Introduction
Melanoma is a skin tumor caused by the malignant
transformation of melanocytes and has an incidence rate of 4% in
the US (1). Malignant melanoma is
commonly characterized by rapid angiogenic growth, tumor cell
apoptosis resistance, and immune tolerance.
The AKT1 gene encodes a serine-threonine protein
kinase called the AKT kinase, which regulates many processes
including metabolism, proliferation, cell survival, growth, and
angiogenesis. High expression of AKT is involved in the resistance
to cell apoptosis in melanoma (2).
AKT has also been implicated in adaptive resistance to treatment.
Although BRAF inhibitor (BRAFi) therapy has shown remarkable
anti-melanoma responses, BRAFi therapy leads to a rebound in
phosphorylated AKT levels, which result in acquired drug resistance
(3).
Estrogen has been demonstrated to be involved in
regulation of the immune system to allow invasion, proliferation,
and migration of tumor cells such as trophoblasts (4). Increasing evidence has shown that
estrogen dependency contributes to the growth of melanoma.
According to results in gender-related differences, melanoma may
act as a hormone-dependent tumor (5–7). The
results also emphasize that melanoma is an estrogen
receptor-positive tumor, whose prognosis is adversely affected by
estrogen (8). In contrast, one
study showed that high expression rates of estrogen receptors have
no significant correlation with the prognosis of conjunctival
melanoma (9). Although the effects
of estrogen on the progression of melanoma are controversial, there
is evidence that estrogen is the co-mediator involved in the growth
of melanoma. Melanoma that is responsive to estrogens is associated
with the superficial spreading melanoma subtype, a type of tumor
with a much better prognosis (6).
Estrogen prevents apoptosis and promotes angiogenesis, allowing
melanoma to become more aggressive (10).
In our investigations, the top hub gene named AKT1
with higher expression in melanoma showed enriched binding sites in
the negative regulation of response to external stimulus, which
adapts cells to changes in the external stimulation for survival.
The AKT1 gene encodes a serine-threonine protein kinase (AKT). The
results revealed that higher expression of AKT primarily induced
the proliferation of melanoma cells. Another finding was that AKT
regulated lipid metabolic process and may be involved in melanoma
progression and promotion of tumor growth through gene enrichment
function analysis. A high association of the estrogen signaling
pathway and the RAP1 signaling pathway with melanoma was shown in
this study. The estrogen signaling pathway modulates immune
tolerance and resistance to cell apoptosis, which contributes to
the growth of melanoma. The RAP1 signaling pathway regulates focal
adhesion (FA) in a negative feedback to cell migration and invasion
in melanoma.
In our study, the top hub gene AKT1 was correlated
with the estrogen signaling and RAP1 signaling pathways which
regulate the growth of melanoma. Screening of the differentially
expressed genes (DEGs) and analysis of the enrichment functions
will provide more insight into the molecular mechanisms of
melanoma. Genes dependent on the kinase signaling transduction
pathways are attractive targets for advanced melanoma therapy.
Materials and methods
Microarray data and preprocessing
The raw gene expression profile GSE3189 (11) was downloaded from the public
database Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/). In total, there
were 7 normal skin and 45 melanoma samples. The corresponding
platform was GPL96 (GeneChip® Human Genome U133 Set
HG-U133A) Affymetrix Human Genome U133A Array which contains
~22,500 human transcripts. The background correction and
normalization of microarray data among microarrays was conducted by
the RMA (Robust Multi-array Averaging) method (12) with the defaulted parameters in the R
bioconductor package 'affy' package. Probe sets were mapped to gene
symbol names using 'Annotate package'. Microarray data were
filtered to extract the most variable probe set for each gene (in R
software using package: genefilter).
DEG screening
In order to identify DEGs, the limma R package
(13) was used to compare the
melanoma samples to the normal skin samples. The raw P-value was
corrected using the Benjamin and Hochberg method (14) to circumvent the multi-test bias. The
fold change value >4 or <0.25 and false discovery rate (FDR)
<0.01 (15) were selected as
cutoff criterion for DEGs.
Construction of the interaction
network
To further analyze these DEGs, we next mapped all
the DEGs to the STRING (Search Tool for the Retrieval of
Interacting Genes) database (16)
to construct an gene-gene interaction network. STRING integrates
other databases to reveal interactions including both direct
(physical) and indirect (functional) associations of the target
genes. A combined score was computed by STRING, which indicates a
higher confidence when more than one type of information supports a
given association.
