Introduction
Hepatocellular carcinoma (HCC) is a highly
aggressive cancer, characterized by the activation of multiple
molecular pathways (1–5). It is the sixth most common cancer and
the third most common cause of cancer-related deaths worldwide
(6–9). China alone accounts for 55% of the
world’s cases due to the high prevalence of chronic hepatitis B
virus (HBV) infection and liver cirrhosis, both significant risk
factors for the disease (10–13).
Currently, radical resection and liver transplantation (LT) offer
the main potentially curative therapeutic modalities. However, the
long-term survival of patients following surgery remains
unsatisfactory due to the high frequency of recurrence due to
metastasis - principally attributed to the presence of microscopic
extrahepatic metastatic foci before surgery (14,15).
Therefore, a better understanding of the molecular mechanisms
underlying HCC recurrence may offer improved diagnostic and
prognostic capabilities in addition to the development of effective
novel therapeutic strategies.
microRNAs, a newly discovered class of non-coding
small RNAs, have been recognized as potential targets for tumor
treatment due to their important roles in carcinogenesis and cancer
progression via binding to specific complementary sites within the
3′ untranslated regions (3’UTR) of their target gene mRNAs
(16–18). Accumulated studies have shown that
miRNAs may function as oncogenes or tumor suppressors in the
tumorigenesis of various human cancers depending on the cellular
contexts and the target genes that they regulate. Among these
functional miRNAs, miR-200a has been demonstrated to function as a
tumor suppressor, and loss of miR-200a expression has been reported
in many cancer types, whereas restoration of miR-200a expression
has been shown to abrogate tumorigenesis (19,20).
For instance, decreased miR-200a has been detected in breast
cancer, whereas enhanced expression of miR-200a was found to induce
growth suppression in breast cancer cells through downregulation of
mitochondrial transcription factor A (TFAM) expression (21). Moreover, miR-200a was observed to
inhibit gastric adenocarcinoma cell proliferation and migration by
suppressing the wnt/β-catenin pathway (22).
However, the role of miR-200a in cell survival and
metastasis of liver cancer and the underlying molecular mechanisms
have not been elucidated. We therefore aimed to ascertain whether
miR-200a may also play an important role in the tumor progression
and metastasis of HCC.
We investigated the gene expression pattern of
miR-200a in HCC and analyzed its effects on facets of HCC biology
including invasion and cell survival. By revealing the functional
implications and molecular mechanisms of miR-200a, we shed crucial
light on how miR-200a functions in HCC pathogenesis.
Materials and methods
Tissue samples, cell lines and cell
transfection
One hundred and fifteen pairs of HCC and their
matched adjacent normal liver tissues were collected. The
clinicopathological characteristics of the patients are shown in
Table I. Specimens of cancer
tissues and their adjacent noncancerous tissues and clinical
information were available from these patients after obtaining
informed consent. All samples were obtained from patients who
underwent LT at Ningbo Yinzhou People’s Hospital [Yinzhou Hospital
Affiliated to the Medical School of Ningbo University and The First
Affiliated Hospital of Zhejiang University School of Medicine
(Hangzhou, China)]. All samples were snap-frozen in liquid nitrogen
and then stored at −80°C for further use. This study was approved
by the Ethics Review Committee of Ningbo Yinzhou People’s Hospital
(Yinzhou Hospital Affiliated to the Medical School of Ningbo
University and The First Affiliated Hospital, School of Medicine,
Zhejiang University).
 | Table ICorrelation between miR-200a
expression and clinicopathological factors in 115 HCC tumors. |
Table I
Correlation between miR-200a
expression and clinicopathological factors in 115 HCC tumors.
