Introduction
Lung cancer is the leading cause of cancer-related
deaths in the majority of countries worldwide and causes 1 million
deaths each year (1–5). According to the pathological
diagnosis, lung cancer can be divided into small cell lung cancer
and non-small cell lung cancer (NSCLC), of which the latter
accounts for ~85% of all diagnosed cases. Although great progress
has been made in the prevention and early diagnosis of lung cancer
by the development of medical technology such as surgery,
radiotherapy, chemotherapy and molecular-targeted therapy, the
overall prognosis of lung cancer is poor and its 5-year survival
rate is only ~15% (6).
The molecular level of tumor occurrence, development
and invasion has become a hotspot of scientific research, and
increasing attention is being focused on the correlation between
tumor metastasis genes and patient prognosis. Numerous studies have
shown that the occurrence and development of lung cancer is a
complex process of multiple gene-factor interactions. Researchers
are seeking molecular markers that may influence the prognosis of
lung cancer.
Metastasis suppressor 1 (MTSS1), also known as
missing in metastasis (MIM), MIM-B, basal cell carcinoma-enriched
gene 4 (BEG4) or KIAA0429 identified by Lee et al as a
transcript missing in the metastasis of bladder cancer (7), is a newly identified actin-binding
protein that is mainly involved in cytoskeletal remodeling, signal
transduction and transcriptional activation, and closely associated
with tumor growth and invasion.
Numerous scholars have thoroughly investigated
MTSS1; Loberg et al found that reduced MIM-A gene expression
in prostate and other cancers may contribute to tumor growth and
development as well as metastasis (8). In 2011, Mustafa et al also
reported that MTSS1 overexpression clearly inhibited prostate
cancer cell metastasis, growth and adherence (9). In a study of breast cancer, Parr and
Jiang reported that patients with reduced MTSS1 levels had poorer
prognosis. High MTSS1 levels correlated with increased patient
overall and disease-free survival. Furthermore, MTSS1
overexpression significantly suppressed the invasive, migratory,
growth and adherence properties of a human breast cancer cell line.
In contrast, MTSS1 knockdown dramatically enhanced these properties
(10).
The prognostic value of MTSS1 has also been
demonstrated in esophageal squamous cell carcinoma, and patients
with high MTSS1 expression levels had a favorable prognosis
compared to those with reduced MTSS1 expression levels (11). A gastric cancer study in a Chinese
population also revealed that the MTSS1 expression levels were
increased in tissues adjacent to carcinoma, but clearly reduced in
cancer tissues, and MTSS1 expression levels were gradually reduced
as histological differentiation degree decreased (12). In the hematopoietic system,
MTSS1-knockout mice had an increased propensity to develop
aggressive B cell lymphomas (13,14).
These studies suggested that MTSS1 acts as a tumor metastasis
suppressor gene in these malignancies, that low MTSS1 expression
levels confer a poorer prognosis and that higher expression levels
correlate with improved overall survival rates. However, in
colorectal cancer, high MTSS1 expression was recently shown to
correlate with poor prognosis (15)
and disease progression in a subset of human melanoma cases
(16).
Therefore, what is the effect of MTSS1 on NSCLC? In
the present study, we chose the NSCLC cell line H1299 and 223
patients with NSCLC [136 with squamous cell carcinoma (SCC) and 87
with adenocarcinoma (ADCA), the two main types of NSCLC] as the
research subjects. The target gene LV-MTSS1 was transfected into
H1299 to analyze the effect of MTSS1 on proliferation, migration
and invasion. The tissue microarray was stained to explore the
correlation between MTSS1 and NSCLC clinicopathological
characteristics and prognosis by statistical analysis.
Materials and methods
Cell culture
NSCLC cell line H1299 was obtained from GeneChem
Co., Ltd. (Shanghai, China). We maintained H1299 in Roswell Park
Memorial Institute (RPMI)-1640 culture medium (Invitrogen Life
Technologies, Carlsbad, CA, USA) supplemented with 10%
heat-inactivated fetal bovine serum (FBS) (HyClone, Logan, UT, USA)
at 37°C with 5% CO2 in a humidified atmosphere. All
experiments were conducted in accordance with the Ethical Standards
of the Declaration of Helsinki and according to national and
international guidelines, and have been approved by the Ethics
Committee of the Affiliated Hospital of Nantong University.
Cell transfection
MTSS1-overexpressing lentivirus (LV-MTSS1) and the
negative control lentivirus (LV-NC) were purchased from GeneChem
Co., Ltd. The highest transfection efficiency was evident when the
virus titer was 1×109 Tu. A total of 2×105
H1299 cells were planted and maintained on a 6-well plate (Corning
Inc., Corning, NY, USA) to achieve 80–90% confluency and then
transfection was carried out according to the manufacturer's
instructions (GeneChem Co., Ltd.). After transfection, cells were
starved for green fluorescent protein (GFP) and fluorescence
detection was performed using an EVOS FL Auto (Invitrogen Life
Technologies).
Quantitative real-time polymerase chain
reaction
Cells in the different groups were cultured in
6-well plates with expansion medium (2 ml/well) at 80–90%
confluency, and total RNA was isolated using a UNIQ-10 Spin Column
RNA Purified kit (Sangon, Shanghai, China). The first strand of
complementary DNA (cDNA) was synthesized using a RevertAid™ First
Strand cDNA Synthesis kit (Fermentas, Burlington, Canada). and was
subsequently subjected to processing by a Corbett RG-6000
polymerase chain reaction system (Qiagen, Dusseldorf, German) using
FastStart Universal SYBR-Green Master Mix (Roche, Basel,
Switzerland). The reactions were optimized by varying the annealing
temperatures at 52–55°C, and the sense and antisense primers were
synthesized as follows: glyceraldehyde 3-phosphate dehydrogenase,
5′-GCAAGTTCAACGGCACAG-3′ and 5′-GCCAGTAGACTCCACGACAT-3′; MTSS1,
5′-GAGGAGATGGAGGCTTGTGA-3′ and 5′-TGGTTGTCTGGGTGCTGTAG-3′.
