Introduction
Hepatocellular carcinoma (HCC) is one of the most
common malignancies worldwide and a common cause of cancer-related
death globally (1). Unfortunately,
the prognosis, early diagnosis and treatment of HCC remain poor.
Few patients diagnosed with HCC are eligible for curative
therapies, including surgical resection, percutaneous ablation and
liver transplantation. Most patients are diagnosed at a late stage
of the disease when potentially curative therapies are least
effective (2,3). Therefore, new therapies are urgently
needed for these patients.
Sorafenib, an oral multikinase inhibitor of several
tyrosine protein kinases and Raf kinases, induces apoptosis in
human leukemia and other malignant cells, and is currently the only
drug categorized as a preferred treatment for advanced primary HCC
(4,5). Investigators have reported that
sorafenib inhibits tumor cell proliferation and promotes apoptosis
by inhibiting several kinases in the mitogen-activated protein
kinase (MAPK) pathway and the translation of pro-survival factors,
such as Mcl-1 and Bcl-2 (6–8). Clinical studies have confirmed the
efficacy of sorafenib during HCC treatment; however, the drug has
several side-effects, and resistance to sorafenib is increasing.
Therefore, it is necessary to clarify the mechanisms of sorafenib
chemoresistance and to identify other therapeutic biomarkers that
might be combined with sorafenib to improve its efficacy.
Cancer cells commonly encounter endoplasmic
reticulum (ER) stress during tumor initiation and progression due
to hypoxia, glucose deprivation, DNA damage or ER calcium depletion
(9). The unfolded protein response
(UPR) activated during ER stress is a compensatory cellular defense
mechanism and an adaptive response that allows tumor cells to
survive under these pathophysiological conditions (10). The UPR in response to
chemotherapeutics is necessary for tumor cells to maintain
malignancy and therapy resistance (11). Identification of the UPR components
that are activated or suppressed in tumors and investigation of the
potential of cancer therapeutics by targeting the UPR may be
potentially useful to inhibit tumor growth and improve cancer
therapy (12,13). IRE1 is the only identified ER stress
sensor in yeast and is essential for the UPR in animals and plants
(14). As an ER transmembrane
protein, IRE1 monitors ER homeostasis through an ER luminal
stress-sensing domain and triggers the UPR through a cytoplasmic
kinase domain and an RNase domain (15). IRE1 activation and attenuation play
a critical role in cell fate decisions during ER stress responses
(16,17). Upon ER stress, IRE1 RNase is
activated through a conformational change, autophosphorylation and
higher-order oligomerization. Activated IRE1 subsequently splices a
26-nucleotide sequence from the mRNA encoding X-box binding protein
1 (XBP1), resulting in the translation of activated/spliced XBP1
(XBP1s). XBP1s is a potent transcription factor and one of the key
regulators of the UPR; it transduces the UPR to the nucleus and
regulates many target genes, including ER chaperones, ER-associated
degradation (ERAD) components and other transcription factors
(18,19). Recent studies have indicated that
XBP1 mRNA splicing activity does not increase progressively under
increasing ER stress intensity and/or duration, and is only
activated during the adaptive/pro-survival phase of ER stress
(20). The IRE1/XBP1 pathway is
important for tumor survival under ER stress (21). The overexpression of XBP1 was found
in various solid tumors, including breast cancer and HCC.
Additionally, prolonged IRE1 signal activation was shown to enhance
cell survival (22). Taken
together, these results suggest that the IRE1/XBP1 axis may play an
important role in the response of tumors to chemotherapeutics and
serve as a key regulator of cell fate.
RACK1 was originally identified as an intercellular
scaffold protein of the Trp-Asp (WD) repeat protein family
(23). RACK1 can recruit and bind
many kinases and receptors, such as PKC and DJ-1, thereby impacting
a wide range of signal transduction pathways (24,25).
RACK1 plays an important role in multiple cellular functions,
including cell growth, migration and differentiation (26,27).
Studies have found that RACK1 is upregulated in several tumor
types, such as breast tumors and HCC (28,29).
Moreover, RACK1 might function as an internal factor that
contributes to stress-mediated chemotherapy resistance (30). However, the regulatory mechanism of
RACK1 in the stress response has remained largely elusive.
A study by Qiu et al (31) showed that RACK1 interacts with IRE1
in a glucose-stimulated or ER stress-responsive manner. This
finding demonstrated that RACK1 is required for IRE1 activation
under ER stress conditions in pancreatic β-cells. However, the
interaction of RACK1 with IRE1 and the role of RACK1 in the IRE1
signaling pathway have not been evaluated in HCC. In the present
study, we showed that RACK1 is overexpressed in both HCC tissue
samples and cell lines, and RACK1 is required for IRE1 signaling
activation under ER stress conditions. This interaction could
contribute to the ability of HCC cells to adapt to stressful
conditions. Our findings provide a new strategy to promote the
chemotherapeutic effects of sorafenib by targeting the IRE1/XBP1
axis.
