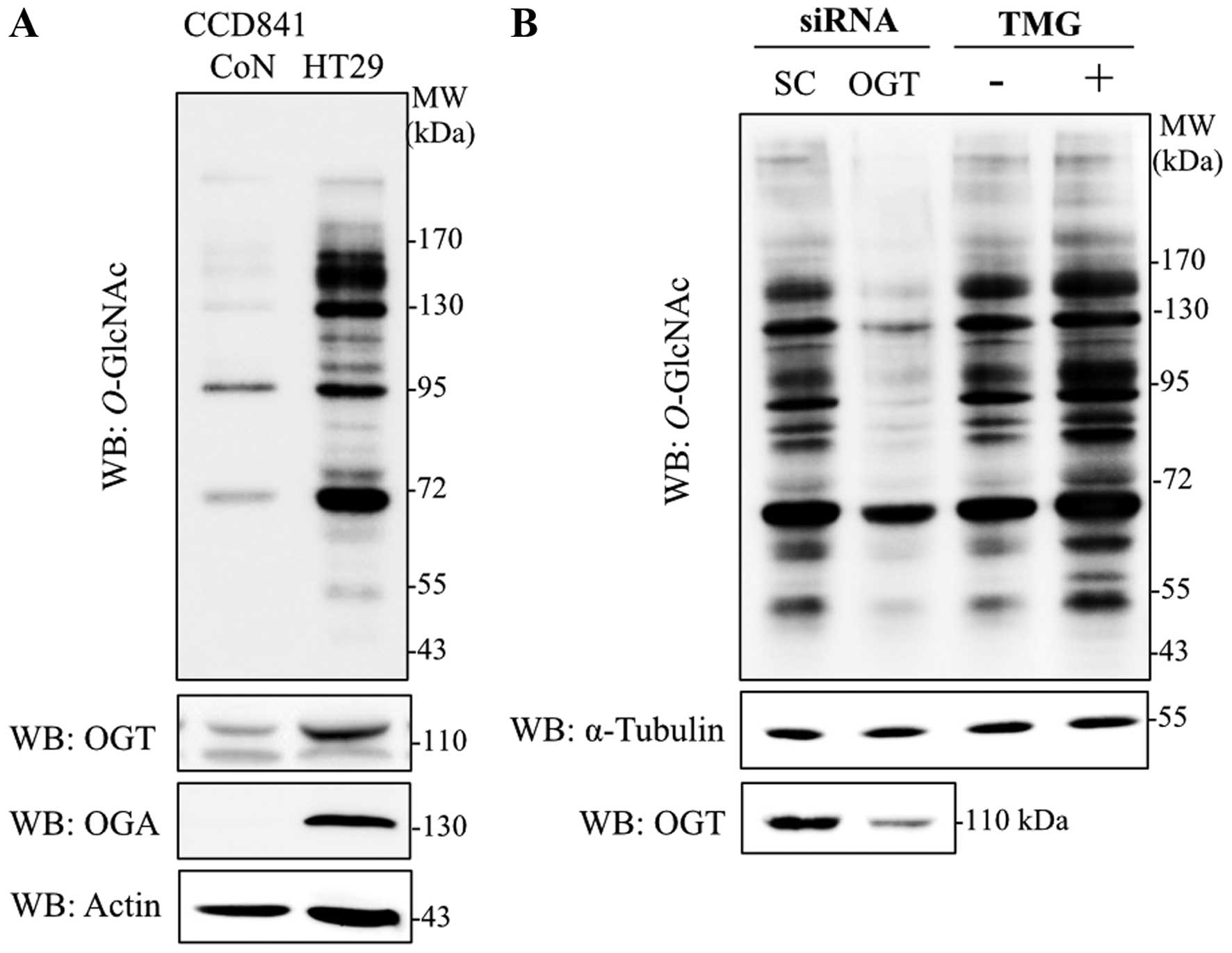
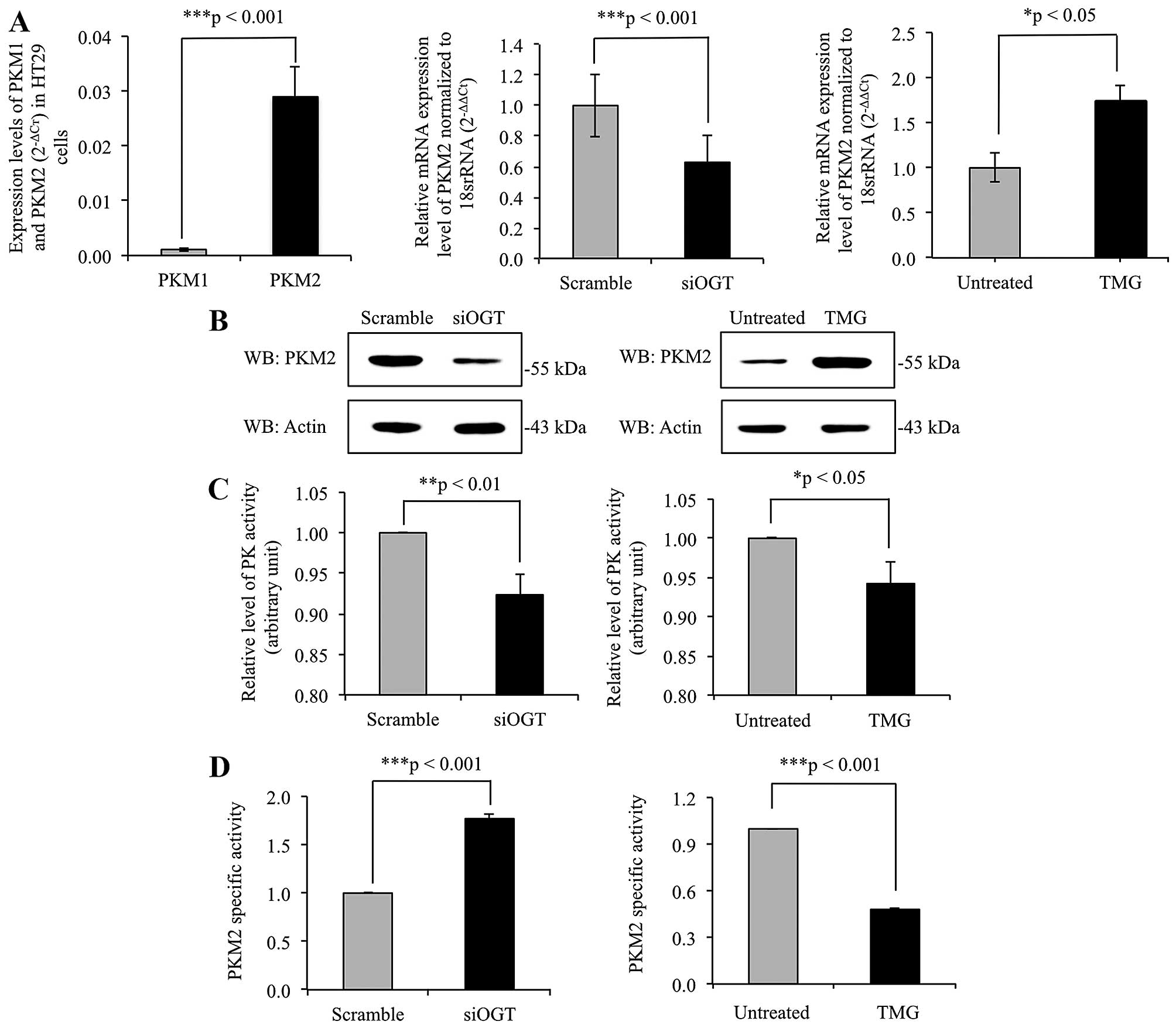
|
1
|
Wong N, Ojo D, Yan J and Tang D: PKM2
contributes to cancer metabolism. Cancer Lett. 356:184–191. 2015.
View Article : Google Scholar
|
|
2
|
Tamada M, Suematsu M and Saya H: Pyruvate
kinase M2: Multiple faces for conferring benefits on cancer cells.
Clin Cancer Res. 18:5554–5561. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Vander Heiden MG, Cantley LC and Thompson
CB: Understanding the Warburg effect: The metabolic requirements of
cell proliferation. Science. 324:1029–1033. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Christofk HR, Vander Heiden MG, Harris MH,
Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL and
Cantley LC: The M2 splice isoform of pyruvate kinase is important
for cancer metabolism and tumour growth. Nature. 452:230–233. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yang W and Lu Z: Regulation and function
of pyruvate kinase M2 in cancer. Cancer Lett. 339:153–158. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mazurek S, Boschek CB, Hugo F and
Eigenbrodt E: Pyruvate kinase type M2 and its role in tumor growth
and spreading. Semin Cancer Biol. 15:300–308. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hanover JA, Krause MW and Love DC: The
hexosamine signaling pathway: O-GlcNAc cycling in feast or famine.
