Introduction
Ovarian cancer is the second most common cancer
among female gynecologic cancers and has become the leading cause
of cancer-related death among females (1). Due to the difficulty in early
detection, 75% of ovarian cancer patients are diagnosed at advanced
stages (stage III or IV) (2). In
stage III or IV, the tumor involves one or both ovaries with
peritoneal metastasis outside the pelvis or distant metastasis to
liver parenchyma or other visceral organs (2,3). Early
invasion and metastasis have been well accepted as the leading
features and main causes of death in ovarian cancer. However,
mechanistic understanding of the metastatic potential of ovarian
cancer remains unclear, and novel targets are yet to be identified
for treating metastatic ovarian cancer.
Lysine-specific demethylase 1 (LSD1/KDM1A/AOF2) is
the first histone demethylase discovered, which specifically
demethylates mono- and dimethylated histone H3 lysine 4 (H3K4) and
histone H3 lysine 9 (H3K9) (4).
LSD1 is frequently overexpressed in lung cancer (5,6),
breast cancer (7), prostate cancer
(8,9), and liver cancer (10). Importantly, overexpression of LSD1
promotes the growth and invasion of various types of cancer cells,
and contributes to human carcinogenesis by regulating the
expression of genes involved in various chromatin-modifying
pathways (6). Conversely,
inhibition of LSD1 was found to suppress cell invasion and
migration in various types of cancers (5,11,12).
Although LSD1 is recently described to be highly expressed in
ovarian cancer (13,14), the biological function of LSD1 in
this cancer remains largely unknown.
Epithelial-messenchymal transition (EMT) is a
process whereby epithelial cells are programmed into mesenchymal
cells (15). EMT is now considered
as the initial and essential step in tumor metastasis. During EMT,
epithelial cells acquire cell motility by reducing cell-cell
junctions, and loss of cell polarity (16,17).
E-cadherin, an epithelial marker, has a crucial role in regulating
cell-cell adhesion and maintenance of tissue architecture (18). Indeed, E-cadherin serves as a
suppressor of cell migration and invasion (19–22).
Transcription factors, including Snail, Slug, Zeb1 and Twist, can
induce EMT by downregulating E-cadherin expression (23–26).
Recent studies show that LSD1 is recruited by the transcription
factor Snail to the promoter of E-cadherin to repress the
expression of the E-cadherin gene consequently contributing to
cancer cell invasion (27,28). Conversely, Ferrari-Amorotti et
al observed that blocking Snail-LSD1 interaction by treatment
with Parnate suppressed the invasiveness of cancer cells (29).
Few studies have reported on how LSD1 induces EMT
and finally contributes to ovarian cancer cell migration. Therefore
in the present study, we examined the effect of LSD1 on cell
migration and invasion using LSD1-knockdown and overexpressing
HO8910 ovarian cancer cells as models. We also examined the
regulatory role of LSD1 in the expression of molecular markers of
EMT. Knockdown of LSD1 reduced cell migration and invasion in the
HO8910 cells, while overexpression of LSD1 stimulated the migration
and invasion of the HO8910 cells. Mechanistic analyses uncovered
that LSD1 promoted cell migration through induction of N-cadherin,
Snail, vimentin, MMP-2 and inhibition of E-cadherin. LSD1
epigenetically regulated the transcription of E-cadherin through
demethylating H3K4 at the E-cadherin promoter. Collectively, these
results suggest that targeting LSD1 may be a novel therapeutic
approach for the treatment of ovarian cancer.
Materials and methods
Cell lines and cell culture
The human ovarian cancer cell line, HO8910, was
kindly provided by Dr Qixiang Shao of Jiangsu University
(Zhenjiang, China). HO8910 cells were cultured in RPMI-1640 medium
supplemented with 10% fetal bovine serum (FBS) (both from Gibco,
Grand Island, NY, USA) at a temperature of 37°C under 5%
CO2. HEK 293T cells were cultured in Dulbecco's modified
Eagle's medium (DMEM; Gibco) containing 10% FBS at a temperature of
37°C under 5% CO2.