Analyzing the topological properties of
the interaction network
Next we analyzed the topological properties of the
interaction network, such as node degree and clustering
coefficient. Node degree is the number of nodes directly connected
to a node, displaying the local centrality of this node in the
network. A higher node degree usually represents a stronger
importance of a node for the stability of the network and a cluster
coefficient usually represents how closely the adjacent nodes are
connected with each other. It has already been determined that most
of the biological networks are subject to scale-free network
property, which means that their degree distribution follows a
power law, and that the clustering coefficient distribution
decreases as the node degree increases (17). These two topological properties were
analyzed based on the Network Analyzer Cytoscape software (18) in the interaction network.
Hub gene identification by Google
PageRank
In order to find the topological hub (important)
genes, we used a weighted Google PageRank method (19,20) to
score all the DEGs in the interaction network. It was first applied
in the Google web-search engine for identifying important web
pages; it measures the importance of a node based on the sum of the
rank of its backlinks (the number of nodes that link to that
particular node). In addition to simply calculating the degree of
each node, the PageRank score measures a gene's importance or
popularity based mainly on the structure of the interaction
network. Therefore, genes with lower degrees, which connect to
other key genes, can also be selected as important genes.
The original PageRank is defined as:
Where Lk is the set of all the adjacent nodes
connected to node k, nj is the number of node k's
adjacent nodes, N is the total number of the nodes in the whole
network, and d is the damping factor, which is usually set to 0.15.
It is noticeable that the rank score of a network is
divided evenly over the nodes to which it links; however, for the
actual gene interaction network, some gene-gene associations are
stronger than the others. Therefore, we modified the proposed
original PageRank with weight information as Weighted PageRank
(WPR):
Where Wj,k is the combined score of the link from gene j
to k, which we obtained from STRING.
Gene Ontology and pathway analysis
ClueGO (21) is a
Cytoscape plugin, which includes precompiled annotation files, such
as GO, KEGG and BioCarta, used to analyze interrelations and
enrichment of terms and functional groups in biological networks.
To explore the biological function and interrelations of the top
ranked important genes, we generated the different cluster of GO
and pathway terms of the selected genes.
Results
Differential gene expression between
melanoma and normal skin samples
The limma R package was used to compare gene
expression profiles of melanoma and normal skin samples. At a fold
change value |log(FC)| >2 [false discovery rate (FDR) <0.01],
a total of 1,310 genes were differentially expressed, including 499
upregulated genes and 811 downregulated genes; Fig. 1 shows the heatmap of all the
differentially expressed genes.
Interaction network construction and
topological property analysis
All the DEGs were mapped to the STRING database to
construct an interaction network. STRING that is linked to other
databases calculated the combined scores of the DEGs in terms of
gene characteristics and spatial structures. The interaction
network of the DEGs is shown in Fig.
2.
We also determined that the topological properties
of the interaction network, such as node degree and clustering
coefficient, are subject to scale-free network property. As shown
in Fig. 3A, from the node degree
distribution in the network, we obtained y =
545.19x(−1.422) (gray line in Fig. 3A) with the power law fitting. The
x-axis stands for the node degrees, which means the number of nodes
directly connected with it, and the y-axis represents the number of
nodes with different degrees. The property implies that the
low-degree nodes belong to very dense sub-graphs and those
sub-graphs are connected to each other through high degree nodes.
Fig. 3B shows the topological
coefficient distribution in the network, and that the clustering
coefficient is able to display the aggregation degree of nodes. The
nodes with a high-clustering coefficient are in the minority, and
the clustering coefficient distributions are mostly located in the
area between 0.1 and 0.4.
