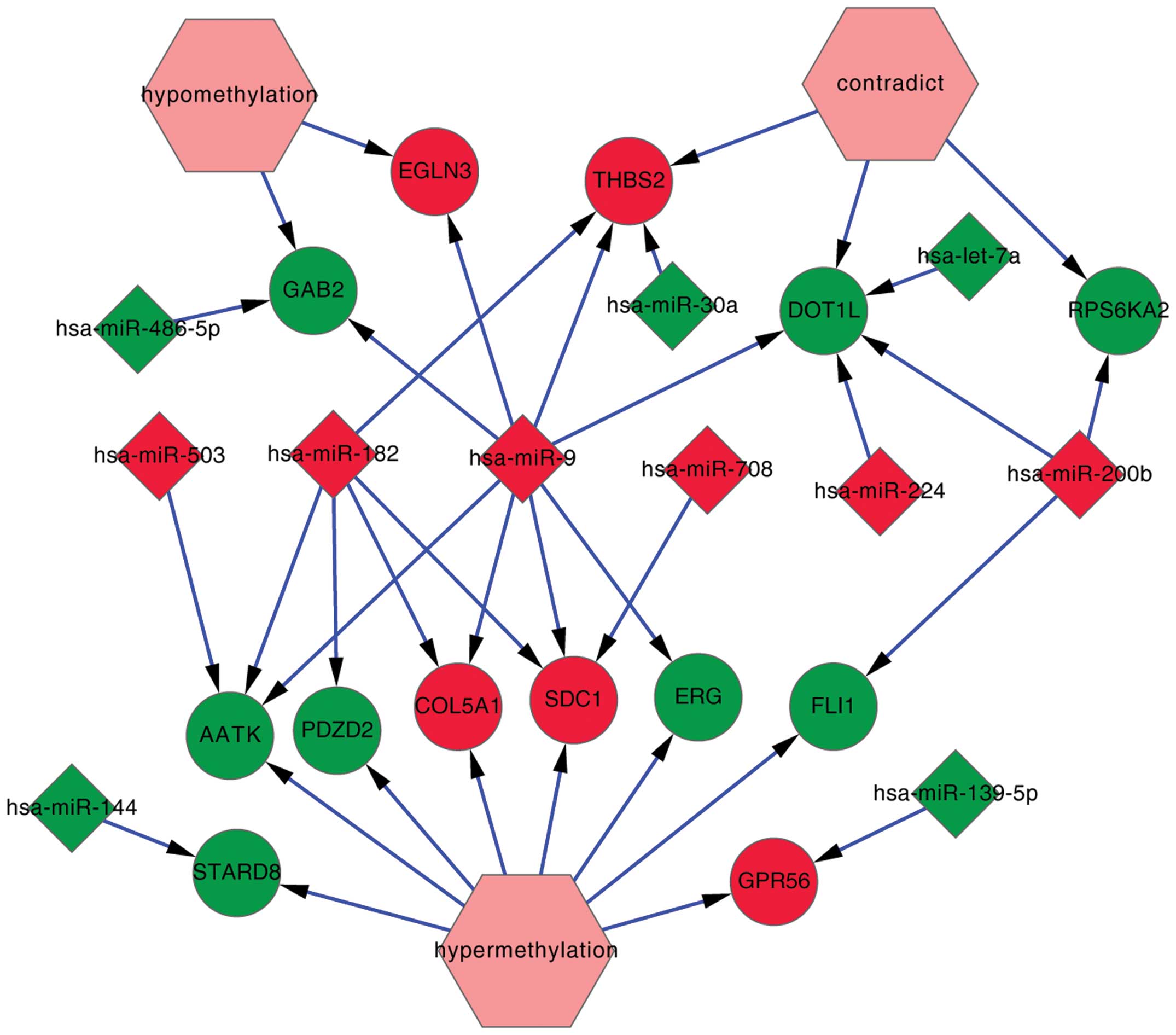
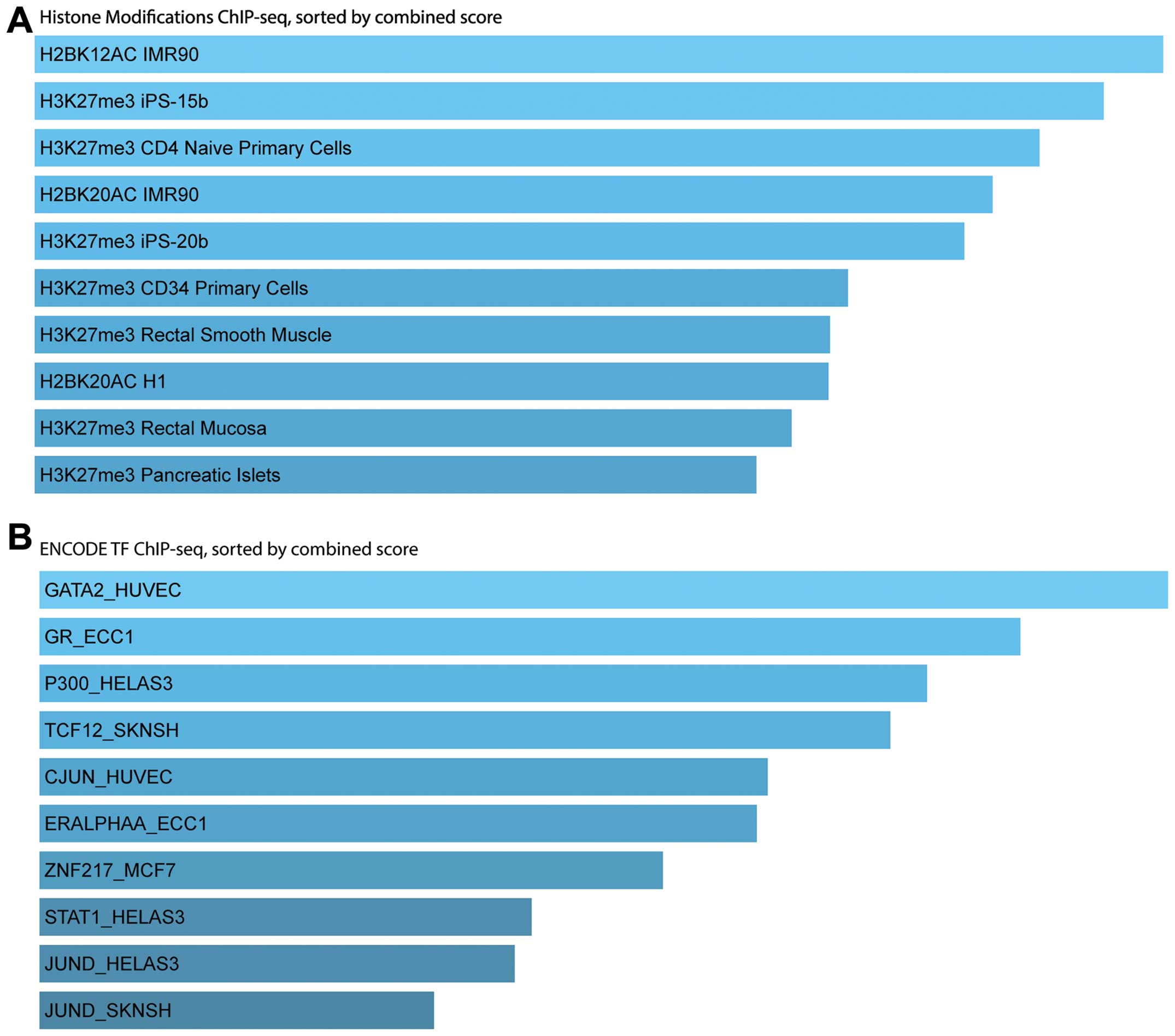
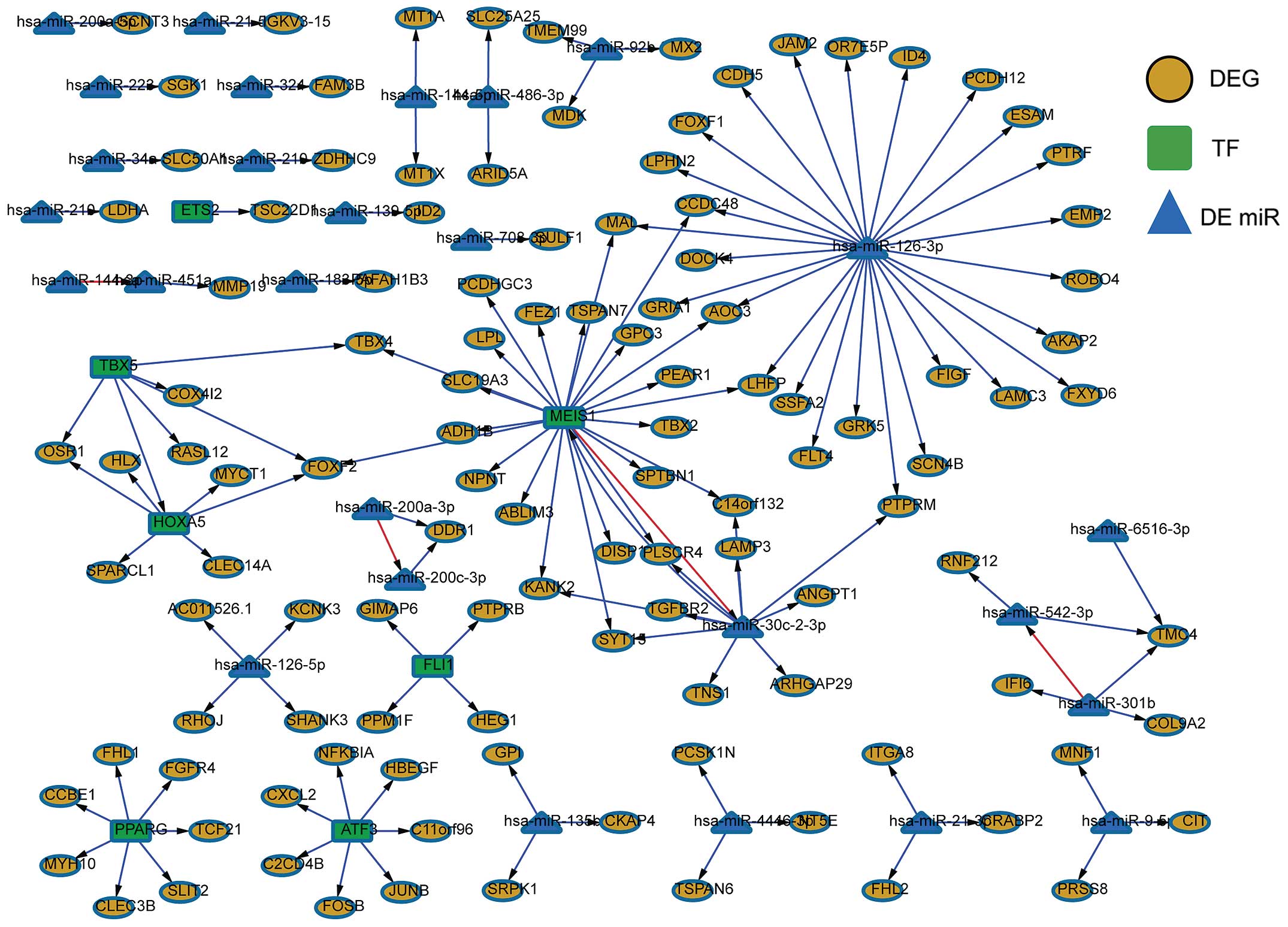
|
1
|
Toh CK, Gao F, Lim WT, Leong SS, Fong KW,
Yap SP, Hsu AA, Eng P, Koong HN, Thirugnanam A, et al:
Never-smokers with lung cancer: Epidemiologic evidence of a
distinct disease entity. J Clin Oncol. 24:2245–2251. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Subramanian J and Govindan R: Lung cancer
in never smokers: A review. J Clin Oncol. 25:561–570. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yano T, Haro A, Shikada Y, Maruyama R and
Maehara Y: Non-small cell lung cancer in never smokers as a
representative ‘non-smoking-associated lung cancer̓: Epidemiology