Antibodies and reagents
The pLKO-Tet-On, pLVX-tight-puro, pHR′-CMV-8.2ΔVPR,
and pHR′-CMV-VSVG vectors were kind gifts from Dr Changdeng Hu
(Purdue University, West Lafayette, in, USA). LSD1, E-cadherin,
Snail, vimentin, N-cadherin and MMP-2 antibodies were purchased
from Cell Signaling Technology Inc. (Danvers, MA, USA). The
α-tubulin and horseradish peroxidase (HRP)-conjugated goat
anti-rabbit antibodies were obtained from Bioworld Technology
(Shanghai, China). Electrochemiluminescence (ECL) reagents were
purchased from Millipore Corp. (Billerica, MA, USA). H3K4me2
antibody was purchased from upstate biotechnology Inc. (Lake
Placid, NY, USA). Polybrene, doxycycline (Dox), puromycin and G418
were purchased from Sigma-Aldrich (St. Louis, MO, USA). The LSD1
inhibitor tranylcypromine (TCP) was obtained from Biomol
International (Plymouth Meeting, PA, USA).
Plasmid constructions and
transfections
For generation of the shRNA-LSD1 plasmid, annealed
short hairpin oligonucleotides (the RNAi Consortium collection
TRCN0000046072; Sigma-Aldrich) targeting CCACGAGTCAAACCTTTATTT in
the coding regions (CDS) of LSD1 were cloned into pLKO-Tet-On by
AgeI and EcoRI sites to produce pLKO-Tet-On-shLSD1 as
described previously (30,31). The constructs were confirmed by DNA
sequencing. All transfections were performed using the
Lipofectamine 2000 reagent (Invitrogen) according to the
manufacturer's instructions.
Establishment of the stable cell lines
(LSD1-knockdown and overexpressing)
To generate lentiviral particles, 293T cells were
seeded in 6-cm dishes and transfected with 2 µg of
pLKO-Tet-On-shLSD1, 1.5 µg of pHR′-CMV-8.2ΔVPR and 0.5
µg of pHR′-CMV-VSVG using Lipofectamine 2000 reagent. The
supernatant containing the lentiviral particles was harvested 24,
48 and 72 h post-transfection, and then centrifuged (124 × g for 5
min) to remove cell debris. HO8910 cells cultured in 6-cm dishes
were infected by adding 1 ml lentiviral supernatant and 3 ml
complete medium containing 8 µg/ml Polybrene. After the
infection (twice), cells were selected with 2.0 µg/ml
puromycin for 3 days and then maintained with 1.0 µg/ml
puromycin for one week.
To generate rTet-repressor expressing (rtTA) cell
line, 293T cells were transfected with 2 µg of pLVX-Tet-On,
1.5 µg of pHR′-CMV-8.2ΔVPR and 0.5 µg of
pHR′-CMV-VSVG using Lipofectamine 2000 reagent. After transfection
(24 h), the viral supernatant was harvested and used to infect
HO8910 cells. After the infection (twice), HO8910 cells were
selected with 200 µg/ml G418 for 1 week. The cells that
survived were stable rtTA. HO8910-rtTA cells were then infected
with the lentiviral particles packaged with pLVX-tight-puro-LSD1.
After infection twice, HO8910-rtTA cells were selected with 2.0
µg/ml of puromycin for 3 days, and then maintained in the
presence of 1.0 µg/ml of puromycin for one week. The
surviving cells were considered as stable clones. The stable clones
were further confirmed by western blot analysis.
RNA extraction and real-time RT-PCR
(qRT-PCR)
Total RNA was isolated from the cells using RNAiso
plus (Takara, Shiga, Japan) and reverse-transcribed using the
PrimeScript RT reagent kit (Takara) to generate cDNAs. Then the
cDNAs were subjected to qRT-PCR as described previously (32). qRT-PCR was performed with SYBR-Green
PCR Master Mix (Takara) on a Bio-Rad CFX96 system (Bio-Rad
Laboratories, Inc., Hercules, CA, USA). The primer sequences used
were: LSD1 (GenBank accession no. NM 015013.3),
5′-CAAGTGTCAATTTGTTCGGG-3′ (forward) and 5′-TTCTTTGGGCTGAGGTACTG-3′
(reverse); and GAPDH (GenBank accession no. NM001256799.1),
5′-GCAAATTCCATGGCACCGTC-3′ (forward) and 5′-TCGCCCCACTTGATTTTGG-3′
(reverse). The relative quantification of mRNA levels was
normalized to levels of GADPH and calculated by comparative
2−ΔΔCt.