Fold-changes in mRNA expression were determined using the
2−ΔΔCt method (17).
Western blot analysis
H1299 cells were lysed by RIPA lysis buffer on ice
and total protein was extracted using a protein extraction kit
(both from Beyotime, Jiangsu, China) according to the
manufacturer's instructions. A bicinchoninic kit (Thermo Fisher
Scientific, Pittsburgh, PA, USA) was used to detect the
concentration of extracted protein. A total of 50 µg of
protein was vertically electrophoresed through sodium
dodecylsulfate-polyacrylamide gels and the separated proteins were
electronically transferred to polyvinylidene difluoride membranes.
Next, the non-specific interactions were blocked by 5% defatted
milk-Tris-buffered saline and Tween-20 solution and incubated at
37°C for 1 h. After being washed, specific antibodies against MTSS1
(1:50; Abcam, San Francisco, CA, USA) and β-actin (1:2,000;
Beyotime) were applied to incubate the membranes at 4°C for 12 h to
detect corresponding proteins. The membranes were then washed and
incubated with horseradish peroxidase-conjugated secondary
antibodies (Bioss, Woburn, MA, USA) for 1 h at 37°C. Immunoblots
were then detected by enhanced chemiluminescence reagents
(Invitrogen Life Technologies) and finally exposed to X-ray films.
Software Quantity One (Bio-Rad, Hercules, CA, USA) was used to
analyze the relative protein expression levels, while β-actin was
introduced as the internal reference.
Cell proliferation assay
Cells in the different groups were seeded onto
96-well plates in 100 µl of RPMI-1640 with 10% FBS at a
density of 2,000 cells/well. Cell viability was determined after
24, 48 and 72 h using a Cell Counting Kit-8 (CCK-8) (Beyotime)
according to the manufacturer's instructions. The optical density
(OD) at 450 nm was detected using a Synergy 2 enzyme mark
instrument (BioTek, Winooski, VT, USA).
Wound healing assay
After transfection, confluent monolayers of cells
were scratched with a 1,000 µl pipette tip to induce a
wound. The wound edges were imaged using a Leica DM IRB inverted
microscope (Leica, Solms, Germany). Images were collected at 0, 24
and 48 h after wounding. Images were quantified by measuring the
number of cells across the initial wounded edge (black line) at
each time-point.
Cell migration and invasion assays
The migration and invasion abilities of the H1299
cells were assessed using Transwell plates with 8-µm pore
membranes (Corning Inc.) according to protocols previously
described (18). Briefly, the H1299
cells were cultured in RPMI-1640 medium for 24 h and the medium was
collected as conditioning medium. For the invasion assay, Matrigel
(0.1 mg/ml; GeneChem Co., Ltd.) was coated on the top surface of
the Transwell chambers. The treated cells were seeded into the
chambers and incubated in a humidified environment with 5%
CO2 at 37°C for 48 h. The cells that passed through the
polyethylene membrane (migrated cells) or through the Matrigel
(invaded cells) were fixed with methanol. Crystal violet staining
was then used to indicate migration or invasion under the EVOS FL
Auto.
Patients and tissue samples
NSCLC tissues were collected from surgical resection
specimens of 223 patients who had not undergone radiotherapy or
chemotherapy in the Affiliated Hospital of Nantong University
between September 2006 and December 2010. Tumor tissues including
136 SCC and 87 ADCA were used to construct a tissue microarray.
Briefly, the tumor of each patient was represented by 2.0-mm cores.
Histotypes were confirmed using hematoxylin and eosin staining. The
Tumor-Node-Metastasis (TNM) staging was according to the 7th
edition of the TNM Staging System for Lung Cancer (19). Written consent was obtained from all
patients before sample analysis, and all investigations were
conducted in accordance with the Ethical Standards according to the
Declaration of Helsinki and according to national and international
guidelines and were approved by the Ethics Committee of the
Affiliated Hospital of Nantong University.
Immunohistochemistry and evaluation of
staining
The immunohistochemical analysis was performed as
previously described (20). Rabbit
anti-MTSS1 polyclonal (1:50; Abcam) antibody was used for detection
and Leica microscopy was used to capture the images. The
immunostaining of these sections was evaluated by two independent
experienced pathologists who were unaware of the
clinicopathological data and patient outcomes. The MTSS1 labeling
score was defined by multiplying the percentage of positive cells
and staining intensity (21). The
percentages of MTSS1-positive cells were scored and placed into
four categories according to staining proportion: <10%=1;
10–50%=2; >50–75%=3; and >75%=4. The MTSS1 staining
intensities were divided into four grades: no staining, 0; weak
staining 1; moderate staining 2; and strong staining 3. MTSS1
positivity was determined using the following formula: Overall
scores = percentage score × intensity score. Overall scores of
<3 and ≥3 were defined as negative and positive, respectively.
All samples were evaluated at a magnification of ×200.
Statistical methods
The data were subjected to Student's t-test or
one-way analysis of variance test. Associations between
clinicopathological variables and MTSS1 were examined by
χ2 tests. Survival curves were calculated by the
Kaplan-Meier method and analyzed by the log-rank test. The
multivariate analysis was performed using a Cox regression model.
P-values <0.05 were considered statistically significant. Data
were analyzed using SPSS 22.0 for Windows (SPSS, Inc., Chicago, IL,
USA).