and clinical features. Int J Clin Oncol. 16:287–293. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Molina JR, Yang P, Cassivi SD, Schild SE
and Adjei AA: Non-small cell lung cancer: Epidemiology, risk
factors, treatment, and survivorship. Mayo Clin Proc. 83:584–594.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Su JL, Shih JY, Yen ML, Jeng YM, Chang CC,
Hsieh CY, Wei LH, Yang PC and Kuo ML: Cyclooxygenase-2 induces
EP1-and HER-2/Neu-dependent vascular endothelial growth factor-C
up-regulation: A novel mechanism of lymphangiogenesis in lung
adenocarcinoma. Cancer Res. 64:554–564. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Skobe M, Hawighorst T, Jackson DG, Prevo
R, Janes L, Velasco P, Riccardi L, Alitalo K, Claffey K and Detmar
M: Induction of tumor lymphangiogenesis by vEGF-C promotes breast
cancer metastasis. Nat Med. 7:192–198. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Heaney AP, Singson R, McCabe CJ, Nelson V,
Nakashima M and Melmed S: Expression of pituitary-tumour
transforming gene in colorectal tumours. Lancet. 355:716–719. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bevilacqua G, Sobel ME, Liotta LA and
Steeg PS: Association of low nm23 RNA levels in human primary
infiltrating ductal breast carcinomas with lymph node involvement
and other histopathological indicators of high metastatic
potential. Cancer Res. 49:5185–5190. 1989.PubMed/NCBI
|
|
9
|
Billah S, Stewart J, Staerkel G, Chen S,
Gong Y and Guo M: EGFR and KRAS mutations in lung carcinoma:
Molecular testing by using cytology specimens. Cancer Cytopathol.