Western blot analysis
Protein lysates were extracted from the cells and
blotted as described previously (33). Equal amounts of soluble proteins
were electrophoresed by SDS-PAGE and transferred to 0.45-µm
PVDF membranes. The membranes were blocked with 5% nonfat-dry milk
for 1 h at room temperature (RT). After incubation with the primary
antibodies against LSD1 (1:1,000), E-cadherin (1:500), Snail
(1:500), vimentin (1:500), N-cadherin (1:500) or MMP-2 (1:500)
overnight at 4°C and with the corresponding secondary antibodies
(1:5,000) for 1 h at RT, the immunoblots were developed by ECL
method.
Migration and invasion assays
For the invasion assay, each Boyden chamber (BD
Biosciences, Bedford, MA, USA) was coated with 60 µl
Matrigel diluted with DMEM (1:30) and incubated at 37°C for 4–6 h.
Cells (1.5×105) were resuspended with DMEM containing
Dox or TCP in the upper chamber. Then, 10% FBS-containing medium
was placed in the lower chamber to act as a chemoattractant. After
a 24-h incubation, the non-invading cells remaining on the upper
surface were removed, and the cells on the lower surface were fixed
with 4% formaldehyde for 30 min, and stained with 0.1% crystal
violet for 15 min. At least 5 fields for each chamber were
photographed (×200 magnification) and counted, and the invading
cells were counted in each field. The cell migration assay was
performed using Boyden chambers without Matrigel coating. All
experiments were performed at least in triplicate.
Chromatin immunoprecipitation (ChIP)
All reagents were provided by Upstate Biotechnology
(EZ-ChIP™ kit 17-371). Cells were fixed with 1% formaldehyde to
cross-link proteins. The reaction was stopped by adding 10X
glycine. Cross-linked cells were washed with PBS twice, pelleted
and resuspended in SDS lysis buffer at a concentration of
1×107 cells/ml. Aliquots of 400 µl were sonicated
with 4–6 sets of 5-sec pulses (32% output) on ice. Then sonicated
lysates were centrifuged and divided into 100 µl aliquots
for each ChIP assay (1×106 cells/IP), and precleared
with protein G-agarose. After incubation with the antibodies
overnight at 4°C, immune complexes were collected with protein
G-agarose, and then washed with low salt immune complex wash
buffer, high salt immune complex wash buffer, and finally TE
buffer. The immune complexes were eluted with 20% SDS, and 1 M
NaHCO3. The crosslinks were reversed overnight at 65°C,
then the DNA was purified using spin columns, and finally subjected
to qRT-PCR. Chromatin eluted from the IPs with IgG and anti-RNA
polymerase were used as the negative and positive control,