Top PageRank gene identification and
function annotation
The Google PageRank method was used to select the
most important genes in the interaction network, and finally the
top 44 genes with highest PageRank scores (PageRank >0.003) were
selected as important genes. As shown in Table I, a high PageRank score is
relatively compared to a high degree. Forty-four genes were
selected as the top hub genes from 1,310 DEGs based on the PageRank
score and are highlighted with red and orange color in the
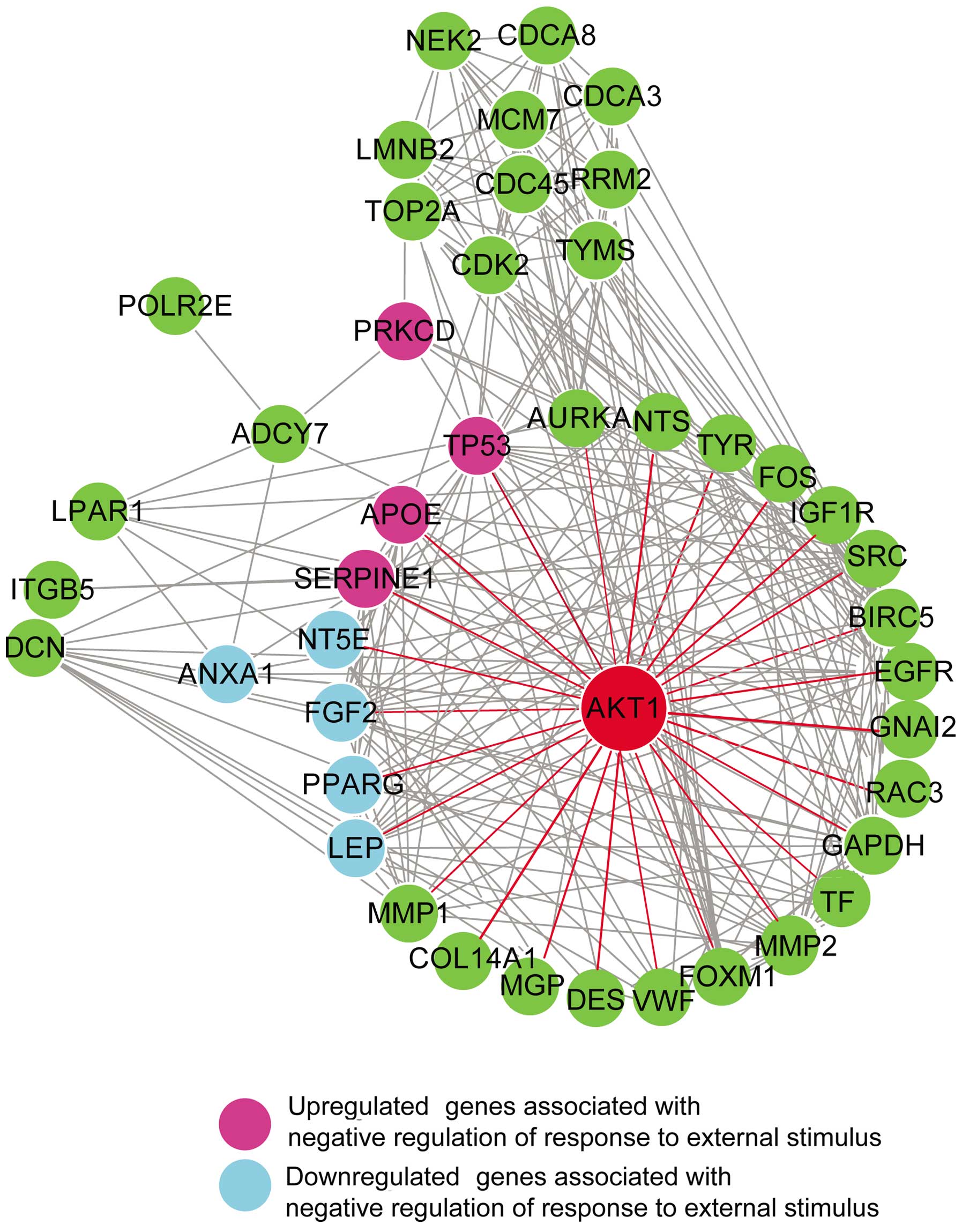
interaction network (Fig. 2).
Unsurprisingly, AKT1 (shown as a red color gene) with upregulated
expression was found to be the top gene associated with melanoma.
One hundred and thirty DEGs including 26 top hub genes corresponded
to AKT1.
 | Table ITop 44 PageRank hub genes and their
topological properties. |
Table I
Top 44 PageRank hub genes and their
topological properties.