| miR-200a
expression | |
|---|
|
| |
|---|
| Variable | Low | High | P-valuea |
|---|
| Age (years) |
| <50 | 25 | 23 | 0.623 |
| ≥50 | 38 | 29 | |
| Gender |
| Female | 9 | 5 | 0.446 |
| Male | 54 | 47 | |
| Preoperative tumor
therapy |
| No | 46 | 41 | 0.468 |
| Yes | 17 | 11 | |
| Tumor size
(cm) |
| ≤5 | 34 | 33 | 0.304 |
| >5 | 29 | 19 | |
| Tumor number |
| Single | 35 | 30 | 0.818 |
| Multiple | 28 | 22 | |
| Tumor
differentiation |
| Well +
moderate | 34 | 29 | 0.847 |
| Poor | 29 | 23 | |
| Preoperative
AFP |
| ≤400 ng/ml | 38 | 32 | 0.894 |
| >400 ng/ml | 25 | 20 | |
| Vascular
invasion |
| None | 34 | 33 | 0.304 |
| Yes | 29 | 19 | |
Eight human HCC cell lines, including HepG2,
SMMC7721, BEL7402, Huh7, HCCLM3, MHCC97L, PLC and normal liver cell
line LO2 were purchased from the American Type Culture Collection
(Manassas, VA, USA), the Shanghai Institute of Cell Biology
(Shanghai, China) and the Liver Cancer Institute of Fudan
University (Shanghai, China). All of the cell lines were cultured
in Dulbecco’s modified Eagle’s medium (DMEM) with 4.5 g/l glucose
(HyClone) supplemented with 10% fetal bovine serum (FBS) (SAFC
Biosciences) and incubated at 37°C in a humidified environment
containing 5% CO2.
Ectopic expression of miR-200a in the cells was
achieved by transfection with miR-200a mimics or inhibitors
(Qiagen, Hilden, Germany) using Lipofectamine 2000 (Invitrogen,
Carlsbad, CA, USA). Cells were plated in 6-well plates and
transfected for 24 or 48 h. Transfected cells were used in further
assays or RNA/protein extraction.
RNA extraction and SYBR-Green
quantitative PCR analysis
Total RNA was extracted from the cells using TRIzol
reagent (Invitrogen). Mature miR-200a expression in the cells was
detected using the miScript SYBR-Green PCR kit (Qiagen). Expression
of RNU6B was used as an endogenous control. MACC1 expression was
measured by SYBR-Green qPCR assay (Takara, Dalian, China). β-actin
and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were used as
internal controls and processed using the 2−ΔΔCt
formula, and then normalized to the internal control. PCR of MACC1
was performed with specific primers: forward, TCGGTCAGGAAGAATTGCAC
and reverse, TTGTGAAGCAAGTCTGGGTCC.
In situ hybridization for miRNA
In situ hybridization for miR-200a was
performed in the formalin-fixed and paraffin-embedded tissue
specimens by using the miRCURY LNA microRNA ISH Optimization kit
(Exiqon, Vedbaek, Denmark). A 5′-DIG and 3′-DIG miRCURY labeled
miR-200a LNA probe (cat. 18094-15; Exiqon) was used. A scrambled
LNA detection probe was used as a negative control (cat. 99004-01;
Exiqon). Briefly, 5 μm-thick paraffin sections were deparaffinized
and treated with proteinase-K (15 μg/ml) at 50°C for 60 min,
followed by stringent washes with 5X standard saline citrate, 1X
saline sodium citrate, and 0.2X saline sodium citrate buffers at
50°C; DIG blocking reagent (Roche, Mannheim, Germany) in maleic
acid buffer containing 2% sheep serum at room temperature for 15
min; and alkaline phosphatase-conjugated anti-digoxigenin (diluted
1:500 in blocking reagent; Roche) at room temperature for 60 min.
Enzymatic development was performed by incubating the slides with
4-nitro-blue tetrazolium and 5-brom-4-chloro-3′-indolylphosphate
substrate (Roche) at 30°C for 2 h at 37°C for 10 min, followed by
nuclear fast red counterstain (Sangon, Shanghai, China) at room
temperature for 1 min. Slides were then dismantled in water,
dehydrated in alcohol solutions and mounted with mounting medium
(Sangon).
Measurement of cell viability, colony
formation assay, invasiveness and migration
Cell viability was measured using the Cell Counting
Kit-8 (Dojindo Laboratories, Kumamoto, Japan). Cells
(0.2×104) transfected with the indicated miRNA or siRNA
for 24 h were seeded in each 96-well plate.
For the colony formation assay, cells were seeded at
200 cells/well in 6-well plates containing complete DMEM on day 0,
which was refreshed twice/week. On day 14, colonies were fixed with
3.7% formaldehyde for 15 min and stained with 1% crystal violet
(Sangon) before quantification.
Cell invasion and migration assays were performed
using the Transwell (Millipore, Billerica, MA, USA) based method.