respectively. Two previously described primers of E-cadherin
promoter for ChIP (34,35) were as follows: E-ca01
5′-GGGCAATACAGGGAGACACA-3′ (forward) and 5′-GGGCTTTTACACTTGGCTGA-3′
(reverse); E-ca02 5′-CACAACAGCATAGGGAGACATT-3′ (forward) and
5′-TGTAGAGCTTCATGGGTTAGTGA-3′ (reverse).
Statistical analysis
All values are presented as the mean ± SEM. The data
were analyzed using the Student's t-test with SPSS 11.5 software
(SPSS Inc.). P-values with a 95% confidence interval were obtained
from at least three independent experiments. A p-value <0.01 was
considered to indicate a statistically significant result.
Results
LSD1 is required for cell migration and
invasion in ovarian cancer cells
To investigate the contribution of LSD1 to the
migration and invasion of ovarian cancer HO8910 cells, we generated
stable LSD1-knockdown (LSD1-KD) clones and LSD1-overexpressing
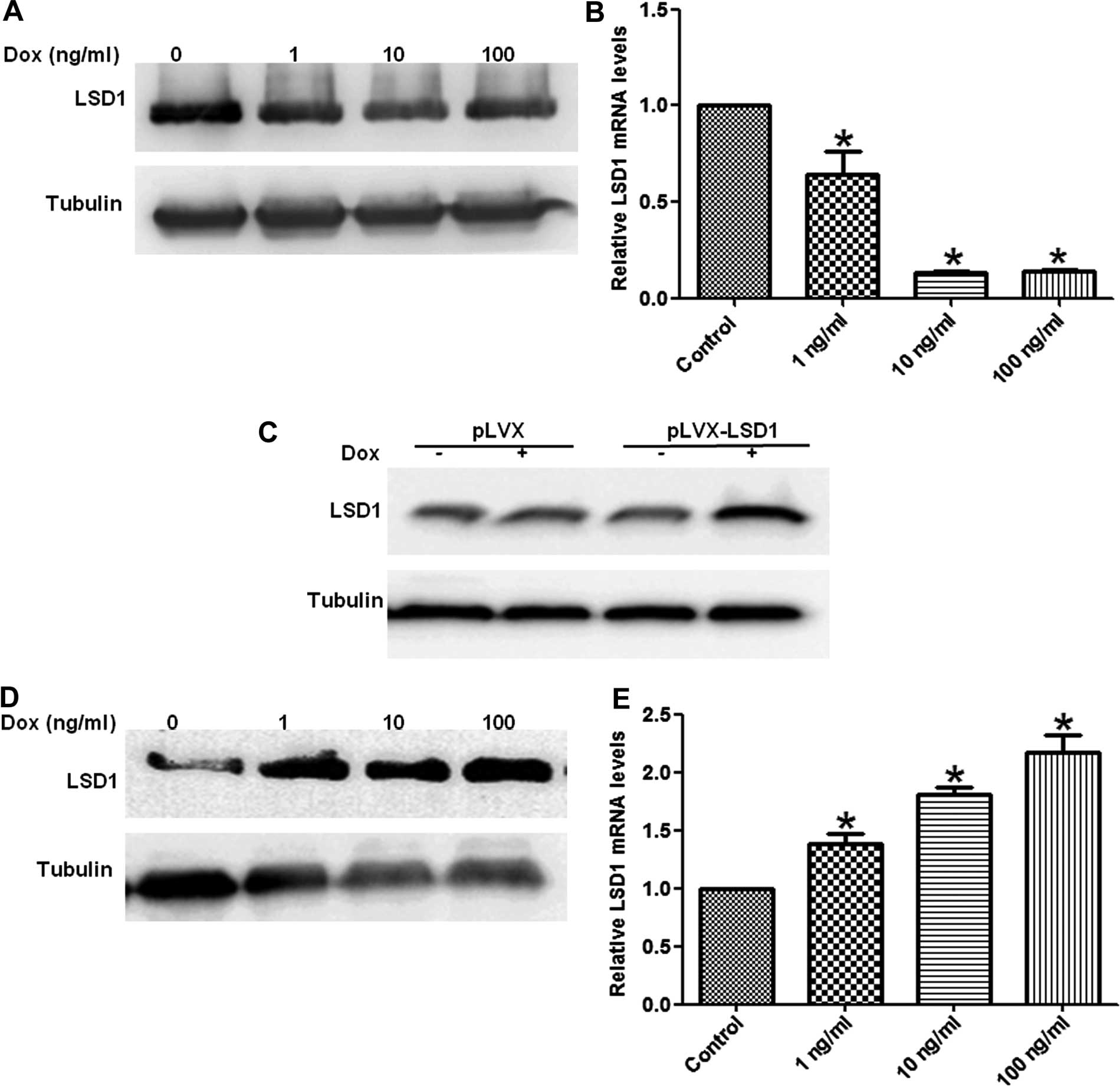
(LSD1-OE) clones from the HO8910 cells. Total RNA and proteins were
extracted from these stable cells treated with increasing doses of
Dox for 24 or 48 h. Our results showed the mRNA and protein
expression of the LSD1 gene was decreased in the LSD1-KD cells in a
dose-dependent manner (Fig. 1A and
B), whereas the levels of LSD1 mRNA and protein expression were
increased in the LSD1-OE cells (Fig.
1C–E).
To understand the effect of LSD1 expression on cell
migration and invasion, we performed Transwell assays to measure
the migratory capacity of these two transfected cell lines. The
LSD1-KD cells displayed less migration and invasion in comparison
with the control (Fig. 2A and B),
whereas the LSD1-OE cells had a higher rate of migration and
invasion as compared to the control (Fig. 2C and D).
To further determine the role of LSD1 in cell
migration, we utilized a known potent inhibitor, TCP (30,36),
to suppress the demethylase activity of LSD1 in HO8910 cells.
Inhibition of LSD1 decreased the migration activity of the HO8910
cells in a dose-dependent manner (Fig.
3A and B). Taken together, these data suggest that LSD1 is
essential for cell migration and invasion in HO8910 ovarian cancer
cells.
LSD1 regulates EMT in ovarian cancer
cells
As epithelial-mesenchymal transition (EMT) is
involved in tumor migration and invasion, we examined the
expression of several EMT markers in the LSD1-KD and LSD1-OE HO8910
cells. We found that knockdown of LSD1 upregulated the expression
of the epithelial marker E-cadherin and downregulated the
expression of the mesenchymal markers N-cadherin, vimentin and
MMP-2 (Fig. 4A). LSD1 knockdown
also caused a decrease in the expression of the transcription
factor Snail (Fig. 4A).