Results
Efficiency of transfection and MTSS1
expression level detection
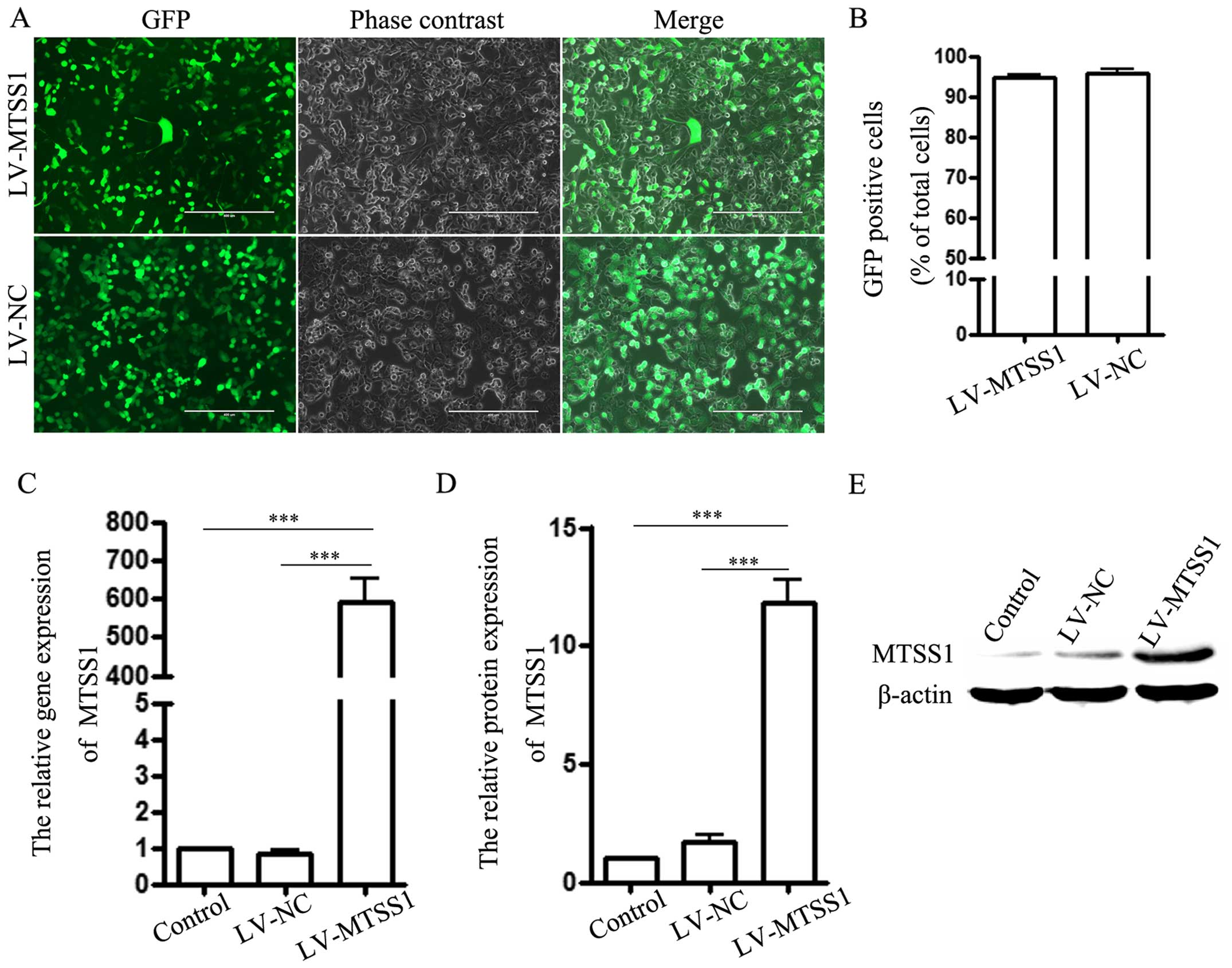
Transfection efficacy was detected using an
immunofluorescence technique. H1299 cells stably expressing
MTSS1-GFP (LV-MTSS1 group) or GFP alone (LV-NC group) were obtained
after lentivirus transfection (Fig.
1A). The transfection efficiency was ~95% (Fig. 1B). Next, real-time polymerase chain
reaction and western blotting were utilized to detect the relative
expression of MTSS1 in each group. The results indicated that MTSS1
expression in H1299 cells (control group) was low and the mRNA and
protein levels of MTSS1 were significantly increased in the
LV-MTSS1 group compared to the LV-NC and the control groups
(Fig. 1C–E). However, there was no
difference between the latter two groups.
Effect of MTSS1 on cell
proliferation
We examined cell proliferation following MTSS1
overexpression in the cells using the CCK-8 assay. The OD value
(450 nm) did not significantly differ among the three groups at 24,
48 or 72 h (Fig. 2). MTSS1 had no
effect on H1299 proliferation.
Effect of MTSS1 on cell migration and
invasion in H1299 cell lines
The Transwell results displayed that the number of
cells that penetrated the membrane in the LV-MTSS1 group was
significantly lower than those of the other two groups in both the
cell migration (Fig. 3A and C) and
cell invasion assays (Fig. 3B and
D). The same result was obtained with the wound healing assay
at 24 and 48 h in the LV-MTSS1 group (Fig. 3E and F). This finding implied that
MTSS1 suppressed cell migration and invasion.