Forty eight hours after plasmid transfection or RNA interference,
the filters coated with Matrigel (BD Bioscience, San Jose, CA, USA)
in the upper compartment were seeded with 0.4×105 or
0.8×105 cells. After 48 or 72 h, invaded cells on the
bottom surface were stained with 0.1% crystal violet (Sangon). The
cell migration assay was performed similarly, except that the cells
were applied to the uncoated filter.
Western blotting
Western blotting was used to detect the expression
of target genes at the protein level. Protein was extracted from
the transfected MHCCLM3 cells using modified RIPA buffer in the
presence of proteinase inhibitor cocktail. Equivalent quantities
(30–50 μg) of protein were separated on 10% SDS-polyacrylamide gels
and transferred to polyvinylidene difluoride membranes. The
membranes were blocked with 5% non-fat milk and then incubated
overnight at 4°C with the appropriate primary antibody at the
dilutions specified by the manufacturer. The membranes were then
washed three times in 10 ml TBST and incubated with the
corresponding horseradish peroxidase (HRP)-conjugated secondary
antibody at a 1:2,000 dilution for 1 h. The bound secondary
antibody was detected using an enhanced chemiluminescence (ECL)
system (Pierce Biotechnology Inc., Rockford, IL, USA). Primary
antibodies were as follows: MACC1 antibody (1:250) and anti-β-actin
antibody (1:1,000) (both from Sigma, St. Louis, MO, USA). β-actin
protein was used as the internal control.
miRNA target prediction
For the prediction of the target genes and the 3’UTR
binding sites by the seed region of miR-200a, the TargetScan
(http://www.targetscan.org/), miRanda
(http:/www.microrna.org/microrna/home.do) and PicTar
(http://pictar.mdc-berlin.de/) databases
were used. The related functions of the targets were also
considered.
3’UTR luciferase reporter plasmid
construction
The 500-bp 3’UTR of human MACC1 complementary DNA
was amplified by polymerase chain reaction with the following
primers: forward, 5′-CCGCTCGAGCACCAGTAAAACAAGGAACTTG-3′ and reverse
primer, 5′-GAATGCGGCCGCTTTACAGAAACAAATGCAATGTTAC-3′. Endonuclease
restriction sites were incorporated in the primers to facilitate
ligation into the luciferase reporter plasmid psiCHECK-2 vector
(Promega, Madison, WI, USA). A psiCHECK-2 construct containing
3’UTR of MACC1 with a mutant seed sequence of miR-200a was also
synthesized (Genepharma, Shanghai, China). All constructs were
verified by DNA sequencing. HEK293 cells were plated in 96-well
plates and then cotransfected with 100 ng constructs with or
without miR-200a. At 48 h after transfection, luciferase activity
was detected using a dual-luciferase reporter assay system
(Promega) and normalized to Renilla activity.
Statistical analysis
The results are expressed as mean ± standard
deviation (SD), as appropriate. Comparisons of continuous data were
analyzed by the Student t-test between two groups, whereas
categorical data were analyzed by the Chi-square test. Statistical
analyses were performed using SPSS for Windows v.16.0 (SPSS, Inc.,
Chicago, IL, USA) and GraphPad Prism 5.0 (GraphPad Software, La
Jolla, CA, USA). P<0.05 was considered to indicate a
statistically significant result.
Results
miR-200a is frequently downregulated in
hepatocellular carcinoma and HCC cell lines
We measured the expression of mature miR-200a in the
human liver tissues and the corresponding HCCs (n=115) using
quantitative RT-PCR (qRT-PCR) with the endogenous control (RNU6B).
We found that miR-200a was markedly decreased in the HCCs
(P<0.01; Fig. 1A). The overall
expression level of miR-200a was decreased ~2.36-fold in the HCC
samples. This result suggests that downregulation of miR-200a may
be a frequent event in HCC.
To corroborate these findings, in situ
hybridization analysis was performed in the HCCs and normal liver
tissues using 5′- and 3′-dig-labeled LNA probes. In situ
hybridization with LNA-modified anti-miR-200a probe also showed a
decrease in the HCC samples. No signal was detected with the
scrambled control demonstrating specificity of the probes (Fig. 1B).