Furthermore, inhibition of LSD1 induced an increase in E-cadherin
expression and a decrease in the expression of N-cadherin,
vimentin, MMP-2 and Snail in a dose-dependent manner (Fig. 4C). On the contrary, overexpression
of LSD1 induced a decrease in E-cadherin expression, with a
concomitant increase in the expression of N-cadherin, Vimentin,
MMP-2 and Snail in the HO8910 cells (Fig. 4B).
LSD1 knockdown increases H3K4me2 levels
at the E-cadherin promoter
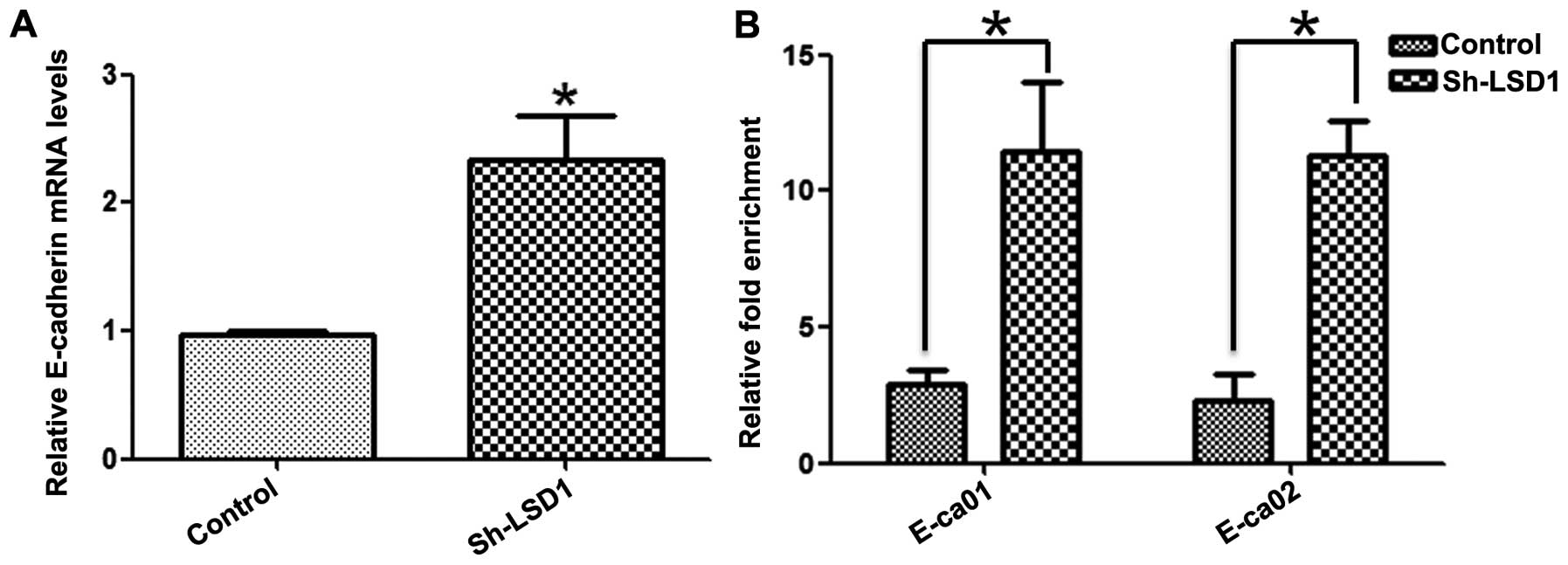
Given that knockdown of LSD1 was accompanied by the
upregulation of E-cadherin at the transcriptional level (Fig. 5A) and inhibition of migration of
ovarian cancer cells (Fig. 2A and
B), we speculated that LSD1 could enhance migration by
downregulating E-cadherin expression via demethylation of H3K4me2,
a major substrate of LSD1 in ovarian cancer cells (30). To confirm this speculation, Chip
assays were performed in the LSD1-KD HO8910 cells incubated with
the anti-H3K4me2 antibody. Quantitative analysis indicated that the
enrichment of H3K4me2 at the promoter of the e-cadherin gene was
significantly higher in the LSD1-KD cells than that in the control
cells (Fig. 5B). Collectively, our
data revealed that the expression of LSD1 caused a decrease in
H3K4me2 levels at the E-cadherin promoter, reduced E-cadherin
expression, and consequently contributed to the migration of HO8910
cells.