MTSS1 expression of tissue
microarray
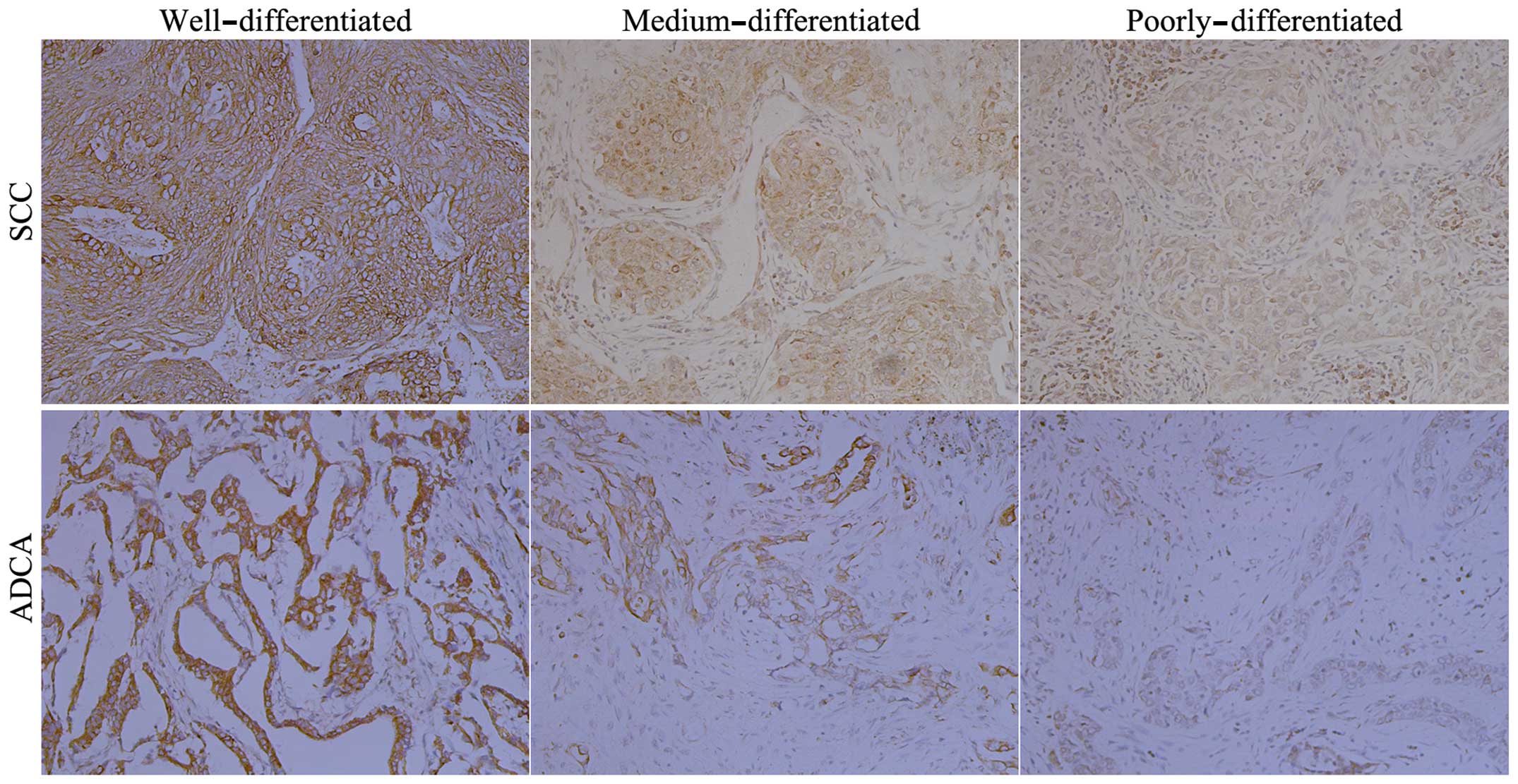
Representative tissue microarray immunohistochemical
staining in NSCLC observed for MTSS1 are shown in Fig. 4. The MTSS1 protein was mainly
located in the cytoplasm and strong staining was detected in
well-differentiated tissues, moderate staining was seen in
medium-differentiated tissues, and weak staining was seen in poorly
differentiated tissues (both SCC and ADCA). The overall MTSS1
expression level scores changed from high to low as the
differentiation degree changed from high to low.
Correlation between MTSS1 expression and
clinicopathological parameters in NSCLC
An overview of MTSS1 expression and
clinicopathological parameters is shown in Table I. MTSS1 expression was significantly
associated with tumor tissue differentiation degree (P<0.05),
TNM stage (P<0.05) and lymph node metastases (P<0.001),
respectively. There was no significant correlation between MTSS1
and variables such as age, gender, tumor histology, tumor size or
smoking history (all P>0.05).
 | Table ICorrelation of MTSS1 expression with
clinicopathological characteristics of the NSCLC cases. |
Table I
Correlation of MTSS1 expression with
clinicopathological characteristics of the NSCLC cases.
| Clinicopathological
characteristics | No. | MTSS1
| P-value |
|---|
| Negative | Positive |
|---|
| Age (years) | | | | 0.879 |
| <60 | 84 | 39 | 45 | |
| ≥60 | 139 | 66 | 73 | |
| Gender | | | | 0.678 |
| Male | 137 | 63 | 74 | |
| Female | 86 | 42 | 44 | |
| Histological
type | | | | 0.415 |
| SCC | 136 | 67 | 69 | |
| ADCA | 87 | 38 | 49 | |
|
Differentiation | | | | 0.010 |
| Well | 28 | 8 | 20 | |
| Medium | 87 | 33 | 54 | |
| Poorly | 108 | 64 | 44 | |
| Tumor size
(cm) | | | | 0.351 |
| <3 | 80 | 41 | 39 | |
| ≥3 | 143 | 64 | 79 | |
| Lymphatic
metastasis | | | | 0.000 |
| N0 | 145 | 48 | 97 | |
| N1+2 | 78 | 57 | 21 | |
| TNM | | | | 0.024 |
| I | 94 | 36 | 58 | |
| II | 51 | 23 | 28 | |
| III+IV | 78 | 46 | 32 | |
| Smoking
history | | | | 0.079 |
| Smoker | 147 | 63 | 84 | |
| Non-smoker | 76 | 42 | 34 | |
Survival analysis
Univariate survival analysis of these 223 patients
revealed that only 27 of the 105 (26%) patients in the
MTSS1-negative group were alive vs. 48 of 118 (38%) in the
MTSS1-positive group (Table II).