Materials and methods
Clinical samples
HCC and adjacent non-tumor (NT) tissue samples were
obtained by surgical resection from 46 patients at our hospital
between June 2011 and April 2013 (see detailed clinicopathological
features in Table I). The HCC and
NT tissues were verified by the classification of the General Rules
for the Clinical and Pathological Study of Primary Liver Cancer
(32). Resected tissues were frozen
immediately at −80°C or fixed in 10% formalin. Informed consent was
obtained from all patients or their guardians for subsequent use of
their resected tissues, and the Ethics Committee of Xijing
Hospital, The Fourth Military Medical University, approved all
aspects of the present study.
 | Table ISummary of the clinicopathological
variables. |
Table I
Summary of the clinicopathological
variables.
|
Characteristics | Data |
|---|
| Patients | 46 |
| Gender | |
| Male | 40 |
| Female | 6 |
| Age (years), range;
median | 37–71; 49.7 |
| Tumor size (cm),
range; median | 0.6–15; 5.2 |
| Clinical stage,
n | |
| I | 2 |
| II | 14 |
| III | 30 |
| IV | 0 |
Plasmid construction and
small-interfering RNAs (siRNAs)
To construct pCDNA3.1(+)-RACK1, the coding sequence
of human RACK1 was amplified by PCR and digested with
HindIII and EcoRI. The RACK1 cDNA was ligated into a
similarly digested pCDNA3.1(+) vector. IRE1 siRNA (sc-40705), RACK1
siRNA (sc-36354) and the non-targeting control siRNA (sc-37007)
were purchased from Santa Cruz Biotechnology (Santa Cruz, CA,
USA).
Cell culture and treatment
The human HCC cell lines HepG2, SMCC7721 and MHCC97
were maintained in our laboratory and cultured in Dulbecco’s
modified Eagle’s medium (DMEM) (HyClone, Logan, UT, USA)
supplemented with 10% heat-inactivated fetal calf serum (Gibco,
Grand Island, NY, USA), 100 U/ml penicillin G and 100 μg/ml
streptomycin at 37°C in a humidified atmosphere containing 5%
CO2. Aliquots of HepG2 or MHCC97 cells were transfected
with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) premixed
with siRNA or a scrambled sequence, following the manufacturer’s
instructions. Successfully transfected cells were obtained after 48
h and analyzed via western blotting.
Protein extraction and western blot
analysis
Tissues and cells were lysed in RIPA lysis buffer
(20 mM Tris-HCl, pH 7.5, 100 mM KCl, 0.1% Nonidet P-40, 1 mM EDTA
and 10% glycerol) containing 1 mM PMSF, 1% protease inhibitor
cocktail (Sigma, St. Louis, MO, USA) and 1% phosphatase inhibitor
cocktail I/II (Sigma) for 20 min at 4°C. Then, the total protein in
the lysates was determined using a bicinchoninic acid (BCA) assay.
Equal amounts of samples were mixed with Laemmli sample buffer and
placed in a boiling waterbath for 5 min. Proteins were then
separated by 10% sodium dodecyl sulfate-polyacrylamide gel
electrophoresis (SDS-PAGE) and transferred to nitrocellulose
membranes (Amersham Biosciences, Uppsala, Sweden). Then, the
membranes were incubated with specific primary antibodies for GRP78
(Bip), IRE1, pT-724 IRE1, RACK1 and XBP1s (Abcam, Cambridge, MA,
USA) overnight at 4°C. Incubation with horseradish peroxidase
(HRP)-coupled secondary antibodies (ProteinTech Group, Chicago, IL,
USA) was performed for 1 h at room temperature. Signals were
detected using enhanced chemiluminescence (Thermo Fisher Scientific
Inc., Rockford, IL, USA) and scanned with a gel imaging system
(Bio-Rad Laboratories, Berkeley, CA, USA). The monoclonal
antibodies anti-GAPDH (Kangcheng, Shanghai, China) or anti-β-actin
(Sigma) were used as controls.
Immunohistochemistry
Immunohistochemical staining for RACK1 was performed
on formalin-fixed, paraffin-embedded specimens using the
streptavidin-peroxidase (SP) method. Histological slides, 4-mm in
thickness, were baked in an oven at 60°C for 30 min, followed by
deparaffinization and rehydration using a graded series of xylene
and ethanol. The slides were then heated in 0.01 M citrate buffer
for 20 min in a microwave oven, and endogenous peroxidase was
blocked with hydrogen peroxide containing 0.3% methanol for 15 min
at room temperature. Subsequently, the sections were placed in a
humidified chamber and incubated for 30 min with 10% goat serum to
block non-specific binding. Afterward, the sections were incubated
overnight at 4°C with a monoclonal anti-RACK1 antibody at a
dilution of 1:100 according to the manufacturer’s specifications.