Discussion
Ovarian cancer is the second most common malignant
gynecologic tumor, and represents the leading cause of
cancer-related death among women worldwide (1). The high mortality rate of ovarian
cancer is caused by tumor metastasis, post-surgical recurrence, and
late detection at advanced stages (3). Ovarian cancer is associated with
multiple risk factors and is currently recognized as both a genetic
and epigenetic disease (37,38).
While the genetic changes in ovarian cancer have been extensively
studied, the contribution of epigenetic alterations to ovarian
cancer progression remains poorly understood. Histone methylation
is a dynamic epigenetic process that has been found to be
associated with cancer, including ovarian cancer (39). LSD1 is a well-characterized
demethylase that can remove methyl groups from H3K4 (4). However, its role and underlying
mechanisms in ovarian cancer are still unclear. In this study, we
showed that LSD1 overexpression induced EMT, migration and invasion
of HO8910 ovarian cancer cells. In contrast, silencing of LSD1
reversed these events in invasive HO8910 cells. We also showed a
mechanistic link between LSD1 and E-cadherin through LSD1-mediated
regulation of H3K4me2, which subsequently leads to the
downregulation of E-cadherin transcription.
Histone demethylases are epigenetic enzymes that can
remove both repressive and activating histone marks. LSD1 family
members are capable of removing the H3K4me2-activating marks and
rendering them potential players in the downregulation of tumor
suppressors (40,41). The putative role of LSD1 as an
oncogene in cancer development is supported by the observation that
LSD1 is highly expressed in ovarian cancer (13,14)
and other malignant tumors (5–10).
LSD1 is reported to play an important role in ovarian cancer cell
proliferation via a Sox2-mediated mechanism (31). Our present study points to a novel
function of LSD1 in ovarian cancer cell migration and invasion
through regulation of EMT.
Recently, the regulation of epigenetic modification
on EMT is a hot topic. Several studies have shown that histone
modifications are involved in Snail-mediated transcriptional
repression of E-cadherin. Peinado et al reported that Snail
induces repressive histone modifications at the E-cadherin promoter
through recruitment of histone deacetylases (HDACs) (42). Recent studies have demonstrated that
Snail recruits LSD1 to the E-cadherin promoter to reduce E-cadherin
expression by removing H3K4me2 (27,28).
In this study, we found that modulation of LSD1 expression alters
the methylation status of H3K4 at the E-cadherin promoter, which in
turn transcriptionally regulates the expression of E-cadherin.
Thus, we conclude that LSD1 transcriptionally downregulates
E-cadherin expression via H3K4 demethylation, and consequently
results in the increased migration and invasion of HO8910
cells.
Taking all these pieces of evidence together, we are
able to show that knockdown of LSD1 impairs the migration and
invasion of HO8910 cells by regulating EMT, while overexpression of
LSD1 has a converse effect on cell migration. By demethylating
H3K4me2 at the E-cadherin promoter, LSD1 downregulates the
E-cadherin expression, and contributes to the metastasis of HO8910
cells. Our results suggest that LSD1 may be a potential therapeutic
target for metastatic ovarian cancer.
Acknowledgments
This study was supported by grants from the National
Natural Science Foundation of China (81170573) and Clinical
Medicine Science & Technology Project of Jiangsu Province of
China (BL2013024).
References
|
1
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. CA Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hennessy BT, Coleman RL and Markman M:
Ovarian cancer. Lancet. 374:1371–1382. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chaffer CL and Weinberg RA: A perspective
on cancer cell metastasis. Science. 331:1559–1564. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shi Y, Lan F, Matson C, Mulligan P,
Whetstine JR, Cole PA, Casero RA and Shi Y: Histone demethylation
mediated by the nuclear amine oxidase homolog LSD1. Cell.
119:941–953. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lv T, Yuan D, Miao X, Lv Y, Zhan P, Shen X
and Song Y: Overexpression of LSD1 promotes proliferation,
migration and invasion in non-small cell lung cancer. PLoS One.
7:e350652012. View Article : Google Scholar
|
|
6
|
Hayami S, Kelly JD, Cho HS, Yoshimatsu M,
Unoki M, Tsunoda T, Field HI, Neal DE, Yamaue H, Ponder BA, et al:
Overexpression of LSD1 contributes to human carcinogenesis through
chromatin regulation in various cancers. Int J Cancer. 128:574–586.