The Kaplan-Meier survival curves showed that decreased or absent
MTSS1 expression was significantly correlated with poor survival
(Fig. 5). When all variables were
compared separately with survival status, only differentiation
stage (P<0.001), TNM stage (P<0.001), lymph node metastases
(P<0.001), MTSS1 (P<0.05) tumor size (P<0.01)
significantly affected postoperative outcome (Table II). The Cox proportional hazards
regression model proved that MTSS1 (P<0.05), differentiation
stage (P<0.05), TNM stage (P<0.001), lymph node metastases
(P<0.01) and tumor size (P<0.05) were independent prognostic
factors in patients with NSCLC (Table
III).
 | Table IISurvival status and
clinicopathological parameters in 223 NSCLC specimens. |
Table II
Survival status and
clinicopathological parameters in 223 NSCLC specimens.
| Clinicopathological
parameters | No. | Survival status
| P-value |
|---|
| Alive | Dead |
|---|
| Age (years) | | | | 0.941 |
| <60 | 84 | 28 | 56 | |
| ≥60 | 139 | 47 | 91 | |
| Gender | | | | 0.139 |
| Male | 137 | 41 | 96 | |
| Female | 86 | 34 | 52 | |
| Histological
type | | | | 0.830 |
| SCC | 136 | 45 | 91 | |
| ADCA | 87 | 30 | 57 | |
|
Differentiation | | | | 0.000 |
| Well | 28 | 18 | 10 | |
| Medium | 87 | 35 | 52 | |
| Poorly | 108 | 22 | 86 | |
| Tumor size
(cm) | | | | 0.003 |
| <3 | 80 | 37 | 43 | |
| ≥3 | 143 | 38 | 105 | |
| Lymphatic
metastasis | | | | 0.000 |
| N0 | 145 | 65 | 80 | |
| N1+2 | 78 | 10 | 68 | |
| TNM | | | | 0.000 |
| I | 94 | 52 | 42 | |
| II | 51 | 16 | 35 | |
| III+IV | 78 | 17 | 71 | |
| MTSS1 | | | | 0.018 |
| Negative | 105 | 27 | 78 | |
| Positive | 118 | 48 | 70 | |
| Smoking
history | | | | 0.287 |
| Smoker | 147 | 53 | 94 | |
| Non-smoker | 76 | 22 | 54 | |
 | Table IIIContribution of various potential
prognostic factors to survival by Cox regression analysis in 223
NSCLC specimens. |
Table III
Contribution of various potential
prognostic factors to survival by Cox regression analysis in 223
NSCLC specimens.
|
Characteristics | Hazard ratio | 95% CI | P-value |
|---|
| Age | 1.282 | 0.891–1.845 | 0.180 |
| Gender | 0.990 | 0.682–1.437 | 0.958 |
| Histological
type | 1.256 | 0.871–1.812 | 0.223 |
|
Differentiation | 0.698 | 0.530–0.919 | 0.010 |
| Tumor size | 1.615 | 1.115–2.338 | 0.011 |
| Lymphatic
metastasis | 1.889 | 1.280–2.788 | 0.001 |
| TNM | 2.014 | 1.619–2.506 | 0.000 |
| MTSS1 | 0.669 | 0.466–0.960 | 0.029 |
| Smoking
history | 0.937 | 0.652–1.347 | 0.725 |
Discussion
Non-small cell lung cancer (NSCLC) remains the
leading cause of death in patients suffering from this malignant
disease, and its cure is rare due to the lack of an effective
screening method and typical early symptoms and as such, many
patients have distant metastases at the time of diagnosis (22). In the present study we aimed to
investigate the effect of MTSS1 on the proliferation, migration and
invasion of H1299 cells as well as the clinical significance of
MTSS1 in NSCLC.
The exact role of MTSS1 in tumor progression remains
under debate. Most of the research to date into gastric, breast,
and prostate cancer has implicated MTSS1 as a tumor suppressor and
that low MTSS1 expression levels confer poorer prognosis while
higher expression levels are correlated with improved overall
survival rates. Our results are consistent with the aforementioned
which revealed that: i) in tissue microarray and clinical
statistical analysis, MTSS1 protein was mainly expressed in the
plasma and that MTSS1 expression was significantly correlated with
tumor differentiation degree (P<0.05), TNM stage (P<0.05) and
lymph node metastasis (P<0.001); ii) in Kaplan-Meier survival
analysis, MTSS1 expression was positively related to patient
survival (P<0.01); and iii) in Cox multivariate survival
analysis, MTSS1 expression (P<0.05), tumor tissue
differentiation (P<0.05), tumor size (P<0.05), TNM stage
(P<0.001) and lymph node metastasis (P<0.01) were independent
factors for NSCLC prognosis.
The role of MTSS1 in cell proliferation differed
among tumor types and cell lines. MIM overexpression decreased the
PC-3 cell proliferation rate in prostate cancer (8), but increased it in head and neck
squamous cell carcinoma. In the present study, the proliferation
ability of the H1299 cells was not significantly suppressed or
promoted by MTSS1 overexpression.
Tumor metastasis is one of the deadliest steps in
cancer progression and currently the most difficult to overcome
(23,24). Tumor metastasis ability is
associated with cell migration and invasion capacity. The wound
healing and Transwell results displayed that cell migration and
invasion abilities in the LV-MTSS1 group were poorer than those in
the other two groups tested. In the clinical statistical analysis,
the positive expression rate of the MTSS1 protein in patients with
NSCLC and lymph node metastasis was 27% (21/78) vs. 67% (97/145) in
patients with NSCLC without lymph node metastasis, showing a
statistically significant difference (P<0.001).
The aforementioned findings suggested that MTSS1
inhibited H1299 cell migration and invasion, and its expression was
negatively correlated with lymph node metastasis. That is, MTSS1
may be able to indicate NSCLC tumor progression and the presence of
lymph node metastasis, which implies that MTSS1 is an
antimetastasis agent (10). Such a
role may be related to actin, as MTSS1 contains three introns and
four exons of 163 bp (2) to 2,903
bp (4) in length over 9.3 kb, and
its protein consists of 356 amino acids (25). MTSS1 has a N-terminal IRSp53/MIM
homology domain and a WASP-homology domain 2 at the C-terminus,
with which MTSS1 can be combined with actin and further influence
cell migration and invasion (26–31).