Primary antibodies were detected using a biotinylated anti-goat IgG
antibody diluted 1:200 followed by SP for 30 min at room
temperature. Afterward, the immunoreaction was visualized using the
Liquid DAB-Plus substrate kit (Zymed Laboratories Inc., San
Francisco, CA, USA) according to the manufacturer’s specifications.
Finally, the sections were rinsed for 10 min in running tap water,
counterstained with hematoxylin staining solution for 5 min,
dehydrated and mounted.
Co-immunoprecipitation
For co-immunoprecipitation analysis, cells were
lysed with lysis buffer, and then the lysates were clarified by
centrifugation at 12,000 rpm for 10 min at 4°C. The supernatants
were mixed with the desired primary antibody overnight at 4°C.
After centrifugation, the immune complexes were captured by mixing
with anti-Flag M2 agarose (Sigma) plus protein G-Sepharose (GE
Healthcare, Buckinghamshire, UK) for 2 h at 4°C in a rotator. When
using the anti-RACK1 antibody in the co-immunoprecipitation assays,
rabbit anti-mouse IgM was included as the bridging antibody. The
beads were subsequently washed three times with washing buffer,
eluted by boiling in 2X SDS loading buffer, and analyzed by
SDS-PAGE and immunoblotting.
Immunofluorescence microscopy
HepG2 and MHCC97 cells on chamber slides were
treated with 10 μg/ml tunicamycin (TM) for 4 h. The slides
were washed twice with PBS. The cells were fixed for 5 min in 4%
paraformaldehyde and permeabilized for 2 min with 0.1% Triton X-100
in PBS. After blocking the cells with 5% donkey serum albumin, the
cells were incubated with a rabbit monoclonal RACK1 antibody at
room temperature for 1 h and washed three times with PBS. Donkey
anti-rabbit secondary antibodies were incubated with the cells for
45 min, and the cells were washed three times with PBS.
Immunofluorescence staining was examined under confocal microscopy
(LSM 700; Zeiss), and images were processed using Volocity software
(Perkin-Elmer, Waltham, MA, USA).
Quantitative real-time PCR analysis
Total RNA was extracted from frozen tissues and cell
lines with TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according
to the manufacturer’s instructions. After reverse transcription by
M-MLV reverse transcriptase (Takara, Shiga, Japan), quantitative
real-time PCR (qRT-PCR) was performed using the SYBR Premix Ex Taq
(Takara) in the StepOne real-time PCR system (Applied Biosystems,
Foster City, CA, USA). qRT-PCR was performed at 95°C for an initial
5 min followed by 30 cycles of denaturation at 95°C for 30 sec,
annealing at 55°C for 30 sec and extension at 72°C for 7 min.
Primer pairs for the target genes were as follows: RACK1, forward
5′-TTCTCCTCTGA CAACCGGCA-3′ and reverse 5′-GCCATCCTTGCCTCCA GAA-3′;
XBP1s, forward 5′-TGCTGAGTCCGCAGCAGG TG-3′ and reverse
5′-GCTGGCAGGCTCTGGGGAAG-3′; and GAPDH, forward
5′-CGGAGTCAACGGATTTGGTC GTAT-3′ and reverse
5′-AGCCTTCTCCATGGTGGTGAAG AC-3′. Data analysis was performed using
the ∆∆Ct method. Expression of the genes of interest was normalized
to the expression of GAPDH.
Evaluation of cell apoptosis
The number of apoptotic cells was determined by
Annexin V-FITC and PI staining. Briefly, the cells were collected,
washed with PBS and incubated with Annexin V-FITC and PI according
to the manufacturer’s instructions (BD Biosciences, San Jose, CA,
USA). Stained cells were analyzed using flow cytometry, and the
assays were performed in triplicate.
Statistical analysis
The data are expressed as the mean ± SD and were
subjected to statistical analysis using SPSS version 17.0 (SPSS
Inc., Chicago, IL, USA). Differences between groups were evaluated
using a two-tailed Student’s t-test or one-way ANOVA, and P<0.05
was considered to indicate a statistically significant result.
Results
Activation of UPR and high expression of
RACK1 are involved in HCC
Activated UPR and upregulated ER chaperones, such as
IRE1 and GRP78, are found in a variety of cancer cells. Therefore,
we first examined the expression of IRE1 and GRP78 via western
blotting. As shown in Fig. 1A, the
expression of IRE1 and GRP78 in the HCC cell lines was much higher
than that in the human hepatic HL-7702 cell line. In addition, the
expression of pIRE1 was higher in the HCC samples compared with
that in their matched NT tissues (Fig.
1B). To determine whether RACK1 is involved in HCC, the
relative mRNA and protein expression of RACK1 was analyzed using
qRT-PCR and western blotting. As shown in Fig. 1A and B, the HCC cell lines showed
high expression of RACK1 at both the mRNA and protein levels.