2011. View Article : Google Scholar
|
|
7
|
Serce N, Gnatzy A, Steiner S, Lorenzen H,
Kirfel J and Buettner R: Elevated expression of LSD1
(Lysine-specific demethylase 1) during tumour progression from
pre-invasive to invasive ductal carcinoma of the breast. BMC Clin
Pathol. 12:132012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wu CY, Hsieh CY, Huang KE, Chang C and
Kang HY: Cryptotanshinone down-regulates androgen receptor
signaling by modulating lysine-specific demethylase 1 function. Int
J Cancer. 131:1423–1434. 2012. View Article : Google Scholar
|
|
9
|
Kahl P, Gullotti L, Heukamp LC, Wolf S,
Friedrichs N, Vorreuther R, Solleder G, Bastian PJ, Ellinger J,
Metzger E, et al: Androgen receptor coactivators lysine-specific
histone demethylase 1 and four and a half LIM domain protein 2
predict risk of prostate cancer recurrence. Cancer Res.
66:11341–11347. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhao Z-K, Yu H-F, Wang D-R, Dong P, Chen
L, Wu WG, Ding WJ and Liu YB: Overexpression of lysine specific
demethylase 1 predicts worse prognosis in primary hepatocellular
carcinoma patients. World J Gastroenterol. 18:6651–6656. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ding J, Zhang ZM, Xia Y, Liao GQ, Pan Y,
Liu S, Zhang Y and Yan ZS: LSD1-mediated epigenetic modification
contributes to proliferation and metastasis of colon cancer. Br J
Cancer. 109:994–1003. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yu Y, Wang B, Zhang K, Lei Z, Guo Y, Xiao
H, Wang J, Fan L, Lan C, Wei Y, et al: High expression of
lysine-specific demethylase 1 correlates with poor prognosis of
patients with esophageal squamous cell carcinoma. Biochem Biophys
Res Commun. 437:192–198. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Konovalov S and Garcia-Bassets I: Analysis
of the levels of lysine-specific demethylase 1 (LSD1) mRNA in human
ovarian tumors and the effects of chemical LSD1 inhibitors in
ovarian cancer cell lines. J Ovarian Res. 6:752013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen C, Ge J, Lu Q, Ping G, Yang C and
Fang X: Expression of lysine-specific demethylase 1 in human
epithelial ovarian cancer. J Ovarian Res. 8:282015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hay ED: An overview of
epithelio-mesenchymal transformation. Acta Anat (Basel). 154:8–20.
1995. View Article : Google Scholar
|
|
16
|
Boyer B, Vallés AM and Edme N: Induction
and regulation of epithelial-mesenchymal transitions. Biochem
Pharmacol. 60:1091–1099. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Thiery JP: Epithelial-mesenchymal
transitions in tumour progression. Nat Rev Cancer. 2:442–454. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Angst BD, Marcozzi C and Magee AI: The
cadherin superfamily. J Cell Sci. 114:625–626. 2001.PubMed/NCBI
|
|
19
|
Hazan RB, Qiao R, Keren R, Badano I and
Suyama K: Cadherin switch in tumor progression. Ann NY Acad Sci.
1014:155–163. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Perl AK, Wilgenbus P, Dahl U, Semb H and
Christofori G: A causal role for E-cadherin in the transition from
adenoma to carcinoma. Nature. 392:190–193. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Birchmeier W, Hulsken J and Behrens J:
E-cadherin as an invasion suppressor. Ciba Foundation Symposium.