| Gene name | Cluster
coefficient | Degree | PageRank |
|---|
| AKT1 | 0.07501491 | 130 | 0.011705631 |
| TP53 | 0.05533826 | 118 | 0.010572723 |
| SRC | 0.07474277 | 102 | 0.009131527 |
| EGFR | 0.07400536 | 99 | 0.008814539 |
| GAPDH | 0.12330199 | 88 | 0.008222844 |
| FOS | 0.11488511 | 78 | 0.007051433 |
| MMP2 | 0.16014027 | 59 | 0.005436854 |
| FGF2 | 0.15136612 | 61 | 0.005407865 |
| DCN | 0.10707804 | 58 | 0.005124292 |
| CDK2 | 0.24784314 | 51 | 0.004830041 |
| PPARG | 0.14441219 | 53 | 0.00479262 |
| TYR | 0.15183673 | 50 | 0.004585681 |
| RAC3 | 0.10544218 | 49 | 0.004534422 |
| BIRC5 | 0.33107089 | 52 | 0.004446444 |
| LEP | 0.21816168 | 43 | 0.004265752 |
| NTS | 0.24615385 | 40 | 0.004015176 |
| SERPINE1 | 0.20512821 | 40 | 0.003705497 |
| FOXM1 | 0.47226174 | 38 | 0.003689789 |
| CDC45 | 0.57936508 | 36 | 0.003612235 |
| VWF | 0.1754386 | 39 | 0.003598718 |
| AURKA | 0.43078627 | 43 | 0.003555397 |
| GNAI2 | 0.26984127 | 36 | 0.003463597 |
| TYMS | 0.52552553 | 37 | 0.003439231 |
| POLR2E | 0.14962121 | 33 | 0.003425802 |
| IGF1R | 0.16642959 | 38 | 0.003385309 |
| MCM7 | 0.54654655 | 37 | 0.003349025 |
| NT5E | 0.21904762 | 36 | 0.003348752 |
| TF | 0.16666667 | 37 | 0.00331314 |
| TOP2A | 0.64171123 | 34 | 0.003299905 |
| ANXA1 | 0.32857143 | 36 | 0.003271953 |
| COL14A1 | 0.25490196 | 34 | 0.003263538 |
| NEK2 | 0.74193548 | 31 | 0.003229935 |
| MGP | 0.29032258 | 32 | 0.003214665 |
| APOE | 0.23172906 | 34 | 0.00320765 |
| PRKCD | 0.10160428 | 34 | 0.00318685 |
| CDCA8 | 0.76344086 | 31 | 0.003183032 |
| LPAR1 | 0.41935484 | 32 | 0.003169619 |
| DES | 0.11711712 | 37 | 0.003168955 |
| ADCY7 | 0.25 | 33 | 0.003111591 |
| MMP1 | 0.25225225 | 37 | 0.003101892 |
| ITGB5 | 0.14795009 | 34 | 0.003067761 |
| LMNB2 | 0.59354839 | 31 | 0.003064281 |
| CDCA3 | 0.84729064 | 29 | 0.003039415 |
| RRM2 | 0.75268817 | 31 | 0.003001865 |
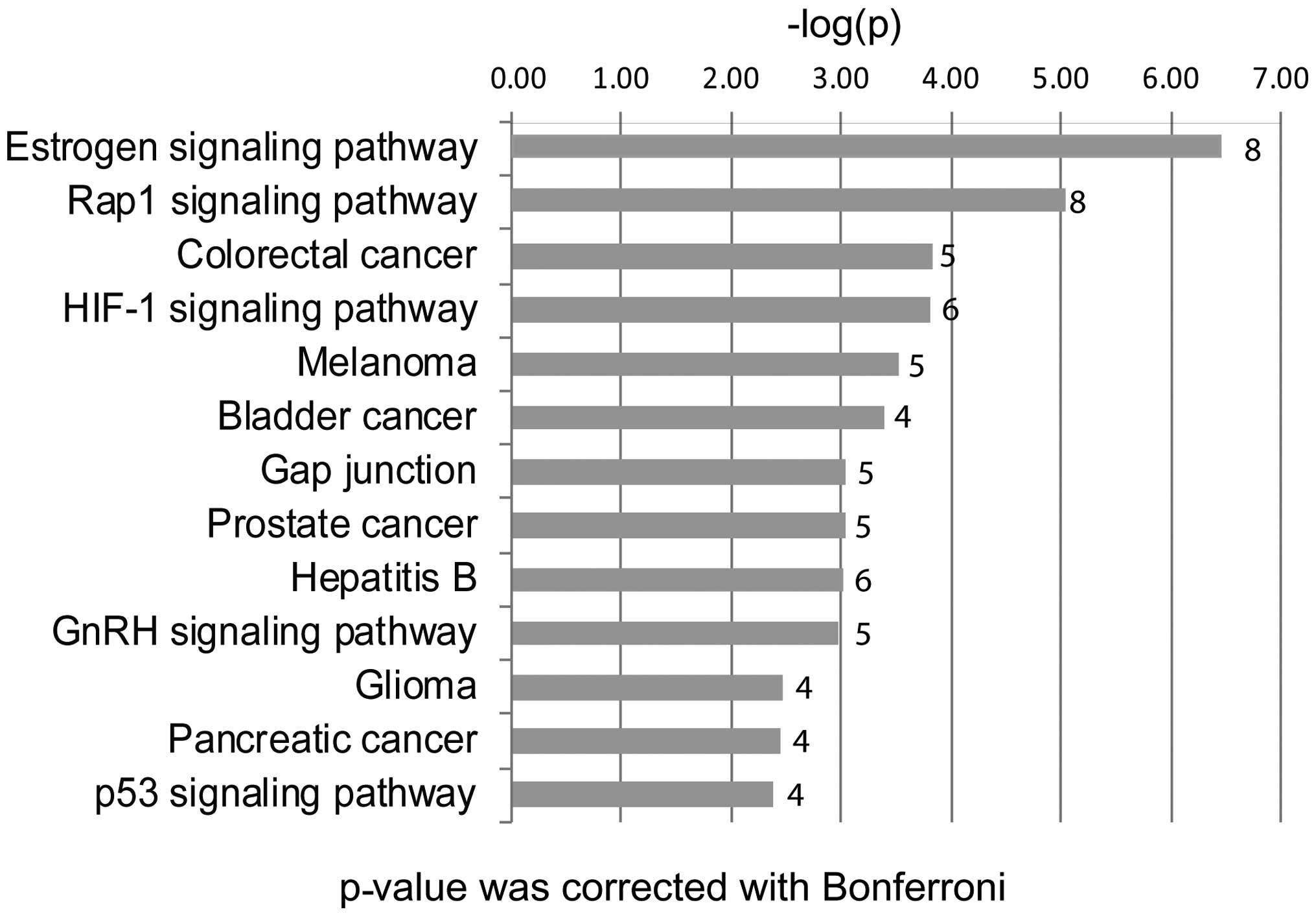
ClueGO (Fig. 4 and
Table II), revealed the enrichment
and interrelation of GO terms and the pathways for these 44 genes,
