Introduction
Tubulin tyrosine ligase like 12 (TTLL12) is
the least characterized and the most divergent member of the TTLL
family, which possesses catalytic functions in tubulin
post-translational modifications (1–3).
However, TTLL12 is said to be a pseudo-enzyme having a
phylogenetically conserved association of two nonfunctional domains
including a SET-like domain and TTL-like domain, for which several
differences exist in the structure, compared with other members of
the TTLL family (4). These two
domains are associated with histone methylation and tubulin
modification, respectively. Recent studies have shown that TTLL12
could affect histone methylation, tubulin modification, mitotic
duration and chromosome ploidy in human larynx cancer cells
(5–8). In addition, there are numerous studies
that have revealed that the TTLL family is closely linked to human
cancer, such as neuroblastomas (9–11). For
example, the TTLL family is often suppressed in human cancer
(10) and is positively correlated
with a poor prognosis in breast cancer (11). In prostate cancer, the level of
TTLL12 expression was found to be increased in the proliferating
layer of benign tissue and during cancer progression to metastasis
(12,13). These findings suggest that TTLL12
may display the same pattern in other types of cancers, and it
could play a crucial role in tumorigenesis and tumor
progression.
In the present study, we found a new transcript
isoform of the TTLL12 gene, namely the 36 amino acid (AA)
plus isoform, with an additional 108-bp nucleotide sequence
inserted between exons 5 and 6 of the wild-type. This new isoform
may be the result of alternative splicing, and we identified it in
several lung cancer cell lines as well as other cancer cell lines.
Notably, some of the lung cancer cell lines presented a much higher
proportion of the 36AA plus isoform compared with the wild-type
TTLL12 transcript, implying the potential role of the new isoform
in lung cancer cells. Moreover, the inserted 36AA was predicted to
be part of a disordered region, which can alter the molecular
structure of the whole protein, and could affect adaptive and
deleterious-on gene expression and function.
Collectively, our discovery of a new transcript
isoform of TTLL12 with an additional 108 bp, predicted to be a
disordered region, provides an alternative prospective to study the
functions of TTLL12 in tumor development and progression. In
addition, our findings may also open a new window to explore
potential targets related to post-translational modification of
tubulin, which may eventually contribute to the development of more
selective agents for cancer therapy.
Materials and methods
Cell culture
Human lung cancer cell lines (H1299, 95-D, SPCA-1,
A549, SK-MES-1, PC-9, H2170 and Hcc-827), human esophageal cancer
cell lines (TE-11 and EC-109), human normal esophageal cell lines
(HET-1A and HEEpiC), human acute monocytic leukemia cell line
(THP-1), and human breast cancer cell line (MCF-7) were purchased
from the Cell Bank of the China Academy of Science (Shanghai,
China). THP-1, EC-109 and all the human lung cancer cell lines were
grown in RPMI-1640 medium (Life Technologies, Carlsbad, CA, USA)
supplemented with 10% fetal bovine serum (FBS; Gemini, Woodland,
CA, USA), 100 IU/ml penicillin and 100 µg/ml streptomycin. TE-11
and MCF-7 cells were grown in Dulbeccos modified Eagles medium
(DMEM) culture medium (Life Technologies) supplemented with 10%
FBS, 100 IU/ml penicillin and 100 µg/ml streptomycin. HET-1A and
HEEpiC were grown in EpiCM-2 complete medium (ScienCell, Carlsbad,
CA, USA) with 100 IU/ml penicillin and 100 µg/ml streptomycin. All
of the cells were cultured at 37°C in a humidified incubator with
5% CO2.
RNA isolation
Total RNA was isolated from the cell cultures using
TRIzol (Life Technologies) according to the manufacturer's
instructions. The total RNA was quantified based on absorbance at
260 nm and the integrity of purified RNA was confirmed by agarose
gel electrophoresis with a 28S/18S ratio not <1. The total RNA
was stored at −80°C.