Furthermore, the RACK1 protein level in the clinical HCC tissues
was evaluated using immunohistochemistry. The expression of RACK1
in the clinical HCC tissues was higher than that in the matched
peritumoral liver tissues (Fig.
1C). Moreover, RACK1 expression was well correlated with the
clinical stage of HCC (Table II).
These results indicate that IRE1 and RACK1 are frequently
upregulated in HCC, suggesting that activated UPR and highly
expressed RACK1 may contribute to the tumori-genesis and
progression during HCC.
 | Table IICorrelation of RACK1 expression in
HCC tissues with clinicopathological features of the 46 HCC
patients. |
Table II
Correlation of RACK1 expression in
HCC tissues with clinicopathological features of the 46 HCC
patients.
| Features | Intensity of
staining
| P-value |
|---|
| N | 1 | 2 | 3 |
|---|
| Age (years) | | | | | 0.668 |
| ≤60 | 35 | 9 | 15 | 11 | |
| >60 | 11 | 3 | 3 | 5 | |
| Gender | | | | | 1.000 |
| Male | 40 | 8 | 18 | 14 | |
| Female | 6 | 1 | 3 | 2 | |
| Tumor size
(cm) | | | | | 0.008 |
| ≤5 | 24 | 7 | 12 | 5 | |
| >5 | 22 | 3 | 4 | 15 | |
| Clinical stage | | | | | 0.028 |
| I-II | 16 | 5 | 5 | 6 | |
| III-IV | 30 | 3 | 6 | 21 | |
Sorafenib induces ER stress-related
apoptosis in HCC
We treated human hepatoma HepG2 cells with sorafenib
at varying doses. The induction of apoptosis was evaluated using
flow cytometry. To determine the activation status of IRE1 after
treatment with sorafenib, the protein expression of pIRE1 and XBP1s
was measured via western blotting. Our results showed that exposure
to 2.5, 5, 10, 15 and 20 μM sorafenib for 24 h increased
cellular apoptosis in a dose-dependent manner compared with the
levels observed in the standard non-stimulated cells (Fig. 2A). Additionally, sorafenib induced
the upregulation of pIRE1 and XBP1s in hepatoma cells (Fig. 2B). Notably, the expression of pIRE1
and XBP1s did not increase progressively. Following low-dose
(<10 μM) treatment with sorafenib, the expression of
pIRE1 and XBP1s increased progressively until reaching a peak and
then declined after high-dose or prolonged treatment with
sorafenib. IRE1 signaling activity is a key step in determining
cell survival after ER stress. To understand the role of IRE1
signaling in sorafenib-induced apoptosis, HepG2 and MHCC97 cells
were transfected with siIRE1 and then exposed to sorafenib. The
flow cytometric analysis of apoptosis revealed that IRE1
siRNA-transfected cells exhibited significantly increased cell
death compared with the control cells (Fig. 2C). These results indicate that
sorafenib induces ER stress-related apoptosis in HCC, and
activation of the IRE1/XBP1 signaling pathway may play a protective
role when HCC cells encounter ER stress in response to
sorafenib.
RACK1 interacts with IRE1 in
sorafenib-stimulated HCC cells
Previous studies identified an interaction between
IRE1 and RACK1 in human embryonic kidney (HEK) 293T cells (31). To determine the interaction of IRE1
and RACK1 under physiological or ER stress conditions, we performed
immunoprecipitation in HepG2 cells treated with TM or sorafenib. TM
is a well-known ER stressor that acts by inhibiting protein N
glycosylation (33). As shown in
Fig. 3A, an increased level of
RACK1 was found in the IRE1 immunoprecipitates from the TM-or
sorafenib-treated cells compared with the level observed in the
control cells. To visualize whether IRE1 and RACK1 co-localize in
the cytoplasm, we examined the subcellular localization of IRE1
relative to RACK1 in the TM-or sorafenib-treated cells using double
immunofluorescence staining. As shown in Fig. 3B, IRE1 aggregated and co-localized
with RACK1 in the cytoplasm after treatment with TM and sorafenib.
In contrast, the co-localization of IRE1 aggregates with RACK1 was
seldom found in the control cells.