MTSS1 expression decline and loss may lead to changes in tyrosine
kinase, which changes the binding capacity of MTSS1 and actin.
Finally, the cytoskeletal changes lead to primary tumor cells loss
and transfer (27,32). The present study showed that the
cytoskeleton structural changes that induce apoptosis can be used
to treat cancer (33–35).
The mechanism involved in the MTSS1 expression
decrease in most tumors may be associated with CpG island DNA
methylation or loss of heterozygosity (LOH) (36). So-called CpG island refers to a
region of DNA in which C stands for cytosine, G stands for guanine,
and p represents the ester bond between them. This DNA contains
large proportions of C and G. In the human genome, dinucleotides
CpG are not evenly distributed. A large non-methylated CpG diploid
is termed a CpG island. Utikal et al (36) reported that a CpG island exists in
the MTSS1 5′-flanking promoter and pointed out that MIM-induced
cell growth inhibition is anchorage-independent, but related to CpG
island DNA methylation within the promoter region. They identified
and cloned the MIM promoter region and found that the main promoter
activity was located at the 5′-flanking sequence CpG island (276
bp). In a gastric cancer study, Yamashita et al (37) also confirmed CpG island DNA
methylation in the promoter region and found that MTSS1 was located
in a genomic region (8q22) with frequent LOH.
LOH means lack of an LOH molecule. A gene on a
chromosome when randomly deleted despite existence of the paired
chromosome, results in unpredictable mutations or deletions.
Chromosome 8 is involved in many human tumors (38,39),
while LOH is a very common genetic alteration in tumor cells
(40–44). MTSS1 expression was higher in normal
gastric mucosa but decreased or absent in gastric cancer tissues,
suggesting a correlation with a high frequency of LOH in the 5′
promoter sequence (37,45). The same results were also reported
about LOH in cancer of the gastroesophageal junction (46), cervical carcinoma (47) and in other progressed tumors which
include transcriptional suppression by DNMT3B (48) or microRNA-mediated effects (49–54).
In summary, MTSS1 can suppress H1299 migration and
invasion and be used as a new independent factor for NSCLC
prognosis. However the exact role of this adaptor molecule that
links intracellular signaling pathways with actin remodeling
(10,55,56)
remains under debate and may likely depend on cell type, molecular
context and disease type (15,16).
Acknowledgments
We would like thank Dr Xuefeng Tan for his support
and assistance with the preparation of this manuscript. The present
study was supported by grants from the University Natural Science
Research Project of Jiangsu Province (grant no. 14KJB310015), and a
project funded by the Natural Foundation of Nantong University
(grant no. 14ZY022).
Abbreviations:
|
MTSS1
|
metastasis suppressor 1
|
|
MIM
|
missing- in-metastasis
|
|
NSCLC
|
non-small cell lung cancer
|
|
BEG4
|
basal cell carcinoma-enriched gene
4
|
|
SCC
|
squamous cell carcinoma
|
|
ADCA
|
adenocarcinoma
|
|
GFP
|
green fluorescent protein
|
|
OD
|
optical density
|
|
CCK-8
|
Cell Counting Kit-8
|
References
|
1
|
Shibuya K and Hiraoka M: Radiation therapy
for lung cancer. Gan To Kagaku Ryoho. 34:544–549. 2007.In Japanese.
PubMed/NCBI
|
|
2
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. CA Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kayser G, Csanadi A, Otto C, Plönes T,
Bittermann N, Rawluk J, Passlick B and Werner M: Simultaneous
multi-antibody staining in non-small cell lung cancer strengthens
diagnostic accuracy especially in small tissue samples. PLoS One.
8:e563332013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kayser G, Sienel W, Kubitz B, Mattern D,
Stickeler E, Passlick B, Werner M and Zur Hausen A: Poor outcome in
primary non-small cell lung cancers is predicted by transketolase
TKTL1 expression. Pathology. 43:719–724. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Schiller JH, Harrington D, Belani CP,
Langer C, Sandler A, Krook J, Zhu J and Johnson DH; Eastern
Cooperative Oncology Group: Comparison of four chemotherapy
regimens for advanced non-small-cell lung cancer. N Engl J Med.
346:92–98. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lee YG, Macoska JA, Korenchuk S and Pienta
KJ: MIM, a potential metastasis suppressor gene in bladder cancer.
Neoplasia. 4:291–294. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Loberg RD, Neeley CK, Adam-Day LL, Fridman
Y, St John LN, Nixdorf S, Jackson P, Kalikin LM and Pienta KJ:
Differential expression analysis of MIM (MTSS1) splice variants and
a functional role of MIM in prostate cancer cell biology. Int J
Oncol. 26:1699–1705. 2005.PubMed/NCBI
|
|
9
|
Mustafa N, Martin TA and Jiang WG:
Metastasis tumour suppressor-1 and the aggressiveness of prostate
cancer cells. Exp Ther Med. 2:157–162. 2011.PubMed/NCBI
|
|
10
|
Parr C and Jiang WG: Metastasis suppressor
1 (MTSS1) demonstrates prognostic value and anti-metastatic
properties in breast cancer. Eur J Cancer. 45:1673–1683. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xie F, Ye L, Chen J, Wu N, Zhang Z, Yang
Y, Zhang L and Jiang WG: The impact of Metastasis Suppressor-1,
MTSS1, on oesophageal squamous cell carcinoma and its clinical
significance. J Transl Med. 9:952011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu K, Wang G, Ding H, Chen Y, Yu G and
Wang J: Downregulation of metastasis suppressor 1(MTSS1) is
associated with nodal metastasis and poor outcome in Chinese
patients with gastric cancer. BMC Cancer. 10:4282010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yu D, Zhan XH, Zhao XF, Williams MS, Carey
GB, Smith E, Scott D, Zhu J, Guo Y, Cherukuri S, et al: Mice
deficient in MIM expression are predisposed to lymphomagenesis.