the cut-off of the P-value corrected with Bonferroni's method for
the GO terms and the KEGG pathway are P<1.00E-4 and
P<1.00E-2, respectively.
 | Table IIGO term enrichment analysis of the
top 44 genes using ClueGO. |
Table II
GO term enrichment analysis of the
top 44 genes using ClueGO.
| GO term | No. of genes | Associated genes
(%) | Term P-value | Term P-value
corrected with Bonferroni's method |
|---|
| Negative regulation
of response to external stimulus | 9 | 5.142857 | 8.32676E-10 | 4.57972E-08 |
| Positive regulation
of lipid metabolic process | 7 | 6.3636365 | 1.70611E-08 | 8.87178E-07 |
| Regulation of lipid
biosynthetic process | 6 | 5.1282053 | 7.02603E-07 | 3.44276E-05 |
| Positive regulation
of phospholipase C activity | 5 | 6.9444447 | 1.42718E-06 | 6.70775E-05 |
| Regulation of
phospholipase C activity | 5 | 6.849315 | 1.52905E-06 | 7.03362E-05 |
By using ClueGo function analysis, negative
regulation of response to external stimulus was most likely
associated with melanoma. As shown in Fig. 5, 9 top hub genes associated with
negative regulation of response to external stimulus were detected,
which included 5 downregulated genes (ANXA1, FGF2, LEP, NT5E and
PPARG; colored in light blue) and 4 upregulated genes (APOE, PRKCD,
SERPINE1 and TP53; colored in purple). Seven of the above 9 genes
are the first neighbors of the top gene, AKT1 gene, except for
ANXA1 and PRKCD.
We found that the 44 candidate genes were
significantly involved in several major pathways, such as the
estrogen signaling pathway, the RAP1 signaling pathway, the p53
signaling pathway, the gap junction and colorectal cancer. Eight of
the 44 top hub genes are associated with the estrogen signaling
pathway, including ADCY7, AKT1, EGFR, FOS, GNAI2, MMP2, PRKCD and
SRC. Nine of the 44 top hub genes were associated with the RAP1
signaling pathway, including ADCY7, AKT1, EGFR, FGF2, GNAI2, IGF1R,
LPAR1, RAC3 and SRC. AKT1 was shown to be involved in both
pathways.
