaling pathway (36–40). Previous studies indicated that
dysregulation of the Wnt/β-catenin signaling pathway is involved in
pancreatic cancer chemoresistance (39). Accumulating research indicates that
Lgr5 is a target of Wnt signaling (5–8). Lgr5
becomes part of the Wnt signaling complex at the membrane level and
enhances Wnt/β-catenin signaling by increasing interactions with
members of the Wnt pathway, including LRP6 and Fzd5 (8). Based on this, we speculate that Lgr5
may influence the sensitivity of gastric cancer cells to
chemotherapeutic drugs via regulation of the Wnt signaling
pathway.
In conclusion, our study demonstrated that elevated
Lgr5 expression in gastric cancer following preoperative
chemotherapy is significantly associated with tumor progression,
poor pathological response to chemotherapy and shorter survival
time. Furthermore, cytological analyses revealed that upregulation
of Lgr5 may increase chemotherapy resistance in gastric cancer
cells, while downregulation of Lgr5 was capable of sensitizing
cells to chemotherapy. Therefore, Lgr5 may be a potential biomarker
for predicting chemotherapeutic sensitivity and prognosis in
gastric cancer patients and may represent a novel therapeutic
target for cancer therapy.
Acknowledgements
This study was supported by grants from the National
Natural Science Foundation of China (nos. 81272698, 81101883 and
81172368), a grant from PLA medical and health research fund
project (no. 11BJZ17), a grant from PLA Medical Technology Key
Project of Scientific Research in 12th Five-Year-Plan (no.
BWS12J049), a grant form the Capital Health Research and
Development of Special (no. 2011-5001-01), and a grant from the
Major Science and Technology Project of ‘National Significant New
Drug Creation’ from the Major Science and Technology of China (no.
2011ZX09307-001-05).
References
|
1
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar
|
|
2
|
Thun MJ, DeLancey JO, Center MM, Jemal A
and Ward EM: The global burden of cancer: priorities for
prevention. Carcinogenesis. 31:100–110. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cunningham D, Allum WH, Stenning SP, et
al: Perioperative chemotherapy versus surgery alone for resectable
gastroesophageal cancer. N Engl J Med. 355:11–20. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Becker K, Langer R, Reim D, et al:
Significance of histopathological tumor regression after
neoadjuvant chemotherapy in gastric adenocarcinomas: a summary of
480 cases. Ann Surg. 253:934–939. 2011. View Article : Google Scholar
|
|
5
|
Van der Flier LG, Sabates-Bellver J, Oving
I, et al: The intestinal Wnt/TCF signature. Gastroenterology.
132:628–632. 2007.PubMed/NCBI
|
|
6
|
Segditsas S, Sieber O, Deheragoda M, et
al: Putative direct and indirect Wnt targets identified through
consistent gene expression changes in APC-mutant intestinal
adenomas from humans and mice. Hum Mol Genet. 17:3864–3875. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yamamoto Y, Sakamoto M, Fujii G, et al:
Overexpression of orphan G-protein-coupled receptor, Gpr49, in
human hepatocellular carcinomas with beta-catenin mutations.
Hepatology. 37:528–533. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Carmon KS, Lin Q, Gong X, Thomas A and Liu
Q: LGR5 interacts and cointernalizes with Wnt receptors to modulate
Wnt/β-catenin signaling. Mol Cell Biol. 32:2054–2064.
2012.PubMed/NCBI
|
|
9
|
Barker N, van Es JH, Kuipers J, et al:
Identification of stem cells in small intestine and colon by marker
gene Lgr5. Nature. 449:1003–1007. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jaks V, Barker N, Kasper M, et al: Lgr5
marks cycling, yet long-lived, hair follicle stem cells. Nat Genet.
40:1291–1299. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Barker N, Huch M, Kujala P, et al:
Lgr5(+ve) stem cells drive self-renewal in the stomach and build
long-lived gastric units in vitro. Cell Stem Cell. 6:25–36.
2010.
|
|
12
|
Simon E, Petke D, Boger C, et al: The
spatial distribution of LGR5+ cells correlates with
gastric cancer progression. PLoS One. 7:e354862012.PubMed/NCBI
|
|
13
|
Takahashi H, Ishii H, Nishida N, et al:
Significance of Lgr5(+ve) cancer stem cells in the colon and
rectum. Ann Surg Oncol. 18:1166–1174. 2011.
|
|
14
|
Kemper K, Prasetyanti PR, De Lau W,
Rodermond H, Clevers H and Medema JP: Monoclonal antibodies against
Lgr5 identify human colorectal cancer stem cells. Stem Cells.