Oncogene. 31:3561–3568. 2012. View Article : Google Scholar :
|
|
14
|
Schemionek M, Kharabi Masouleh B, Klaile
Y, Krug U, Hebestreit K, Schubert C, Dugas M, Büchner T, Wörmann B,
Hiddemann W, et al: Identification of the adapter molecule MTSS1 as
a potential oncogene-specific tumor suppressor in acute myeloid
leukemia. PLoS One. 10:e01257832015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang D, Xu MR, Wang T, Li T and Zhu J:
MTSS1 overexpression correlates with poor prognosis in colorectal
cancer. J Gastrointest Surg. 15:1205–1212. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mertz KD, Pathria G, Wagner C, Saarikangas
J, Sboner A, Romanov J, Gschaider M, Lenz F, Neumann F, Schreiner
W, et al: MTSS1 is a metastasis driver in a subset of human
melanomas. Nat Commun. 5:34652014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pfaffl MW: A new mathematical model for
relative quantification in real-time RT-PCR. Nucleic Acids Res.
29:e452001. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen K, Zhang S, Ji Y, Li J, An P, Ren H,
Liang R, Yang J and Li Z: Baicalein inhibits the invasion and
metastatic capabilities of hepatocellular carcinoma cells via
down-regulation of the ERK pathway. PLoS One. 8:e729272013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Goldstraw P: The 7th Edition of TNM in
Lung Cancer: what now? J Thorac Oncol. 4:671–673. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tischler V, Pfeifer M, Hausladen S,
Schirmer U, Bonde AK, Kristiansen G, Sos ML, Weder W, Moch H,
Altevogt P, et al: L1CAM protein expression is associated with poor
prognosis in non-small cell lung cancer. Mol Cancer. 10:1272011.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li JC, Yang XR, Sun HX, Xu Y, Zhou J, Qiu
SJ, Ke AW, Cui YH, Wang ZJ, Wang WM, et al: : Up-regulation of
Krüppel-like factor 8 promotes tumor invasion and indicates poor
prognosis for hepatocellular carcinoma. Gastroenterology.
139:2146–2157.e12. 2010. View Article : Google Scholar
|
|
22
|
Pao W: New approaches to targeted therapy
in lung cancer. Proc Am Thorac Soc. 9:72–73. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ishii K, Kinami S, Funaki K, Fujita H,
Ninomiya I, Fushida S, Fujimura T, Nishimura G and Kayahara M:
Detection of sentinel and non-sentinel lymph node micrometastases
by complete serial sectioning and immunohistochemical analysis for
gastric cancer. J Exp Clin Cancer Res. 27:72008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Guan-Zhen Y, Ying C, Can-Rong N, Guo-Dong
W, Jian-Xin Q and Jie-Jun W: Reduced protein expression of
metastasis-related genes (nm23, KISS1, KAI1 and p53) in lymph node
and liver metastases of gastric cancer. Int J Exp Pathol.
88:175–183. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mattila PK, Salminen M, Yamashiro T and
Lappalainen P: Mouse MIM, a tissue-specific regulator of
cytoskeletal dynamics, interacts with ATP-actin monomers through
its C-terminal WH2 domain. J Biol Chem. 278:8452–8459. 2003.
View Article : Google Scholar
|
|
26
|
Mattila PK, Pykäläinen A, Saarikangas J,
Paavilainen VO, Vihinen H, Jokitalo E and Lappalainen P:
Missing-in-metastasis and IRSp53 deform PI(4,5)P2-rich
membranes by an inverse BAR domain-like mechanism. J Cell Biol.
176:953–964. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lee SH, Kerff F, Chereau D, Ferron F, Klug
A and Dominguez R: Structural basis for the actin-binding function
of missing-in-metastasis. Structure. 15:145–155. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Suetsugu S, Murayama K, Sakamoto A,
Hanawa-Suetsugu K, Seto A, Oikawa T, Mishima C, Shirouzu M,
Takenawa T and Yokoyama S: The RAC binding domain/IRSp53-MIM
homology domain of IRSp53 induces RAC-dependent membrane
deformation. J Biol Chem. 281:35347–35358. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Millard TH, Bompard G, Heung MY, Dafforn
TR, Scott DJ, Machesky LM and Fütterer K: Structural basis of
filopodia formation induced by the IRSp53/MIM homology domain of
human IRSp53. EMBO J. 24:240–250. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bompard G, Sharp SJ, Freiss G and Machesky
LM: Involvement of Rac in actin cytoskeleton rearrangements induced
by MIM-B. J Cell Sci. 118:5393–5403. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Pollard TD and Borisy GG: Cellular
motility driven by assembly and disassembly of actin filaments.
Cell. 112:453–465. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhong C, Kinch MS and Burridge K:
Rho-stimulated contractility contributes to the fibroblastic
phenotype of Ras-transformed epithelial cells. Mol Biol Cell.
8:2329–2344. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Scita G, Confalonieri S, Lappalainen P and
Suetsugu S: IRSp53: Crossing the road of membrane and actin
dynamics in the formation of membrane protrusions. Trends Cell
Biol. 18:52–60. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Maecker HT, Todd SC and Levy S: The
tetraspanin superfamily: Molecular facilitators. FASEB J.
11:428–442. 1997.PubMed/NCBI
|
|
35
|
Kelley LC, Shahab S and Weed SA: Actin
cytoskeletal mediators of motility and invasion amplified and
overexpressed in head and neck cancer. Clin Exp Metastasis.