119:111–117. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ju YS, Lee WC, Shin JY, Lee S, Bleazard T,
Won JK, Kim YT, Kim JI, Kang JH and Seo JS: A transforming KIF5B
and RET gene fusion in lung adenocarcinoma revealed from
whole-genome and transcriptome sequencing. Genome Res. 22:436–445.
2012. View Article : Google Scholar :
|
|
11
|
Togashi Y, Soda M, Sakata S, Sugawara E,
Hatano S, Asaka R, Nakajima T, Mano H and Takeuchi K: KLC1-ALK: A
novel fusion in lung cancer identified using a formalin-fixed
paraffin-embedded tissue only. PLoS One. 7:e313232012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kim SC, Jung Y, Park J, Cho S, Seo C, Kim
J, Kim P, Park J, Seo J, Kim J, et al: A high-dimensional,
deep-sequencing study of lung adenocarcinoma in female
never-smokers. PLoS One. 8:e555962013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bolger AM, Lohse M and Usadel B:
Trimmomatic: A flexible trimmer for Illumina sequence data.
Bioinformatics. 30:2114–2120. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Langmead B and Salzberg SL: Fast
gapped-read alignment with Bowtie 2. Nat Methods. 9:357–359. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li H, Handsaker B, Wysoker A, Fennell T,
Ruan J, Homer N, Marth G, Abecasis G and Durbin R: 1000 Genome
Project Data Processing Subgroup: The sequence alignment/map format
and SAMtools. Bioinformatics. 25:2078–2079. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lienhard M, Grimm C, Morkel M, Herwig R
and Chavez L: MEDIPS: Genome-wide differential coverage analysis of
sequencing data derived from DNA enrichment experiments.
Bioinformatics. 30:284–286. 2014. View Article : Google Scholar :
|
|
17
|
Quinlan AR and Hall IM: BEDTools: A
flexible suite of utilities for comparing genomic features.
Bioinformatics. 26:841–842. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Trapnell C, Pachter L and Salzberg SL:
TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics.
25:1105–1111. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Anders S, Reyes A and Huber W: Detecting
differential usage of exons from RNA-seq data. Genome Res.
22:2008–2017. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Trapnell C, Williams BA, Pertea G,
Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ and Pachter
L: Transcript assembly and quantification by RNA-Seq reveals
unannotated transcripts and isoform switching during cell
differentiation. Nat Biotechnol. 28:511–515. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang WC, Lin FM, Chang WC, Lin KY, Huang
HD and Lin NS: miRExpress: Analyzing high-throughput sequencing
data for profiling microRNA expression. BMC Bioinformatics.
10:3282009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Anders S, McCarthy DJ, Chen Y, Okoniewski
M, Smyth GK, Huber W and Robinson MD: Count-based differential
expression analysis of RNA sequencing data using R and
Bioconductor. Nat Protoc. 8:1765–1786. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Huang DW, Sherman BT, Tan Q, Kir J, Liu D,
Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC, et al: DAvID
Bioinformatics Resources: Expanded annotation database and novel
algorithms to better extract biology from large gene lists. Nucleic
Acids Res. 35(Web Server issue): W169–W175. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Vergoulis T, Vlachos IS, Alexiou P,
Georgakilas G, Maragkakis M, Reczko M, Gerangelos S, Koziris N,
Dalamagas T and Hatzigeorgiou AG: TarBase 6.0: Capturing the
exponential growth of miRNA targets with experimental support.