We also investigated expression of miR-200a in 7 HCC
cell lines and the normal liver cell line LO2. Except for the PLC
cells, a low level of miR-200a was found in most of the HCC cell
lines when compared with the expression in the LO2 cells. Among
them, HCCLM3 and HepG2 cells were selected for subsequent
investigation (Fig. 1C).
miR-200a inhibits tumor cell growth and
metastasis in HCC
To investigate the effects of miR-200a on tumor
biology, HCCLM3 and HepG2 cells were then transfected with miR-200a
mimics and inhibitors or their respective controls and the
efficiency of miR-200a and anti-miR-200a was validated (Fig. 1D).
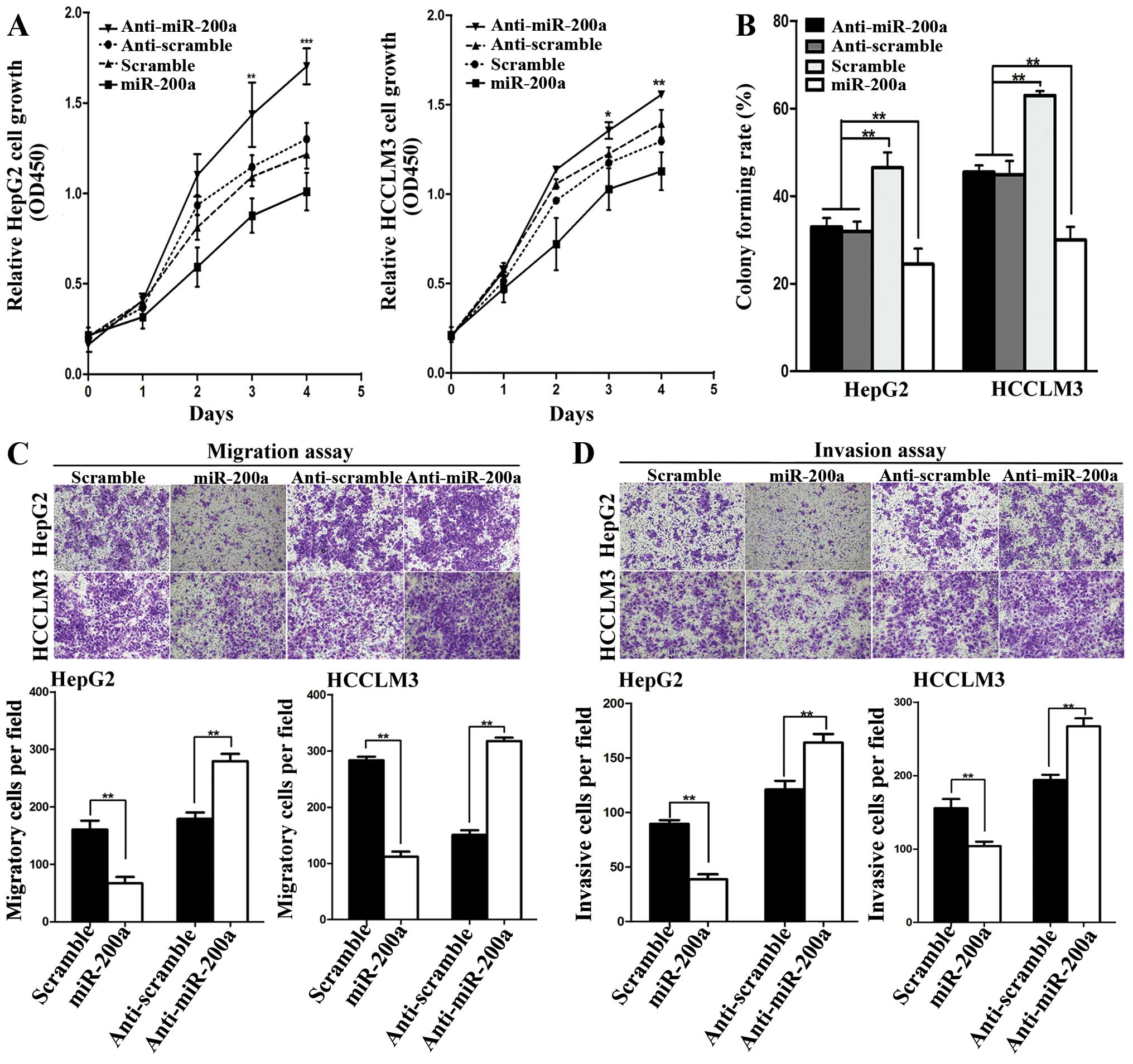
To clarify the effect of miR-200a on tumor growth,
cancer cell proliferation with CCK-8 further demonstrated that
ectopic miR-200a markedly inhibited the cell viability of the
HCCLM3 and HepG2 cells as compared with those transfected with the
scramble control (P<0.01; Fig.