Discussion
Melanoma is a high-risk skin cancer characterized by
atypical melanocyte proliferation and invasion. It is resistant to
apoptosis and contributes to the growth of melanoma, which leads to
metastatic melanoma, an untreatable condition.
In our study, we identified 1,310 DEGs in the gene
expression profile GSE3189 (11)
from the public genomics database Gene Expression Omnibus. After
PageRank analysis of the interaction network, AKT1 as well as 43
other genes were selected as the top hub genes from 1,310 DEGs
based on the Page-rank score (Table
I).
We also analyzed the functions of the top hub genes
by using ClueGo function analysis (Table II). The high significant enrichment
functions of the genes associated with negative regulation of
response to external stimulus were detected. Negative responses to
external stimulus caused cells to adapt to changes in external
stimulation for survival. Tumor cells effectuate this adaptive
behavior in order to escape immune system attack by inducing immune
tolerance and resisting apoptosis. We found that 9 top hub genes
were highly associated with melanoma and had an effect on the
negative regulation of response to external stimulus. Seven genes
highly corresponded to the top one, AKT1 gene.
The AKT1 gene, which encodes a serine-threonine
protein kinase named AKT kinase, was found to have higher
expression in our study. The higher expression of AKT as well as
PI3K primarily induced the proliferation of melanoma cells
(22) suggesting that AKT is
negatively correlated with autophagy to resist apoptosis. According
to recent studies, basal autophagy is down-modulated in primary
melanomas, and autophagy inhibition specifically targets the
metastatic melanoma cells (2).
Blocking the higher AKT activity in primary melanoma is sufficient
to promote autophagy.
The RAS-RAF-MEK-MAPK and PI3K-AKT pathways play
major roles in the regulation of proliferation and survival. The
mutation of the BRAF gene is common in metastatic melanomas, which
triggers the activation of the mitogen-activated protein kinase
(MAPK) pathway-induced cell proliferation and survival (23). Depletion of BRAF as well as the BRAF
gene mutation results in significantly reduced cell proliferation
through inhibition of extracellular signal-regulated kinase 1/2
(ERK1/2) activation and mitogen-activated protein kinase 1/2
(MEK1/2) activation (24).
Therapies using BRAFi have shown a dramatic clinical efficacy in
melanoma; however, the efficacy of BRAFi is short-lived due to
acquired drug resistance (25,26).
In order to restore BRAFi sensitivity, inhibitors of the PI3K-AKT
pathway as well as a mitogen-activated protein kinase inhibitor
(MEKi) are required for BRAFi resistance (27). BRAFi treatment was found to lead to
rebound levels of phosphatidylinositol (3,4,5)-trisphosphate
(PIP3) and p-AKT, which participate in melanoma survival.
MEK-dependent PTEN expression was found to limit PIP3 phosphate
accumulation and AKT signaling (2).
Thus, MAPK pathway inhibition enhances the PI3K-AKT signaling
pathway and melanoma drug resistance.
An important finding was that the significantly
enriched functions of melanoma top hub genes were associated with
positive regulation of the lipid metabolic process and regulation
of the lipid biosynthetic process.
Specifically, the AKT1 gene is involved in the
aforementioned two functions suggesting that AKT regulates lipid
metabolic processes and may be involved in melanoma progression and
promotion of tumor growth. Recent research found that adipocytes
promote melanoma cell growth by activating AKT (22), which provides evidence to confirm
our hypothesis.
Through the KEGG pathway analysis, we detected that
the most relevant pathway linked to melanoma is the estrogen
signaling pathway and a secondary relative pathway named the RAP1
signaling pathway. The AKT1 gene is also involved in these two
pathways in our study.
The most significant pathway was the estrogen
signaling pathway, which is highly associated with melanoma
according to our study. Augmenting the efficacy of the estrogen
signaling pathway to affect the growth of melanoma requires its
efficacy to focus on immune tolerance as well as apoptosis
resistance. The estrogen signaling pathway modulates immune
tolerance by inducing IL-10 secretion and inhibiting TNF-α
secretion in T cells (27).
Estradiol-17β (E2) at low concentrations causes the immune system
to regulate immune tolerance to a non-self antigen to maintain the
progression of melanoma. Estrogen can also downregulate the release
of pro-inflammatory cytokines by inhibiting transcription NF-κB
activation, which causes resistance to cell apoptosis (27).