Nucleic Acids Res. 40:D222–D229. 2012. View Article : Google Scholar :
|
|
26
|
Vlachos IS, Kostoulas N, Vergoulis T,
Georgakilas G, Reczko M, Maragkakis M, Paraskevopoulou MD,
Prionidis K, Dalamagas T and Hatzigeorgiou AG: DIANA miRPath v.2.0:
Investigating the combinatorial effect of microRNAs in pathways.
Nucleic Acids Res. 40:W498–W504. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lachmann A, Xu H, Krishnan J, Berger SI,
Mazloom AR and Ma’ayan A: ChEA: Transcription factor regulation
inferred from integrating genome-wide ChIP-X experiments.
Bioinformatics. 26:2438–2444. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Consortium EP: ENCODE Project Consortium:
The ENCODE (ENCyclopedia Of DNA Elements) Project. Science.
306:636–640. 2004. View Article : Google Scholar
|
|
29
|
Huang GT, Athanassiou C and Benos PV:
mirConnX: Condition-specific mRNA-microRNA network integrator.
Nucleic Acids Res. 39(suppl): W416–W423. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang WC, Shyh-Chang N, Yang H, Rai A,
Umashankar S, Ma S, Soh BS, Sun LL, Tai BC, Nga ME, et al: Glycine
decarboxylase activity drives non-small cell lung cancer
tumor-initiating cells and tumorigenesis. Cell. 148:259–272. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Joshi M, Liu X and Belani CP: Taxanes,
past, present, and future impact on non-small cell lung cancer.
Anticancer Drugs. 25:571–583. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Joseph P, Bowman B, Galban S and
Rehemtulla A: A Kras driven murine model elucidates oncogenic role
of FADD in lung cancer. Department of Radiation Oncology,
University of Michigan; Ann Arbor, MI: 2013, Honor’s Bachelor’s
Thesis. http://deepblue.lib.umich.edu/bitstream/handle/2027.42/98916/parijo.pdf?sequence=1.
|
|
33
|
Rauch TA, Wang Z, Wu X, Kernstine KH,
Riggs AD and Pfeifer GP: DNA methylation biomarkers for lung
cancer. Tumour Biol. 33:287–296. 2012. View Article : Google Scholar
|
|
34
|
Reddy ES, Rao VN and Papas TS: The erg
gene: A human gene related to the ets oncogene. Proc Natl Acad Sci
USA. 84:6131–6135. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Thoms JA, Birger Y, Foster S, Knezevic K,
Kirschenbaum Y, Chandrakanthan V, Jonquieres G, Spensberger D, Wong
JW, Oram SH, et al: ERG promotes T-acute lymphoblastic leukemia and
is transcriptionally regulated in leukemic cells by a stem cell
enhancer. Blood. 117:7079–7089. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Leyten GH, Hessels D, Jannink SA, Smit FP,
de Jong H, Cornel EB, de Reijke TM, Vergunst H, Kil P, Knipscheer
BC, et al: Prospective multicentre evaluation of PCA3 and
TMPRSS2-ERG gene fusions as diagnostic and prognostic urinary
biomarkers for prostate cancer. Eur Urol. 65:534–542. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Durkin ME, Ullmannova V, Guan M and
Popescu NC: Deleted in liver cancer 3 (DLC-3), a novel Rho
GTPase-activating protein, is downregulated in cancer and inhibits
tumor cell growth. Oncogene. 26:4580–4589. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Mokarram P, Kumar K, Brim H,
Naghibalhossaini F, Saberi-firoozi M, Nouraie M, Green R, Lee E,
Smoot DT and Ashktorab H: Distinct high-profile methylated genes in
colorectal cancer. PLoS One. 4:e70122009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Bornstein P: Thrombospondins function as
regulators of angiogenesis. J Cell Commun Signal. 3:189–200. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Brezillon S, Zeltz C, Schneider L, Terryn
C, Vuillermoz B, Ramont L, Perrau C, Pluot M, Diebold MD, Radwanska
A, et al: Lumican inhibits B16F1 melanoma cell lung metastasis. J
Physiol Pharmacol. 60(Suppl 4): 15–22. 2009.
|
|
41
|
Barski A, Cuddapah S, Cui K, Roh TY,
Schones DE, Wang Z, Wei G, Chepelev I and Zhao K: High-resolution
profiling of histone methylations in the human genome. Cell.