Biochim Biophys Acta. 1800:80–95. 2010. View Article : Google Scholar :
|
|
8
|
Lazarus MB, Nam Y, Jiang J, Sliz P and
Walker S: Structure of human O-GlcNAc transferase and its complex
with a peptide substrate. Nature. 469:564–567. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chaiyawat P, Netsirisawan P, Svasti J and
Champattanachai V: Aberrant O-GlcNAcylated proteins: New
perspectives in breast and colorectal cancer. Front Endocrinol
(Lausanne). 5:1932014.
|
|
10
|
Willems L, Jacque N, Jacquel A, Neveux N,
Maciel TT, Lambert M, Schmitt A, Poulain L, Green AS, Uzunov M, et
al: Inhibiting glutamine uptake represents an attractive new
strategy for treating acute myeloid leukemia. Blood. 122:3521–3532.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mi W, Gu Y, Han C, Liu H, Fan Q, Zhang X,
Cong Q and Yu W: O-GlcNAcylation is a novel regulator of lung and
colon cancer malignancy. Biochim Biophys Acta. 1812:514–519. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Phueaouan T, Chaiyawat P, Netsirisawan P,
Chokchaichamnankit D, Punyarit P, Srisomsap C, Svasti J and
Champattanachai V: Aberrant O-GlcNAc-modified proteins expressed in
primary colorectal cancer. Oncol Rep. 30:2929–2936. 2013.PubMed/NCBI
|
|
13
|
Butkinaree C, Park K and Hart GW: O-linked
beta-N-acetylglu-cosamine (O-GlcNAc): Extensive crosstalk with
phosphorylation to regulate signaling and transcription in response
to nutrients and stress. Biochim Biophys Acta. 1800:96–106. 2010.
View Article : Google Scholar
|
|
14
|
Dias WB, Cheung WD and Hart GW:
O-GlcNAcylation of kinases. Biochem Biophys Res Commun.
422:224–228. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Champattanachai V, Netsirisawan P,
Chaiyawat P, Phueaouan T, Charoenwattanasatien R,
Chokchaichamnankit D, Punyarit P, Srisomsap C and Svasti J:
Proteomic analysis and abrogated expression of O-GlcNAcylated
proteins associated with primary breast cancer. Proteomics.
13:2088–2099. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Srisomsap C, Sawangareetrakul P,
Subhasitanont P, Panichakul T, Keeratichamroen S, Lirdprapamongkol
K, Chokchaichamnankit D, Sirisinha S and Svasti J: Proteomic
analysis of cholangiocarcinoma cell line. Proteomics. 4:1135–1144.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tit-Oon P, Chokchaichamnankit D,
Khongmanee A, Sawangareetrakul P, Svasti J and Srisomsap C:
Comparative secretome analysis of cholangiocarcinoma cell line in
three-dimensional culture. Int J Oncol. 45:2108–2116.
2014.PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
19
|
Hu P, Shimoji S and Hart GW: Site-specific
interplay between O-GlcNAcylation and phosphorylation in cellular
regulation. FEBS Lett. 584:2526–2538. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang Z, Gucek M and Hart GW: Cross-talk
between GlcNAcylation and phosphorylation: Site-specific
phosphorylation dynamics in response to globally elevated O-GlcNAc.
Proc Natl Acad Sci USA. 105:13793–13798. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Dias WB, Cheung WD, Wang Z and Hart GW:
Regulation of calcium/calmodulin-dependent kinase IV by O-GlcNAc
modification. J Biol Chem. 284:21327–21337. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Luo B, Soesanto Y and McClain DA: Protein
modification by O-linked GlcNAc reduces angiogenesis by inhibiting
Akt activity in endothelial cells. Arterioscler Thromb Vasc Biol.
28:651–657. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Robles-Flores M, Meléndez L, García W,
Mendoza-Hernández G, Lam TT, Castañeda-Patlán C and
González-Aguilar H: Post-translational modifications on protein
kinase c isozymes. Effects of epinephrine and phorbol esters.
Biochim Biophys Acta. 1783:695–712. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Renwick SB, Snell K and Baumann U: The
crystal structure of human cytosolic serine
hydroxymethyltransferase: A target for cancer chemotherapy.
Structure. 6:1105–1116. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Amelio I, Cutruzzolá F, Antonov A,
Agostini M and Melino G: Serine and glycine metabolism in cancer.