Increasing evidence shows that the estrogen
signaling pathway plays an important role in maintaining
self-tolerance and modulating tolerance to non-self antigens, which
contributes to melanoma proliferation. Estrogen was found to exert
a proliferative effect on melanocytes and block cell cycle
progression in the G1 phase, which led to the development of
hyperpigmentation and to melanoma (28). Estrogen also prevents apoptosis.
Estrogen when linked to an NOS inhibitor exerted significantly
higher anti-proliferation to induce prominent apoptosis in melanoma
cells (10). Angiolymphoid
hyperplasia is also driven by estrogen-promoting tumor growth
(10,29).
The PI3K-PTEN-AKT and MAPK pathways are also
involved in the estrogen signaling pathway. Tamoxifen, an
anti-estrogen agent, was found to suppress phosphorylated ERK1/2
and AKT, thereby inhibiting mouse melanoma cell migration,
invasion, and metastasis (30).
This may explain why estrogen affects the growth of melanoma.
In our study, we also detected that the RAP1
signaling pathway was associated with melanoma, and regulates FA in
a negative feedback mechanism to mediate cell migration and
invasion in melanoma. RAP1-GTP-interacting adaptor molecule (RIAM)
is an adapter protein involved in FA dynamics, and its depletion
leads to defective melanoma cell migration and invasion through
inhibition of the MEK-ERK pathway (31).
In summary, our study highlighted the top PageRank
hub gene AKT1 and its correlation with the estrogen signaling and
RAP1 signaling pathways to alter the proliferation and apoptosis of
melanoma cells. Analysis of the enrichment functions of genes
associated with melanoma show promise in elucidating the exact
mechanisms of melanoma and to bring about advancements to a full
potential in novel targeted cancer therapy.
References
|
1
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. CA Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Maes H, Martin S, Verfaillie T and
Agostinis P: Dynamic interplay between autophagic flux and Akt
during melanoma progression in vitro. Exp Dermatol. 23:101–106.
2014. View Article : Google Scholar
|
|
3
|
Shi H, Hong A, Kong X, Koya RC, Song C,
Moriceau G, Hugo W, Yu CC, Ng C, Chodon T, et al: A novel AKT1
mutant amplifies an adaptive melanoma response to BRAF inhibition.
Cancer Discov. 4:69–79. 2014. View Article : Google Scholar :
|
|
4
|
Holtan SG, Creedon DJ, Haluska P and
Markovic SN: Cancer and pregnancy: parallels in growth, invasion,
and immune modulation and implications for cancer therapeutic
agents. Mayo Clin Proc. 84:985–1000. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Miller JG and Mac Neil S: Gender and
cutaneous melanoma. Br J Dermatol. 136:657–665. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Enninga EA, Holtan SG, Creedon DJ, Dronca
RS, Nevala WK, Ognjanovic S and Markovic SN: Immunomodulatory
effects of sex hormones: requirements for pregnancy and relevance
in melanoma. Mayo Clin Proc. 89:520–535. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Janik ME, Bełkot K and Przybyło M: Is
oestrogen an important player in melanoma progression? Contemp
Oncol (Pozn). 18:302–306. 2014.
|
|
8
|
Stevenson S and Thornton J: Effect of
estrogens on skin aging and the potential role of SERMs. Clin
Interv Aging. 2:283–297. 2007.PubMed/NCBI
|
|
9
|
Bredow L, Stutzel L, Bohringer D, Gundlach
E, Reinhard T and Auw-Haedrich C: Progesterone and estrogen
receptors in conjunctival melanoma and nevi. Graefes Arch Clin Exp
Ophthalmol. 252:359–365. 2014. View Article : Google Scholar
|
|
10
|
Roy S, Reddy BS, Sudhakar G, Kumar JM and
Banerjee R: 17β-estradiol-linked nitro-L-arginine as simultaneous
inducer of apoptosis in melanoma and tumor-angiogenic vascular
endothelial cells. Mol Pharm. 8:350–359. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Talantov D, Mazumder A, Yu JX, Briggs T,
Jiang Y, Backus J, Atkins D and Wang Y: Novel genes associated with
malignant melanoma but not benign melanocytic lesions. Clin Cancer
Res. 11:7234–7242. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hipfel R, Garbe C and Schittek B: RNA
isolation from human skin tissues for colorimetric differential
display. J Biochem Biophys Methods. 37:131–135. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kerr MK: Linear models for microarray data
analysis: hidden similarities and differences. J Comput Biol.