189:124–136; discussion 136–141, 174–126. 1995.PubMed/NCBI
|
|
22
|
Batlle E, Sancho E, Francí C, Domínguez D,
Monfar M, Baulida J and García De Herreros A: The transcription
factor snail is a repressor of E-cadherin gene expression in
epithelial tumour cells. Nat Cell Biol. 2:84–89. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kang Y and Massagué J:
Epithelial-mesenchymal transitions: twist in development and
metastasis. Cell. 118:277–279. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kato Y, Yashiro M, Noda S, Tendo M,
Kashiwagi S, Doi Y, Nishii T, Matsuoka J, Fuyuhiro Y, Shinto O, et
al: Establishment and characterization of a new hypoxia-resistant
cancer cell line, OCUM-12/Hypo, derived from a scirrhous gastric
carcinoma. Br J Cancer. 102:898–907. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang J, Mani SA, Donaher JL, Ramaswamy S,
Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A and
Weinberg RA: Twist, a master regulator of morphogenesis, plays an
essential role in tumor metastasis. Cell. 117:927–939. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Waldmann J, Feldmann G, Slater EP, Langer
P, Buchholz M, Ramaswamy A, Saeger W, Rothmund M and Fendrich V:
Expression of the zinc-finger transcription factor Snail in
adrenocortical carcinoma is associated with decreased survival. Br
J Cancer. 99:1900–1907. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lin Y, Wu Y, Li J, Dong C, Ye X, Chi YI,
Evers BM and Zhou BP: The SNAG domain of Snail1 functions as a
molecular hook for recruiting lysine-specific demethylase 1. EMBO
J. 29:1803–1816. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lin T, Ponn A, Hu X, Law BK and Lu J:
Requirement of the histone demethylase LSD1 in Snai1-mediated
transcriptional repression during epithelial-mesenchymal
transition. Oncogene. 29:4896–4904. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ferrari-Amorotti G, Fragliasso V, Esteki
R, Prudente Z, Soliera AR, Cattelani S, Manzotti G, Grisendi G,
Dominici M, Pieraccioli M, et al: Inhibiting interactions of lysine
demethylase LSD1 with snail/slug blocks cancer cell invasion.
Cancer Res. 73:235–245. 2013. View Article : Google Scholar :
|
|
30
|
Shao G, Wang J, Li Y, Liu X, Xie X, Wan X,
Yan M, Jin J, Lin Q, Zhu H, et al: Lysine-specific demethylase 1
mediates epidermal growth factor signaling to promote cell
migration in ovarian cancer cells. Sci Rep. 5:153442015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang X, Lu F, Wang J, Yin F, Xu Z, Qi D,
Wu X, Cao Y, Liang W, Liu Y, et al: Pluripotent stem cell protein
Sox2 confers sensitivity to LSD1 inhibition in cancer cells. Cell
Reports. 5:445–457. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shao GB, Wang J, Zhang LP, Wu CY, Jin J,
Sang JR, Lu HY, Gong AH, Du FY and Peng WX: Aging alters histone H3
lysine 4 methylation in mouse germinal vesicle stage oocytes.
Reprod Fertil Dev. 27:419–426. 2015. View
Article : Google Scholar
|
|
33
|
Zhang L, Wang J, Pan Y, Jin J, Sang J,
Huang P and Shao G: Expression of histone H3 lysine 4 methylation
and its demethylases in the developing mouse testis. Cell Tissue
Res. 358:875–883. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li LC, Okino ST, Zhao H, Pookot D, Place
RF, Urakami S, Enokida H and Dahiya R: Small dsRNAs induce
transcriptional activation in human cells. Proc Natl Acad Sci USA.
103:17337–17342. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ting AH, Schuebel KE, Herman JG and Baylin
SB: Short double-stranded RNA induces transcriptional gene
silencing in human cancer cells in the absence of DNA methylation.
Nat Genet. 37:906–910. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sun G, Alzayady K, Stewart R, Ye P, Yang
S, Li W and Shi Y: Histone demethylase LSD1 regulates neural stem
cell proliferation. Mol Cell Biol. 30:1997–2005. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Verma M, Seminara D, Arena FJ, John C,
Iwamoto K and Hartmuller V: Genetic and epigenetic biomarkers in
cancer: improving diagnosis, risk assessment, and disease
stratification. Mol Diagn Ther. 10:1–15. 2006. View Article : Google Scholar
|
|
38
|
Seeber LM and van Diest PJ: Epigenetics in
ovarian cancer. Methods Mol Biol. 863:253–269. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
He Y, Korboukh I, Jin J and Huang J:
Targeting protein lysine methylation and demethylation in cancers.
Acta Biochim Biophys Sin (Shanghai). 44:70–79. 2012. View Article : Google Scholar
|
|
40
|
Chen Y, Jie W, Yan W, Zhou K and Xiao Y:
Lysine-specific histone demethylase 1 (LSD1): a potential molecular
target for tumor therapy. Crit Rev Eukaryot Gene Expr. 22:53–59.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lim S, Janzer A, Becker A, Zimmer A,
Schüle R, Buettner R and Kirfel J: Lysine-specific demethylase 1
(LSD1) is highly expressed in ER-negative breast cancers and a
biomarker predicting aggressive biology. Carcinogenesis.
31:512–520. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Peinado H, Ballestar E, Esteller M and
Cano A: Snail mediates E-cadherin repression by the recruitment of
the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex. Mol Cell
Biol. 24:306–319. 2004. View Article : Google Scholar :
|