Trends Biochem Sci. 39:191–198. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jain M, Nilsson R, Sharma S, Madhusudhan
N, Kitami T, Souza AL, Kafri R, Kirschner MW, Clish CB and Mootha
VK: Metabolite profiling identifies a key role for glycine in rapid
cancer cell proliferation. Science. 336:1040–1044. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Schweizer J, Bowden PE, Coulombe PA,
Langbein L, Lane EB, Magin TM, Maltais L, Omary MB, Parry DA,
Rogers MA, et al: New consensus nomenclature for mammalian
keratins. J Cell Biol. 174:169–174. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chou CF, Smith AJ and Omary MB:
Characterization and dynamics of O-linked glycosylation of human
cytokeratin 8 and 18. J Biol Chem. 267:3901–3906. 1992.PubMed/NCBI
|
|
29
|
Owens DW and Lane EB: The quest for the
function of simple epithelial keratins. Bioessays. 25:748–758.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Omary MB, Ku NO, Liao J and Price D:
Keratin modifications and solubility properties in epithelial cells
and in vitro. Subcell Biochem. 31:105–140. 1998.
|
|
31
|
Srikanth B, Vaidya MM and Kalraiya RD:
O-GlcNAcylation determines the solubility, filament organization,
and stability of keratins 8 and 18. J Biol Chem. 285:34062–34071.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Rossbach O, Hung LH, Khrameeva E,
Schreiner S, König J, Curk T, Zupan B, Ule J, Gelfand MS and
Bindereif A: Cross-linking-immunoprecipitation (iCLIP) analysis
reveals global regulatory roles of hnRNP L. RNA Biol. 11:146–155.
2014. View Article : Google Scholar :
|
|
33
|
Liu G, Razanau A, Hai Y, Yu J, Sohail M,
Lobo VG, Chu J, Kung SK and Xie J: A conserved serine of
heterogeneous nuclear ribonucleoprotein L (hnRNP L) mediates
depolarization-regulated alternative splicing of potassium
channels. J Biol Chem. 287:22709–22716. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Shimi T, Butin-Israeli V, Adam SA,
Hamanaka RB, Goldman AE, Lucas CA, Shumaker DK, Kosak ST, Chandel
NS and Goldman RD: The role of nuclear lamin B1 in cell
proliferation and senescence. Genes Dev. 25:2579–2593. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hocevar BA, Burns DJ and Fields AP:
Identification of protein kinase C (PKC) phosphorylation sites on
human lamin B. Potential role of PKC in nuclear lamina structural
dynamics. J Biol Chem. 268:7545–7552. 1993.PubMed/NCBI
|
|
36
|
Fiume R, Ramazzotti G, Teti G, Chiarini F,
Faenza I, Mazzotti G, Billi AM and Cocco L: Involvement of nuclear
PLCbeta1 in lamin B1 phosphorylation and G2/M cell cycle
progression. FASEB J. 23:957–966. 2009. View Article : Google Scholar
|
|
37
|
Yi W, Clark PM, Mason DE, Keenan MC, Hill
C, Goddard WA III, Peters EC, Driggers EM and Hsieh-Wilson LC:
Phosphofructokinase 1 glycosylation regulates cell growth and
metabolism. Science. 337:975–980. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Dombrauckas JD, Santarsiero BD and Mesecar
AD: Structural basis for tumor pyruvate kinase M2 allosteric
regulation and catalysis. Biochemistry. 44:9417–9429. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Mazurek S: Pyruvate kinase type M2: A key
regulator of the metabolic budget system in tumor cells. Int J
Biochem Cell Biol. 43:969–980. 2011. View Article : Google Scholar
|
|
40
|
Ido-Kitamura Y, Sasaki T, Kobayashi M, Kim
HJ, Lee YS, Kikuchi O, Yokota-Hashimoto H, Iizuka K, Accili D and
Kitamura T: Hepatic FoxO1 integrates glucose utilization and lipid
synthesis through regulation of Chrebp O-glycosylation. PLoS One.
7:e472312012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Schäfer D, Hamm-Künzelmann B and Brand K:
Glucose regulates the promoter activity of aldolase A and pyruvate
kinase M2 via dephosphorylation of Sp1. FEBS Lett. 417:325–328.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Jackson SP and Tjian R: O-glycosylation of
eukaryotic transcription factors: Implications for mechanisms of
transcriptional regulation. Cell. 55:125–133. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Haltiwanger RS, Grove K and Philipsberg
GA: Modulation of O-linked N-acetylglucosamine levels on nuclear
and cytoplasmic proteins in vivo using the peptide
O-GlcNAc-beta-N-acetylglucosaminidase inhibitor
O-(2-acetamido-2-deoxy-D-glucopyranosylidene)amino-N-
phenylcarbamate. J Biol Chem. 273:3611–3617. 1998. View Article : Google Scholar : PubMed/NCBI
|