2A). However, knockdown of miR-200a with miR-200a inhibitors
restored the malignant cell proliferation (P<0.01; Fig. 2A). In addition, ectopic miR-200a
expression significantly suppressed colony forming rates both in
the HCCLM3 and HepG2 cells. Conversely, colony forming rates were
increased by knocking down miR-200a expression (P<0.01; Fig. 2B). To elucidate the role of miR-200a
in HCC metastasis, the effects of miR-200a on the migration and
invasion of HCC cells were analyzed initially in vitro.
Transwell assays showed that both the migratory and invasive
abilities of the HCC cells were suppressed by miR-200a
overexpression and the metastatic potential was enhanced when
cellular miR-200a was neutralized by anti-miR-200a (P<0.01;
Fig. 2C and D). Collectively, the
in vitro studies suggest the suppressive effects of miR-200a
on tumor cell growth and metastasis in HCC.
miR-200a targets the 3’UTR of the MACC1
transcript and downregulates its expression
We then explored the molecular mechanism behind the
inhibitory function of miR-200a on HCC growth and metastasis.
Putative targets of miR-200a were predicted with TargetScan. Among
these, MACC1 was chosen for further validation. The sequence of the
predicted miR-200a binding sites and the MACC1 3’UTR segments
containing the miR-200a complementary sequence are shown (Fig. 3A). qRT-PCR and western blotting
further demonstrated that overexpression of miR-200a markedly
suppressed the endogenous mRNA and protein levels of MACC1
respectively, whereas inhibition of miR-200a increased the
expression of MACC1, in the HCCLM3 cells (P<0.05; Fig. 3B and C). The miR-200a or
anti-miR-200a constructs were then cotransfected with the
dual-luciferase reporter vector into HEK293 cells. A
dual-luciferase reporter assay revealed that the cotransfection of
miR-200a significantly inhibited the activity of the firefly
luciferase reporter with wild-type 3’UTR of MACC1, whereas this
effect was abrogated when the predicted 3’UTR binding site was
mutated (P<0.05; Fig. 3D). These
data indicate that miR-200a may negatively regulate MACC1
expression by directly targeting its 3’UTR.
Decreased miR-200a expression is
correlated with poor prognosis in HCC patients following liver
transplantation
To explore whether miR-200a may be a candidate
biomarker for predicting the clinical outcome of HCC patients
following LT, we examined the expression of miR-200a in 115 HCC
samples. Patients were segregated into high or low expression
groups on the basis of receiver operating characteristic analysis.
Following clinicopathological correlation analysis, the clinical
characteristics not directly correlated with the expression of
miR-200a included age, gender, tumor differentiation, vascular
invasion, preoperative α-fetoprotein (AFP) level, tumor number and
tumor size (Table I).
Univariate analysis revealed that vascular invasion,
preoperative serum AFP level (>400 ng/ml), multiple tumors and
tumor size (>5 cm) and the expression level of miR-200a
(miR-200ahigh) were predictors for overall survival (OS)
and cumulative recurrence (Table
II). HCC patients with low miR-200a expression had a
significantly worse prognosis than those with high expression of
miR-200a (Table II). The 1-, 3-
and 5-year cumulative recurrence rates of miR-200a-low HCC were
much higher than those of miR-200a-high HCC (P<0.001; Fig. 4A). The 1-, 3- and 5-year overall
survival rates were significantly lower for patients with
miR-200a-low HCC than for patients with miR-200a-high HCC (P=0.037;
Fig. 4B).
 | Table IImiR-200a expression is an independent
prognostic factor for HCC patients following LT. |
Table II
miR-200a expression is an independent
prognostic factor for HCC patients following LT.
| Cumulative
recurrence | Overall
survival |
|---|
|
|
|
|---|
| Variable | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Univariate
analysisa |
| Age (year) (≥50
vs. <50) | 0.546
(0.355–0.841) | 0.006 | 0.808
(0.521–1.253) | 0.341 |
| Gender (male vs.