RACK1 is essential for the activation of
IRE1 signaling
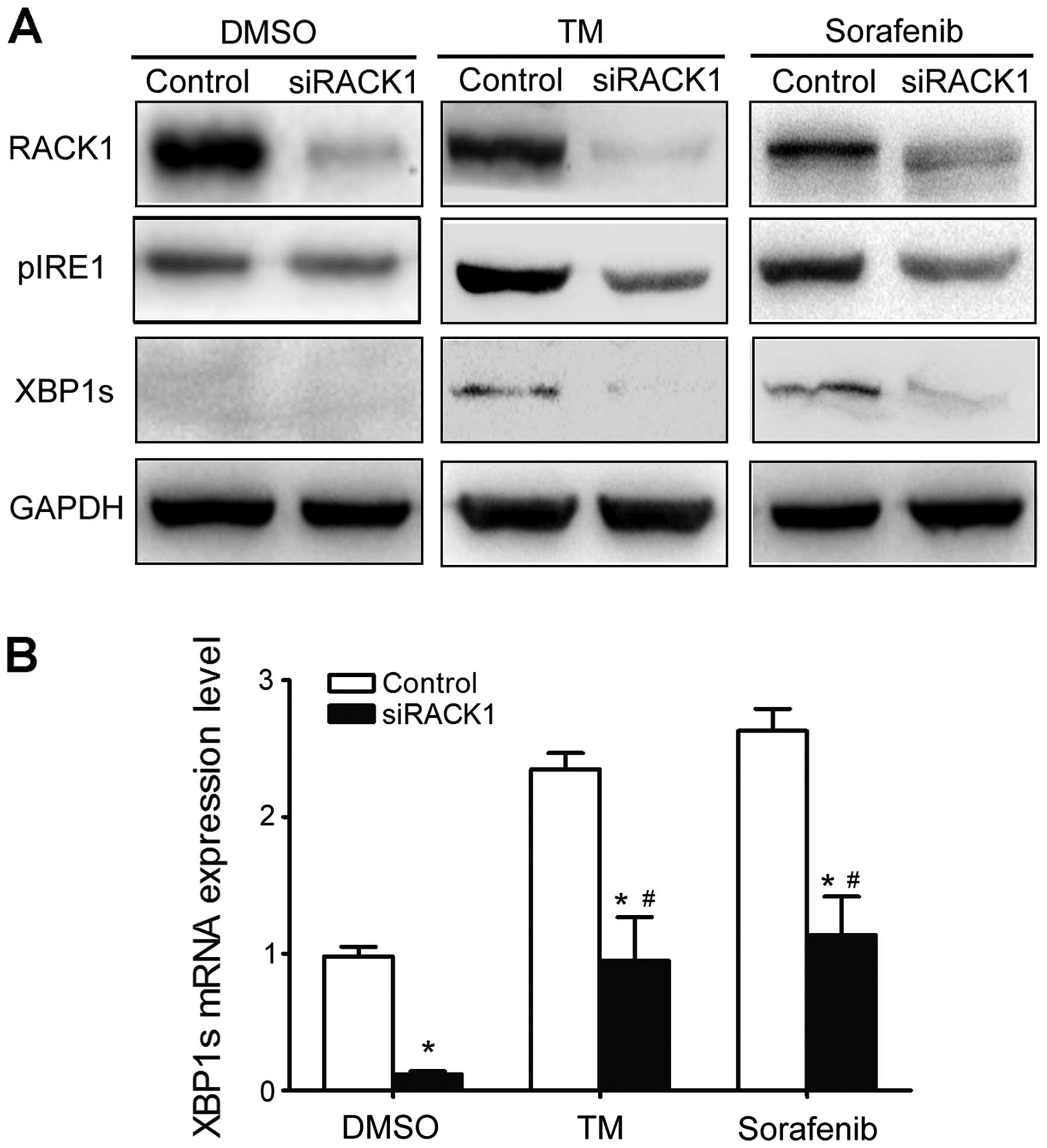
To investigate whether the RACK1/IRE1 interaction
affects activation of the UPR, RACK1 protein expression was knocked
down with siRNA specific for RACK1. Western blot analysis revealed
that siRACK1 significantly suppressed the basal level of RACK1.
Knockdown of RACK1 suppressed the phosphorylation of IRE1 in
response to TM or sorafenib compared with that of cells transfected
with a scrambled negative control (Fig.
4A). Moreover, reduced pIRE1 expression under the condition of
RACK1 knockdown was associated with decreased XBP1 mRNA splicing
and XBP1s expression (Fig. 4B).
These results indicate that RACK1 may exert distinct actions in the
regulation of IRE1 signaling in response to ER stress.
RACK1 modulates sorafenib-induced
apoptosis of HCC cells
Several reports have implicated IRE1 in pathways
associated with apoptosis resistance. Since RACK1 is required for
the activation of IRE1 signaling under ER stress, it appeared
likely that RACK1 was also essential for the protective effect of
the UPR. To test this hypothesis, HepG2 and MHCC97 cells were
transfected with pGFP-RACK1 or si-RACK1 and then exposed to
sorafenib. The apoptotic effects of sorafenib were determined using
flow cytometric assays. The results demonstrated that the
overexpression of RACK1 protected HCC cells from sorafenib-induced
apoptosis (Fig. 5A, left
histograms). In contrast, we observed that sorafenib induced
significant apoptosis in HCC cells transfected with si-RACK1
(Fig. 5B, left histograms). As XBP1
splicing is characteristic of UPR activation, we next evaluated the
levels of the spliced form of XBP1 using qRT-PCR. At the mRNA
level, XBP1s was significantly upregulated in RACK1
gain-of-function cells (Fig. 5A,
right histograms). However, XBP1s was reduced in the RACK1
knockdown cells (Fig. 5B, right
histograms). These findings indicate that RACK1 is involved in the
protective effect of UPR against sorafenib-induced apoptosis.