30:2378–2386. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wu XS, Xi HQ and Chen L: Lgr5 is a
potential marker of colorectal carcinoma stem cells that correlates
with patient survival. World J Surg Oncol. 10:2442012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
McClanahan T, Koseoglu S, Smith K, et al:
Identification of overexpression of orphan G protein-coupled
receptor GPR49 in human colon and ovarian primary tumors. Cancer
Biol Ther. 5:419–426. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tanese K, Fukuma M, Yamada T, et al:
G-protein-coupled receptor GPR49 is up-regulated in basal cell
carcinoma and promotes cell proliferation and tumor formation. Am J
Pathol. 173:835–843. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Becker L, Huang Q and Mashimo H: Lgr5, an
intestinal stem cell marker, is abnormally expressed in Barrett’s
esophagus and esophageal adenocarcinoma. Dis Esophagus. 23:168–174.
2010.PubMed/NCBI
|
|
19
|
Nakata S, Campos B, Bageritz J, et al:
LGR5 is a marker of poor prognosis in glioblastoma and is required
for survival of brain cancer stem-like cells. Brain Pathol.
23:60–72. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Vermeulen L, Todaro M, de Sousa Mello F,
et al: Single-cell cloning of colon cancer stem cells reveals a
multi-lineage differentiation capacity. Proc Natl Acad Sci USA.
105:13427–13432. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Becker L, Huang Q and Mashimo H:
Immunostaining of Lgr5, an intestinal stem cell marker, in normal
and premalignant human gastrointestinal tissue.
ScientificWorldJournal. 8:1168–1176. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Saigusa S, Inoue Y, Tanaka K, et al:
Clinical significance of LGR5 and CD44 expression in locally
advanced rectal cancer after preoperative chemoradiotherapy. Int J
Oncol. 41:1643–1652. 2012.PubMed/NCBI
|
|
23
|
Saigusa S, Inoue Y, Tanaka K, et al:
Significant correlation between LKB1 and LGR5 gene expression and
the association with poor recurrence-free survival in rectal cancer
after preoperative chemoradiotherapy. J Cancer Res Clin Oncol.
139:131–138. 2013. View Article : Google Scholar
|
|
24
|
Ozkan M, Akbudak IH, Deniz K, et al:
Prognostic value of excision repair cross-complementing gene 1
expression for cisplatin-based chemotherapy in advanced gastric
cancer. Asian Pac J Cancer Prev. 11:181–185. 2010.PubMed/NCBI
|
|
25
|
Oken MM, Creech RH, Tormey DC, et al:
Toxicity and response criteria of the Eastern Cooperative Oncology
Group. Am J Clin Oncol. 5:649–655. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Becker K, Mueller JD, Schulmacher C, et
al: Histomorphology and grading of regression in gastric carcinoma
treated with neoadjuvant chemotherapy. Cancer. 98:1521–1530. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xi HQ, Wu XS, Wei B and Chen L: Aberrant
expression of EphA3 in gastric carcinoma: correlation with tumor
angiogenesis and survival. J Gastroenterol. 47:785–794. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xi HQ, Zhao P and Han WD:
Clinicopathological significance and prognostic value of LRP16
expression in colorectal carcinoma. World J Gastroenterol.
16:1644–1648. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Matsubara J, Yamada Y, Nakajima TE, et al:
Clinical significance of insulin-like growth factor type 1 receptor
and epidermal growth factor receptor in patients with advanced
gastric cancer. Oncology. 74:76–83. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wood LD, Calhoun ES, Silliman N, et al:
Somatic mutations of GUCY2F, EPHA3, and NTRK3 in human cancers. Hum
Mutat. 27:1060–1061. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT
method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lordick F, Stein HJ, Peschel C and Siewert
JR: Neoadjuvant therapy for oesophagogastric cancer. Br J Surg.
91:540–551. 2004. View
Article : Google Scholar
|
|
33
|
Al-Hajj M: Cancer stem cells and oncology
therapeutics. Curr Opin Oncol. 19:61–64. 2007.PubMed/NCBI
|
|
34
|
Dirks PB: Cancer: stem cells and brain
tumours. Nature. 444:687–688. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
35
|
Setoguchi T, Taga T and Kondo T: Cancer
stem cells persist in many cancer cell lines. Cell Cycle.
3:414–415. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Haq R and Zanke B: Inhibition of apoptotic
signaling pathways in cancer cells as a mechanism of chemotherapy
resistance. Cancer Metastasis Rev. 17:233–239. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Gu L, Findley HW, Zhu N and Zhou M:
Endogenous TNFalpha mediates cell survival and chemotherapy
resistance by activating the PI3K/Akt pathway in acute
lymphoblastic leukemia cells. Leukemia. 20:900–904. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Mahon KL, Henshall SM, Sutherland RL and
Horvath LG: Pathways of chemotherapy resistance in
castration-resistant prostate cancer. Endocr Relat Cancer.
18:R103–R123. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Cui J, Jiang W, Wang S, Wang L and Xie K:
Role of Wnt/β-catenin signaling in drug resistance of pancreatic
cancer. Curr Pharm Des. 18:2464–2471. 2012.
|
|
40
|
Zhang H, Zhang X, Wu X, et al:
Interference of Frizzled 1 (FZD1) reverses multidrug resistance in
breast cancer cells through the Wnt/β-catenin pathway. Cancer Lett.
323:106–113. 2012.PubMed/NCBI
|