female) | 2.161
(0.834–5.095) | 0.117 | 2.654
(0.970–7.261) | 0.057 |
| Preoperative
treatment (yes vs. no) | 1.196
(0.747–1.916) | 0.457 | 1.025
(0.617–1.703) | 0.924 |
| Tumor size (>5
vs. ≤5 cm) | 3.934
(2.527–6.123) | <0.001 | 3.070
(1.960–4.809) | <0.001 |
| Tumor number
(multiple vs. single) | 2.595
(1.637–4.114) | <0.001 | 2.356
(1.468–3.779) | <0.001 |
| Tumor
differentiation (poor vs. well + moderate) | 1.485
(0.947–2.265) | 0.066 | 1.043
(0.672–1.620) | 0.851 |
| Preoperative AFP,
ng/ml (>400 vs. ≤400) | 2.261
(1.440–3.548) | <0.001 | 1.820
(1.151–2.878) | 0.010 |
| Vascular invasion
(yes vs. none) | 2.995
(1.959–4.580) | <0.001 | 2.310
(1.491–3.579) | <0.001 |
|
miR-200ahigh vs.
miR-200alow | 1.858
(1.204–2.867) | 0.005 | 1.613
(1.035–2.514) | 0.035 |
| Multivariate
analysisa |
| Age (year) (≥50
vs. <50) | - | 0.153 | - | - |
| Tumor size (>5
vs. ≤5 cm) | 3.350
(2.123–5.288) | <0.001 | 2.719
(1.725–4.286) | <0.001 |
| Tumor number
(multiple vs. single) | - | 0.141 | - | 0.150 |
| Preoperative AFP,
ng/ml (>400 vs. ≤400) | 1.789
(1.130–2.833) | 0.013 | 1.624
(1.020–2.587) | 0.041 |
| Vascular invasion
(yes vs. none) | 2.267
(1.474–3.485) | <0.001 | 1.853
(1.192–2.880) | 0.006 |
|
miR-200ahigh vs.
miR-200alow | 3.444
(1.707–6.948) | 0.001 | 3.001
(1.509–5.970) | 0.002 |
Cox multivariate analysis (Table II) revealed that in addition to
vascular invasion, a high preoperative AFP level (>400 ng/ml)
and large tumor size (>5 cm), miR-200a overexpression in HCC was
an independent prognostic factor for predicting tumor recurrence
(P<0.001) and OS (P<0.001) in HCC patients following LT.
Discussion
The development of early diagnostic and effective
therapeutic strategies for HCC is an important issue for scientists
and clinicians. Currently, there has been limited information
regarding the differentiation states of HCC, although some
information concerning HCC may be obtainable from surgical
specimens. However, only 10–20% of HCC patients are suitable for
surgical operations, making it difficult to develop an effective
and comprehensive therapeutic strategy for all HCC patients. In
addition, a surgical operation is not indicated for treating HCC
patients with metastatic tumors. A new biomarker of HCC progression
may be helpful for early diagnosis or to determine a suitable
solution for HCC therapy. Recently, miRNAs have become useful as
prognostic and diagnostic clinical biomarkers. Determining whether
the expression of specific miRNAs is correlated with the
progressive state of HCC may be valuable for clinical
applications.
miRNAs may function as critical regulators in the
tumorigenesis of various human cancers, and loss of miR-200a
expression has been reported in breast cancer, ovarian cancer and
pancreatic cancer (21,23–27).
However, the expression pattern and its role in cell survival and
metastasis of primary hepatocytes and the underlying mechanism have
not been elucidated. In the present study, we identified that
miR-200a was markedly downregulated in HCC and exerted a
suppressive effect on tumor cell growth and metastasis. In
addition, we found that miR-200a suppressed tumor growth and
metastasis in HCC by directly targeting MACC1 through binding to a
specific complementary site within its 3’UTR.
The miR-200 family has been linked to tumor
progression and metastasis in several carcinomas. Members of the
miR-200 family are downregulated during progression and migration
of ovarian and breast cancer, as well as in bladder, pancreatic,
prostate and esophageal carcinoma. Furthermore, in several of these
entities, downregulation of miR-200 is associated with reduced
overall survival and poor response to chemotherapy (28–35).
In the present study, we found that HCC patients with low miR-200a
expression had significantly worse prognosis than those with high
expression of miR-200a.