Discussion
Sorafenib is one of the preferred systemic therapies
in the treatment of advanced HCC. However, little is known
concerning the mechanism by which sorafenib induces cell death and
the drug resistance gained in response to HCC treatment. Activation
of the UPR by the IRE1 signaling pathway is a key component of cell
survival decisions in response to ER stress (34). We observed the activation of IRE1 in
the sorafenib-treated HCC cells, thereby indicating that sorafenib
induces the UPR and ER stress-related apoptosis. The combination of
sorafenib and an agent that targets the UPR is a promising
therapeutic strategy for the treatment of HCC.
RACK1, a multifaceted scaffolding protein,
participates in various signaling pathways and plays an important
role in multiple cellular functions. RACK1 has been associated with
the migration and proliferation of various types of tumors,
including HCC. Consistent with recent reports, our findings showed
that RACK1 is highly expressed in HCC tissues and cell lines,
indicating that RACK1 may be involved in the tumorigenesis and
prognosis of HCC. We discovered that the levels of RACK1 and IRE1
were markedly increased in the sorafenib-treated HCC cells, and we
also indicated that RACK1 interacts with IRE1 intercellularly and
that the two are co-localized in the cytoplasm by immunoprecipation
and immunofluorescence. Knockdown of RACK1 expression severely
repressed the phosphorylation of IRE1 in response to TM or
sorafenib and led to inhibition of the UPR. These results raise the
possibility that RACK1 and IRE1 work as a complex to mediate
activation of the IRE1 signaling pathway during sorafenib-induced
UPR, thereby modulating the ER stress-induced apoptosis of HCC
cells after exposure to sorafenib.
Activation of the UPR is an adaptive response that
allows cells to survive prolonged ER stress conditions in various
solid tumors (35). The IRE1
signaling pathway functions as a switch between cytoprotective and
proapoptotic outcomes in response to ER stress, and the activation
of IRE1 plays a crucial role in the UPR to enhance cell survival
(34). Our results indicate that
sorafenib treatment triggers the UPR and ER stress-related
apoptosis in HCC cells, given the essential role of IRE1 activation
in governing the switch from adaptive UPR to ER stress-associated
apoptosis in sorafenib-treated HCC cells. It is important to
understand how RACK1 affects the actions of other IRE1 interactors,
such as XBP1, CHOP and Bcl-2. Activated IRE1 can function as a
transmembrane kinase to control the splicing of XBP1 mRNA. The
activation of IRE1 in various temporal patterns during ER stress
influences the cell’s ultimate fate, whereas XBP1 mRNA splicing is
only activated during the adaptive phase of ER stress. Previous
reports indicate that XBP1 expression is increased in certain solid
tumors in association with chemotherapy resistance (36,37).
In the present study, we found that sorafenib was able to stimulate
the UPR in HCC cells. A low-dose and short treatment with sorafenib
was able to enhance the expression of IRE1 and XBP1 mRNA splicing,
which may play a protective role and improve cell survival. We also
observed that when cells encounter severe or prolonged stress, IRE1
becomes constitutively active while XBP1 mRNA splicing is
downregulated. When the expression of IRE1 was knocked down using
siIRE1, XBP1 mRNA splicing decreased, and IRE1 siRNA-transfected
cells exhibited a significant increase in cell death following
sorafenib treatment. These results indicate that activation of the
UPR protects cells from ER stress induced by sorafenib, and
targeting the IRE1/XBP1 axis is a promising antitumor strategy.
Recent studies (27,29)
have demonstrated that RACK1 plays a context-dependent role in
tumorigenesis and is responsible for stress-mediated chemotherapy
resistance. Our results demonstrated that RACK1 modulated
sorafenib-induced apoptosis by regulating IRE1 activity, and the
overexpression of RACK1 promoted the phosphorylation of IRE1 and
inhibited apoptosis induced by sorafenib. In contrast, knockdown of
RACK1 significantly enhanced the lethality of sorafenib to HCC
cells. These results raise the possibility that RACK1 forms a
complex with IRE1 following treatment with sorafenib, and may
influence the activation state of the IRE1/XBP1 axis, thereby
alleviating the ER stress induced by sorafenib. Thus, we
hypothesize that RACK1 may be a potential molecular targeted cancer
treatment.
Taken together, the present study confirmed that
activation of the IRE1 signaling pathway is involved in
sorafenib-induced UPR. Our data raise the possibility that the
RACK1/IRE1 complex might contribute to activation of the UPR in HCC
cells. Targeting RACK1 is a potential adjuvant therapeutic strategy
in combination with sorafenib for advanced HCC.