25:289–304. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Utikal J, Gratchev A, Muller-Molinet I,
Oerther S, Kzhyshkowska J, Arens N, Grobholz R, Kannookadan S and
Goerdt S: The expression of metastasis suppressor MIM/MTSS1 is
regulated by DNA methylation. Int J Cancer. 119:2287–2293. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yamashita S, Tsujino Y, Moriguchi K,
Tatematsu M and Ushijima T: Chemical genomic screening for
methylation-silenced genes in gastric cancer cell lines using
5-aza-2′-deoxycytidine treatment and oligonucleotide microarray.
Cancer Sci. 97:64–71. 2006. View Article : Google Scholar
|
|
38
|
Mazurenko NN, Bliev AIu, Bidzhieva BA,
Peskov DIu, Snigur NV, Savinova EB and Kiselev FL: Loss of
heterozygosity at chromosome 6 as a marker of early genetic
alterations in cervical intraepithelial neoplasias and
microinvasive carcinomas. Mol Biol. 40:436–447. 2006.In Russian.
View Article : Google Scholar
|
|
39
|
Mark HF, Feldman D, Samy M, Sun C, Das S,
Mark S and Lathrop J: Assessment of chromosome 8 copy number in
cervical cancer by fluorescent in situ hybridization. Exp Mol
Pathol. 66:157–162. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ocádiz R, Sauceda R, Salcedo M, Ortega V,
Rodríguez H, Gordillo C, Chávez P and Gariglio P: Occurrence of
human papillomavirus type 16 DNA sequences and c-myc oncogene
alterations in uterine-cervix carcinoma. Arch Invest Med.
20:355–362. 1989.
|
|
41
|
Ocadiz R, Sauceda R, Cruz M, Graef AM and
Gariglio P: High correlation between molecular alterations of the
c-myc oncogene and carcinoma of the uterine cervix. Cancer Res.
47:4173–4177. 1987.PubMed/NCBI
|
|
42
|
Kersemaekers AM, Fleuren GJ, Kenter GG,
Van den Broek LJ, Uljee SM, Hermans J and Van de Vijver MJ:
Oncogene alterations in carcinomas of the uterine cervix:
Overexpression of the epidermal growth factor receptor is
associated with poor prognosis. Clin Cancer Res. 5:577–586.
1999.PubMed/NCBI
|
|
43
|
Zhang A, Månér S, Betz R, Angström T,
Stendahl U, Bergman F, Zetterberg A and Wallin KL: Genetic
alterations in cervical carcinomas: Frequent low-level
amplifications of oncogenes are associated with human
papillomavirus infection. Int J Cancer. 101:427–433. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wang Y, Zheng E and Ke Y: Studies of loss
of heterozygosity (LOH) in Chinese human gastric cancer tissues.
Zhonghua Zhong Liu Za Zhi. 20:116–118. 1998.In Chinese.
|
|
45
|
Jones PA and Laird PW: Cancer epigenetics
comes of age. Nat Genet. 21:163–167. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
van Duin M, van Marion R, Vissers KJ, Hop
WC, Dinjens WN, Tilanus HW, Siersema PD and van Dekken H:
High-resolution array comparative genomic hybridization of
chromosome 8q: Evaluation of putative progression markers for
gastroesophageal junction adenocarcinomas. Cytogenet Genome Res.
118:130–137. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Bhattacharya N, Singh RK, Mondal S, Roy A,
Mondal R, Roychowdhury S and Panda CK: Analysis of molecular
alterations in chromosome 8 associated with the development of
uterine cervical carcinoma of Indian patients. Gynecol Oncol.
95:352–362. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Fan H, Chen L, Zhang F, Quan Y, Su X, Qiu
X, Zhao Z, Kong KL, Dong S, Song Y, et al: MTSS1, a novel target of
DNA methyltransferase 3B, functions as a tumor suppressor in
hepatocellular carcinoma. Oncogene. 31:2298–2308. 2012. View Article : Google Scholar
|
|
49
|
Hirata H, Ueno K, Shahryari V, Deng G,
Tanaka Y, Tabatabai ZL, Hinoda Y and Dahiya R: MicroRNA-182-5p
promotes cell invasion and proliferation by down regulating FOXF2,
RECK and MTSS1 genes in human prostate cancer. PLoS One.
8:e555022013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Liu S, Guo W, Shi J, Li N, Yu X, Xue J, Fu
X, Chu K, Lu C, Zhao J, et al: MicroRNA-135a contributes to the
development of portal vein tumor thrombus by promoting metastasis
in hepatocellular carcinoma. J Hepatol. 56:389–396. 2012.
View Article : Google Scholar
|
|
51
|
Liu Z, Liu J, Segura MF, Shao C, Lee P,
Gong Y, Hernando E and Wei JJ: MiR-182 overexpression in
tumourigenesis of high-grade serous ovarian carcinoma. J Pathol.
228:204–215. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Wang J, Li J, Shen J, Wang C, Yang L and
Zhang X: MicroRNA-182 downregulates metastasis suppressor 1 and
contributes to metastasis of hepatocellular carcinoma. BMC Cancer.
12:2272012. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Pasternak K, Nowacka O, Wróbel D,
Pieszyński I, Bryszewska M and Kujawa J: Influence of MLS laser
radiation on erythrocyte membrane fluidity and secondary structure
of human serum albumin. Mol Cell Biochem. 388:261–267. 2014.
View Article : Google Scholar :
|
|
54
|
Zhou W, Li X, Liu F, Xiao Z, He M, Shen S
and Liu S: MiR-135a promotes growth and invasion of colorectal
cancer via metastasis suppressor 1 in vitro. Acta Biochim Biophys
Sin. 44:838–846. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Machesky LM and Johnston SA: MIM: A
multifunctional scaffold protein. J Mol Med. 85:569–576. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Xie F, Ye L, Ta M, Zhang L and Jiang WG:
MTSS1: A multifunctional protein and its role in cancer invasion
and metastasis. Front Biosci. 3:621–631. 2011. View Article : Google Scholar
|