In conclusion, our study revealed the inhibitory
effect of miR-200a on MACC1 in HCC and partly elucidated a
potential molecular mechanism by which miR-200a participates in
tumor aggressiveness. These findings suggest that miR-200a may be
recognized as a novel potential biomarker to predict the survival
of patients with HCCs following LT.
References
|
1
|
Shiraha H, Yamamoto K and Namba M: Human
hepatocyte carcinogenesis (Review). Int J Oncol. 42:1133–1138.
2013.PubMed/NCBI
|
|
2
|
Finn RS: Emerging targeted strategies in
advanced hepatocellular carcinoma. Semin Liver Dis. 33(Suppl 1):
S11–S19. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Whittaker S, Marais R and Zhu AX: The role
of signaling pathways in the development and treatment of
hepatocellular carcinoma. Oncogene. 29:4989–5005. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Carr BI: Some new approaches to the
management of hepatocellular carcinoma. Semin Oncol. 39:369–373.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hamed O, Kimchi ET, Sehmbey M, Gusani NJ,
Kaifi JT and Staveley-O’Carroll K: Impact of genetic targets on
cancer therapy: hepatocellular cancer. Adv Exp Med Biol. 779:67–90.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Marrero JA: Multidisciplinary management
of hepatocellular carcinoma: where are we today? Semin Liver Dis.
33(Suppl 1): S3–S10. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Alves RC, Alves D, Guz B, Matos C, Viana
M, Harriz M, Terrabuio D, Kondo M, Gampel O and Polletti P:
Advanced hepatocellular carcinoma. Review of targeted molecular
drugs. Ann Hepatol. 10:21–27. 2011.PubMed/NCBI
|
|
8
|
Herszényi L and Tulassay Z: Epidemiology
of gastrointestinal and liver tumors. Eur Rev Med Pharmacol Sci.
14:249–258. 2010.PubMed/NCBI
|
|
9
|
Hussain SA, Ferry DR, El-Gazzaz G, Mirza
DF, James ND, McMaster P and Kerr DJ: Hepatocellular carcinoma. Ann
Oncol. 12:161–172. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tanaka M, Katayama F, Kato H, Tanaka H,
Wang J, Qiao YL and Inoue M: Hepatitis B and C virus infection and
hepatocellular carcinoma in China: a review of epidemiology and
control measures. J Epidemiol. 21:401–416. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
El-Serag HB: Hepatocellular carcinoma: an
epidemiologic view. J Clin Gastroenterol. 35(5 Suppl 2): S72–S78.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lai EC and Lau WY: The continuing
challenge of hepatic cancer in Asia. Surgeon. 3:210–215. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yuen MF, Hou JL and Chutaputti A; Asia
Pacific Working Party on Prevention of Hepatocellular Carcinoma.
Hepatocellular carcinoma in the Asia pacific region. J
Gastroenterol Hepatol. 24:346–353. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Grossman EJ and Millis JM: Liver
transplantation for non-hepatocellular carcinoma malignancy:
Indications, limitations, and analysis of the current literature.
Liver Transpl. 16:930–942. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kaido T, Mori A, Ogura Y, Hata K,
Yoshizawa A, Iida T and Uemoto S: Living donor liver
transplantation for recurrent hepatocellular carcinoma after liver
resection. Surgery. 151:55–60. 2012. View Article : Google Scholar
|
|
16
|
Ambros V, Bartel B, Bartel DP, Burge CB,
Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths-Jones S,
Marshall M, Matzke M, Ruvkun G and Tuschl T: A uniform system for
microRNA annotation. RNA. 9:277–279. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Iorio MV and Croce CM: microRNA
involvement in human cancer. Carcinogenesis. 33:1126–1133. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Schetter AJ and Harris CC: Alterations of
microRNAs contribute to colon carcinogenesis. Semin Oncol.
38:734–742. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen Y, Zhang L and Hao Q: Candidate
microRNA biomarkers in human epithelial ovarian cancer: systematic
review profiling studies and experimental validation. Cancer Cell
Int. 13:862013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Feng B, Wang R and Chen LB: Review of
miR-200b and cancer chemosensitivity. Biomed Pharmacother.