Acknowledgments
The present study is supported by grants from the
Natural Science Foundation of China (nos. 81070363, 81030010 and
81101818).
Abbreviations:
|
HCC
|
hepatocellular carcinoma
|
|
ER
|
endoplasmic reticulum
|
|
UPR
|
unfolded protein response
|
|
XBP-1
|
X-box binding protein 1
|
|
TM
|
tunicamycin
|
|
ERAD
|
ER associated degradation
|
|
GRP78
|
78-kDa glucose-regulated protein
|
|
IRE1
|
inositol-requiring enzyme 1
|
|
RACK1
|
receptor for activated C kinase 1
|
References
|
1
|
Ferlay J; ISME: GLOBOCAN 2012: Estimated
Cancer Incidence, Mortality and Prevalence Worldwide in 2012.
http://globocan.iarc.fr/Default.aspx.
IARC CancerBase; 2013
|
|
2
|
Flores A and Marrero JA: Emerging trends
in hepatocellular carcinoma: Focus on diagnosis and therapeutics.
Clin Med Insights Oncol. 8:71–76. 2014.PubMed/NCBI
|
|
3
|
Peck-Radosavljevic M: Drug therapy for
advanced-stage liver cancer. Liver Cancer. 3:125–131. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Llovet JM, Ricci S, Mazzaferro V, Hilgard
P, Gane E, Blanc JF, de Oliveira AC, Santoro A, Raoul JL, Forner A,
et al SHARP Investigators Study Group: Sorafenib in advanced
hepatocellular carcinoma. N Engl J Med. 359:378–390. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gauthier A and Ho M: Role of sorafenib in
the treatment of advanced hepatocellular carcinoma: An update.
Hepatol Res. 43:147–154. 2013. View Article : Google Scholar :
|
|
6
|
Wilhelm SM, Carter C, Tang L, Wilkie D,
McNabola A, Rong H, Chen C, Zhang X, Vincent P, McHugh M, et al:
BAY 43-9006 exhibits broad spectrum oral antitumor activity and
targets the RAF/MEK/ERK pathway and receptor tyrosine kinases
involved in tumor progression and angiogenesis. Cancer Res.
64:7099–7109. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu L, Cao Y, Chen C, Zhang X, McNabola A,
Wilkie D, Wilhelm S, Lynch M and Carter C: Sorafenib blocks the
RAF/MEK/ERK pathway, inhibits tumor angiogenesis, and induces tumor
cell apoptosis in hepatocellular carcinoma model PLC/PRF/5. Cancer
Res. 66:11851–11858. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rahmani M, Davis EM, Bauer C, Dent P and
Grant S: Apoptosis induced by the kinase inhibitor BAY 43-9006 in
human leukemia cells involves down-regulation of Mcl-1 through
inhibition of translation. J Biol Chem. 280:35217–35227. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang WA, Groenendyk J and Michalak M:
Endoplasmic reticulum stress associated responses in cancer.
Biochim Biophys Acta. 1843:2143–2149. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li X, Zhang K and Li Z: Unfolded protein
response in cancer: The physician’s perspective. J Hematol Oncol.
4:82011. View Article : Google Scholar
|
|
11
|
Vandewynckel YP, Laukens D, Geerts A,
Bogaerts E, Paridaens A, Verhelst X, Janssens S, Heindryckx F and
Van Vlierberghe H: The paradox of the unfolded protein response in
cancer. Anticancer Res. 33:4683–4694. 2013.PubMed/NCBI
|
|
12
|
Zhang XD, Hersey P, Tay KH, Tseng H, Jiang
CC and Dong L: Adaptation to ER stress as a mechanism of resistance
of melanoma to treatment. Current Management of Malignant Melanoma.
Cao M: InTech, Rijeka; Croatia: 2011
|
|
13
|
Suh DH, Kim MK, Kim HS, Chung HH and Song
YS: Unfolded protein response to autophagy as a promising druggable
target for anticancer therapy. Ann NY Acad Sci. 1271:20–32. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gardner BM and Walter P: Unfolded proteins
are Ire1-activating ligands that directly induce the unfolded
protein response. Science. 333:1891–1894. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Schröder M and Kaufman RJ: ER stress and
the unfolded protein response. Mutat Res. 569:29–63. 2005.
View Article : Google Scholar
|
|
16
|
Han D, Lerner AG, Vande Walle L, Upton JP,
Xu W, Hagen A, Backes BJ, Oakes SA and Papa FR: IRE1alpha kinase
activation modes control alternate endoribonuclease outputs to
determine divergent cell fates. Cell. 138:562–575. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hassler J, Cao SS and Kaufman RJ: IRE1, a
double-edged sword in pre-miRNA slicing and cell death. Dev Cell.
23:921–923. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tirosh B, Iwakoshi NN, Glimcher LH and
Ploegh HL: Rapid turnover of unspliced Xbp-1 as a factor that
modulates the unfolded protein response. J Biol Chem.