10:891–901. 2003. View Article : Google Scholar
|
|
14
|
Benjamini Y, Drai D, Elmer G, Kafkafi N
and Golani I: Controlling the false discovery rate in behavior
genetics research. Behav Brain Res. 125:279–284. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Reiner A, Yekutieli D and Benjamini Y:
Identifying differentially expressed genes using false discovery
rate controlling procedures. Bioinformatics. 19:368–375. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Franceschini A, Szklarczyk D, Frankild S,
Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering
C, et al: STRING v9.1: protein-protein interaction networks, with
increased coverage and integration. Nucleic Acids Res.
41:D808–D815. 2013. View Article : Google Scholar :
|
|
17
|
Assenov Y, Ramírez F, Schelhorn SE,
Lengauer T and Albrecht M: Computing topological parameters of
biological networks. Bioinformatics. 24:282–284. 2008. View Article : Google Scholar
|
|
18
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: a
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Page L, Brin S, Motwani R and Winograd T:
The PageRank Citation Ranking: Bringing Order to the Web. Technical
Report. Stanford InfoLab; 1999, http://ilpubs.stanford.edu:8090/422/.
|
|
20
|
Griffiths TL, Steyvers M and Firl A:
Google and the mind: predicting fluency with PageRank. Psychol Sci.
18:1069–1076. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bindea G, Mlecnik B, Hackl H, Charoentong
P, Tosolini M, Kirilovsky A, Fridman WH, Pagès F, Trajanoski Z and
Galon J: ClueGO: A Cytoscape plug-in to decipher functionally
grouped gene ontology and pathway annotation networks.
Bioinformatics. 25:1091–1093. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kwan HY, Fu X, Liu B, Chao X, Chan CL, Cao
H, Su T, Tse AK, Fong WF and Yu ZL: Subcutaneous adipocytes promote
melanoma cell growth by activating the Akt signaling pathway: role
of palmitic acid. J Biol Chem. 289:30525–30537. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Davies H, Bignell GR, Cox C, Stephens P,
Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W,
et al: Mutations of the BRAF gene in human cancer. Nature.
417:949–954. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Calipel A, Mouriaux F, Glotin AL, Malecaze
F, Faussat AM and Mascarelli F: Extracellular signal-regulated
kinase-dependent proliferation is mediated through the protein
kinase A/B-Raf pathway in human uveal melanoma cells. J Biol Chem.
281:9238–9250. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chapman PB, Hauschild A, Robert C, Haanen
JB, Ascierto P, Larkin J, Dummer R, Garbe C, Testori A, Maio M, et
al: Improved survival with vemurafenib in melanoma with BRAF V600E
mutation. N Engl J Med. 364:2507–2516. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lo RS: Combinatorial therapies to overcome
B-RAF inhibitor resistance in melanomas. Pharmacogenomics.
13:125–128. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zang YC, Halder JB, Hong J, Rivera VM and
Zhang JZ: Regulatory effects of estriol on T cell migration and
cytokine profile: inhibition of transcription factor NF-kappa B. J
Neuroimmunol. 124:106–114. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ribeiro MP, Silva FS, Paixão J, Santos AE
and Custódio JB: The combination of the antiestrogen endoxifen with
all-trans-retinoic acid has anti-proliferative and anti-migration
effects on melanoma cells without inducing significant toxicity in
non-neoplasic cells. Eur J Pharmacol. 715:354–362. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shifren JL, Tseng JF, Zaloudek CJ, Ryan
IP, Meng YG, Ferrara N, Jaffe RB and Taylor RN: Ovarian steroid
regulation of vascular endothelial growth factor in the human
endometrium: implications for angiogenesis during the menstrual
cycle and in the pathogenesis of endometriosis. J Clin Endocrinol
Metab. 81:3112–3118. 1996.PubMed/NCBI
|
|
30
|
Matsuoka H, Tsubaki M, Yamazoe Y, Ogaki M,
Satou T, Itoh T, Kusunoki T and Nishida S: Tamoxifen inhibits tumor
cell invasion and metastasis in mouse melanoma through suppression
of PKC/MEK/ERK and PKC/PI3K/Akt pathways. Exp Cell Res.
315:2022–2032. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Coló GP, Hernández-Varas P, Lock J,
Bartolomé RA, Arellano-Sánchez N, Strömblad S and Teixidó J: Focal
adhesion disassembly is regulated by a RIAM to MEK-1 pathway. J
Cell Sci. 125:5338–5352. 2012. View Article : Google Scholar : PubMed/NCBI
|