129:823–837. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hussain M, Rao M, Humphries AE, Hong JA,
Liu F, Yang M, Caragacianu D and Schrump DS: Tobacco smoke induces
polycomb-mediated repression of Dickkopf-1 in lung cancer cells.
Cancer Res. 69:3570–3578. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chen X, Song N, Matsumoto K, Nanashima A,
Nagayasu T, Hayashi T, Ying M, Endo D, Wu Z and Koji T: High
expression of trimethylated histone H3 at lysine 27 predicts better
prognosis in non-small cell lung cancer. Int J Oncol. 43:1467–1480.
2013.PubMed/NCBI
|
|
44
|
Bogush TA, Dudko EA, Beme AA, Bogush EA,
Kim AI, Polotsky BE, Tjuljandin SA and Davydov MI: Estrogen
receptors, antiestrogens, and non-small cell lung cancer.
Biochemistry (Mosc). 75:1421–1427. 2010. View Article : Google Scholar
|
|
45
|
Mak P, Chang C, Pursell B and Mercurio AM:
Estrogen receptor β sustains epithelial differentiation by
regulating prolyl hydroxylase 2 transcription. Proc Natl Acad Sci
USA. 110:4708–4713. 2013. View Article : Google Scholar
|
|
46
|
Zhao G, Nie Y, Lv M, He L, Wang T and Hou
Y: ERβ-mediated estradiol enhances epithelial mesenchymal
transition of lung adenocarcinoma through increasing transcription
of midkine. Mol Endocrinol. 26:1304–1315. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Schlossmacher G, Stevens A and White A:
Glucocorticoid receptor-mediated apoptosis: Mechanisms of
resistance in cancer cells. J Endocrinol. 211:17–25. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Koutsimpelas D, Pongsapich W, Heinrich U,
Mann S, Mann WJ and Brieger J: Promoter methylation of MGMT, MLH1
and RASSF1A tumor suppressor genes in head and neck squamous cell
carcinoma: Pharmacological genome demethylation reduces
proliferation of head and neck squamous carcinoma cells. Oncol Rep.
27:1135–1141. 2012.PubMed/NCBI
|
|
49
|
Abe M, Hamada J, Takahashi O, Takahashi Y,
Tada M, Miyamoto M, Morikawa T, Kondo S and Moriuchi T: Disordered
expression of HOX genes in human non-small cell lung cancer. Oncol
Rep. 15:797–802. 2006.PubMed/NCBI
|
|
50
|
Svingen T and Tonissen KF: Altered HOX
gene expression in human skin and breast cancer cells. Cancer Biol
Ther. 2:518–523. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Argiropoulos B, Yung E and Humphries RK:
Unraveling the crucial roles of Meis1 in leukemogenesis and normal
hematopoiesis. Genes Dev. 21:2845–2849. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Hatcher CJ, Kim M-S, Mah CS, Goldstein MM,
Wong B, Mikawa T and Basson CT: TBX5 transcription factor regulates
cell proliferation during cardiogenesis. Dev Biol. 230:177–188.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Aherne NJ, Rangaswamy G and Thirion P:
Prostate cancer in a male with Holt-Oram syndrome: First clinical
association of the TBX5 mutation. Case Rep Urol.
2013:4053432013.PubMed/NCBI
|
|
54
|
Miko E, Margitai Z, Czimmerer Z, Várkonyi
I, Dezso B, Lányi A, Bacsó Z and Scholtz B: miR-126 inhibits
proliferation of small cell lung cancer cells by targeting SLC7A5.
FEBS Lett. 585:1191–1196. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zhou H, Xu X, Xun Q, Yu D, Ling J, Guo F,
Yan Y, Shi J and Hu Y: microRNA-30c negatively regulates
endometrial cancer cells by targeting metastasis-associated gene-1.
Oncol Rep. 27:807–812. 2012.
|