281:5852–5860. 2006. View Article : Google Scholar
|
|
19
|
Yoshida H, Matsui T, Yamamoto A, Okada T
and Mori K: XBP1 mRNA is induced by ATF6 and spliced by IRE1 in
response to ER stress to produce a highly active transcription
factor. Cell. 107:881–891. 2001. View Article : Google Scholar
|
|
20
|
Maurel M, Chevet E, Tavernier J and Gerlo
S: Getting RIDD of RNA: IRE1 in cell fate regulation. Trends
Biochem Sci. 39:245–254. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chitnis N, Pytel D and Diehl JA:
UPR-inducible miRNAs contribute to stressful situations. Trends
Biochem Sci. 38:447–452. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lin JH, Li H, Yasumura D, Cohen HR, Zhang
C, Panning B, Shokat KM, Lavail MM and Walter P: IRE1 signaling
affects cell fate during the unfolded protein response. Science.
318:944–949. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ron D, Chen CH, Caldwell J, Jamieson L,
Orr E and Mochly-Rosen D: Cloning of an intracellular receptor for
protein kinase C: A homolog of the beta subunit of G proteins. Proc
Natl Acad Sci USA. 91:839–843. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Adams DR, Ron D and Kiely PA: RACK1, A
multifaceted scaffolding protein: Structure and function. Cell
Commun Signal. 9:222011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ma J, Wu R, Zhang Q, Wu JB, Lou J, Zheng
Z, Ding JQ and Yuan Z: DJ-1 interacts with RACK1 and protects
neurons from oxidative-stress-induced apoptosis. Biochem J.
462:489–497. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
McCahill A, Warwicker J, Bolger GB,
Houslay MD and Yarwood SJ: The RACK1 scaffold protein: A dynamic
cog in cell response mechanisms. Mol Pharmacol. 62:1261–1273. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wu J, Meng J, Du Y, Huang Y, Jin Y, Zhang
J, Wang B, Zhang Y, Sun M and Tang J: RACK1 promotes the
proliferation, migration and invasion capacity of mouse
hepatocellular carcinoma cell line in vitro probably by PI3K/Rac1
signaling pathway. Biomed Pharmacother. 67:313–319. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cao XX, Xu JD, Liu XL, Xu JW, Wang WJ, Li
QQ, Chen Q, Xu ZD and Liu XP: RACK1: A superior independent
predictor for poor clinical outcome in breast cancer. Int J Cancer.
127:1172–1179. 2010. View Article : Google Scholar
|
|
29
|
Ruan Y, Sun L, Hao Y, Wang L, Xu J, Zhang
W, Xie J, Guo L, Zhou L, Yun X, et al: Ribosomal RACK1 promotes
chemoresistance and growth in human hepatocellular carcinoma. J
Clin Invest. 122:2554–2566. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
Arimoto K, Fukuda H, Imajoh-Ohmi S, Saito
H and Takekawa M: Formation of stress granules inhibits apoptosis
by suppressing stress-responsive MAPK pathways. Nat Cell Biol.
10:1324–1332. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Qiu Y, Mao T, Zhang Y, Shao M, You J, Ding
Q, Chen Y, Wu D, Xie D, Lin X, et al: A crucial role for RACK1 in
the regulation of glucose-stimulated IRE1alpha activation in
pancreatic beta cells. Sci Signal. 3:ra72010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liver Cancer Study Group of Japan: The
General Rules for the Clinical and Pathological Study of Primary
Liver Cancer. 2nd English edition. Kanehara & Co, Ltd; Tokyo:
pp. 13–14. 2003
|
|
33
|
Rojas R, Segovia C, Trombert AN, Santander
J and Manque P: The effect of tunicamycin on the glucose uptake,
growth, and cellular adhesion in the protozoan parasite Crithidia
fasciculata. Curr Microbiol. 69:541–548. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chen Y and Brandizzi F: IRE1: ER stress
sensor and cell fate executor. Trends Cell Biol. 23:547–555. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li X, Zhang K and Li Z: Unfolded protein
response in cancer: The physician’s perspective. J Hematol Oncol.
4:82011. View Article : Google Scholar
|
|
36
|
Fujimoto T, Onda M, Nagai H, Nagahata T,
Ogawa K and Emi M: Upregulation and overexpression of human X-box
binding protein 1 (hXBP-1) gene in primary breast cancers. Breast
Cancer. 10:301–306. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Shuda M, Kondoh N, Imazeki N, Tanaka K,
Okada T, Mori K, Hada A, Arai M, Wakatsuki T, Matsubara O, et al:
Activation of the ATF6, XBP1 and grp78 genes in human
hepatocellular carcinoma: A possible involvement of the ER stress
pathway in hepatocarcinogenesis. J Hepatol. 38:605–614. 2003.
View Article : Google Scholar : PubMed/NCBI
|