66:397–402. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yao J, Zhou E, Wang Y, Xu F, Zhang D and
Zhong D: microRNA-200a inhibits cell proliferation by targeting
mitochondrial transcription factor A in breast cancer. DNA Cell
Biol. 33:291–300. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cong N, Du P, Zhang A, Shen F, Su J, Pu P,
Wang T, Zjang J, Kang C and Zhang Q: Downregulated microRNA-200a
promotes EMT and tumor growth through the wnt/β-catenin pathway by
targeting the E-cadherin repressors ZEB1/ZEB2 in gastric
adenocarcinoma. Oncol Rep. 29:1579–1587. 2013.PubMed/NCBI
|
|
23
|
Kobayashi M, Salomon C, Tapia J, Illanes
SE, Mitchell MD and Rice GE: Ovarian cancer cell invasiveness is
associated with discordant exosomal sequestration of Let-7 miRNA
and miR-200. J Transl Med. 12:42014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lam SS, Mak AS, Yam JW, Cheung AN, Ngan HY
and Wong AS: Targeting estrogen-related receptor alpha inhibits
epithelial-to-mesenchymal transition and stem cell properties of
ovarian cancer cells. Mol Ther. 22:743–751. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu L, Zou J, Wang Q, Yin FQ, Zhang W and
Li L: Novel microRNAs expression of patients with chemotherapy
drug-resistant and chemotherapy-sensitive epithelial ovarian
cancer. Tumour Biol. 35:7713–7717. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lu Y, Lu J, Li X, Zhu H, Fan X, Zhu S,
Wang Y, Guo Q, Wang L, Huang Y, Zhu M and Wang Z: MiR-200a inhibits
epithelial-mesenchymal transition of pancreatic cancer stem cell.
BMC Cancer. 14:852014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sun Q, Zou X, Zhang T, Shen J, Yin Y and
Xiang J: The role of miR-200a in vasculogenic mimicry and its
clinical significance in ovarian cancer. Gynecol Oncol.
132:730–738. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Banyard J, Chung I, Wilson AM, Vetter G,
Le Béchec A, Bielenberg DR and Zetter BR: Regulation of epithelial
plasticity by miR-424 and miR-200 in a new prostate cancer
metastasis model. Sci Rep. 3:31512013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cao Q, Lu K, Dai S, Hu Y and Fan W:
Clinicopathological and prognostic implications of the miR-200
family in patients with epithelial ovarian cancer. Int J Clin Exp
Pathol. 7:2392–2401. 2014.PubMed/NCBI
|
|
30
|
Kolesnikoff N, Attema JL, Roslan S, Bert
AG, Schwarz QP, Gregory PA and Goodall GJ: Specificity protein 1
(Sp1) maintains basal epithelial expression of the miR-200 family:
implications for epithelial-mesenchymal transition. J Biol Chem.
289:11194–11205. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Koutsaki M, Spandidos DA and Zaravinos A:
Epithelial-mesenchymal transition-associated miRNAs in ovarian
carcinoma, with highlight on the miR-200 family: prognostic value
and prospective role in ovarian cancer therapeutics. Cancer Lett.
351:173–181. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Song F1, Yang D, Liu B, Guo Y, Zheng H, Li
L, Wang T, Yu J, Zhao Y, Niu R, Liang H, Winkler H, Zhang W, Hao X
and Chen K: Integrated microRNA network analyses identify a
poor-prognosis subtype of gastric cancer characterized by the
miR-200 family. Clin Cancer Res. 20:878–889. 2014. View Article : Google Scholar
|
|
33
|
Truong HH, Xiong J, Ghotra VP, Nirmala E,
Haazen L, Le Dévédec SE, Balcioğlu HE, He S, Snaar-Jagalska BE,
Vreugdenhil E, Meerman JH, van de Water B and Danen EH: β1 integrin
inhibition elicits a prometastatic switch through the
TGFβ-miR-200-ZEB network in E-cadherin-positive triple-negative
breast cancer. Sci Signal. 7:ra152014. View Article : Google Scholar
|
|
34
|
Wang CH, Chen CL, More SV, Hsiao PW, Hung
WC and Li WS: The tetraindole SK228 reverses the
epithelial-to-mesenchymal transition of breast cancer cells by
up-regulating members of the miR-200 family. PLoS One.
9:e1010882014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang CM, Liu R, Wang L, Nascimento L,
Brennan VC and Yang WH: SUMOylation of FOXM1B alters its
transcriptional activity on regulation of miR-200 family and JNK1
in MCF7 human breast cancer cells. Int J Mol Sci. 15:10233–10251.
2014. View Article : Google Scholar : PubMed/NCBI
|