3′-Rapid amplification of cDNA ends
(3′-RACE)
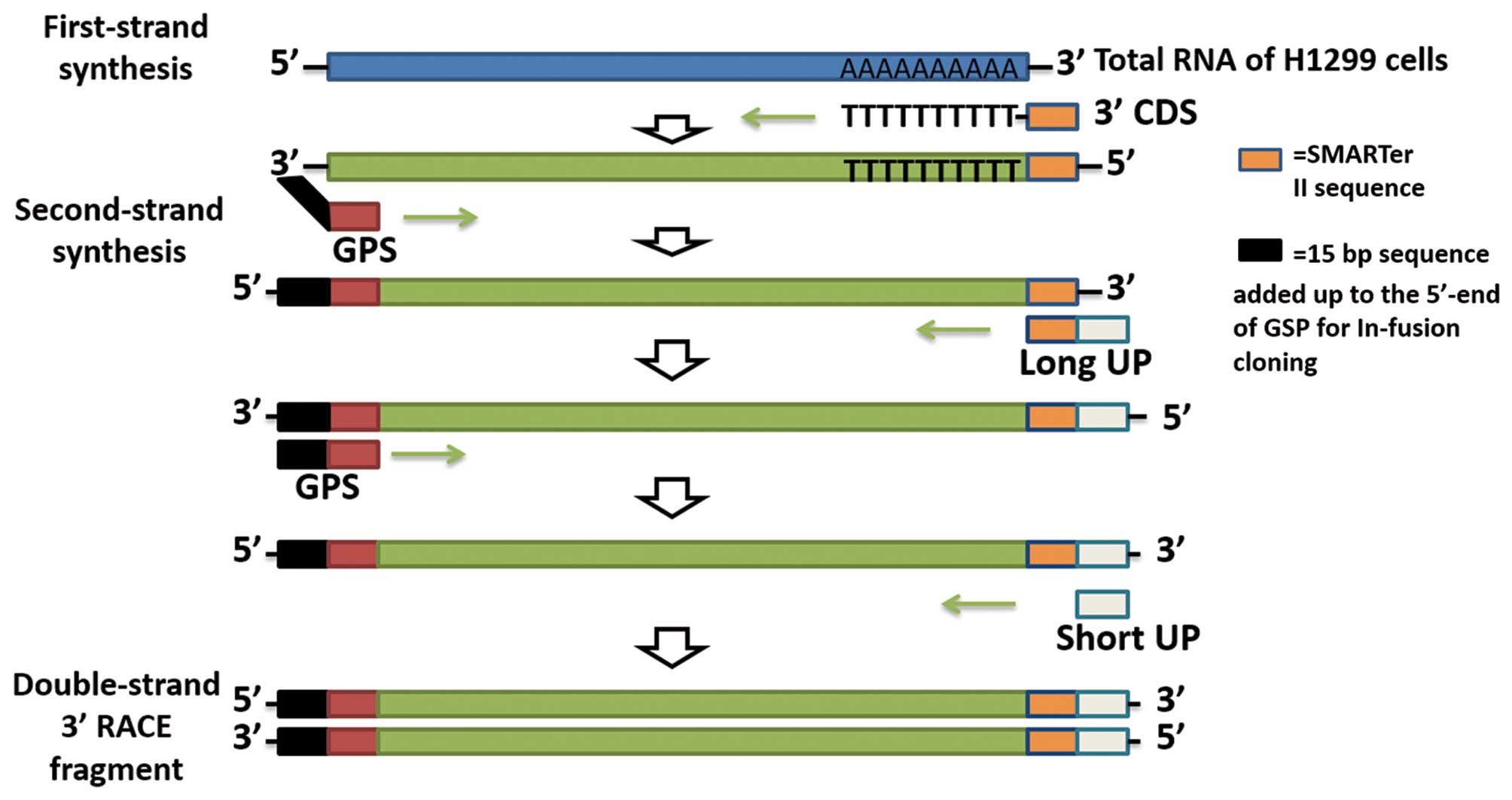
As displayed in Fig.
1, the full length of TTLL12 cDNA was amplified by 3′-RACE
(SMARTer RACE 3′ kit; Takara, Dalian, China). First-strand cDNA
synthesis from total RNA of H1299 was performed using a traditional
reverse transcription procedure, but with a special oligo(dT)
primer: [3′-RACE CDS primer, 5′-AAGCAGTGGTATCAACGCAGAGTAC (T)
30VN-3′; N=A, C, G or T; V=A, G, or C]. The first-strand cDNA
synthesis reaction products were diluted with 10 µl Tricine-EDTA
Buffer (Takara). The diluted first-strand cDNA was used as template
and second-strand synthesis was amplified with a 3′ gene-specific
primer (3′ GSP, 5′-GATTACGCCAAGCTTAGAGCACACAGACGGCGCGGGTG-3′) and
universal primer mix A (UPM; long,
5′-CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGT-3′ and short,
5′-CTAATACGACTCACTATAGGGC-3′). The 3′-RACE DNA samples were
electrophoresed on an agarose gel. The position of the desired
fragment was located under UV light and the products were extracted
with the NuceloSpin Gel and PCR Clean-Up kit (Takara).
In-fusion cloning of RACE
products
The 3′-RACE products were cloned into a pUC19 vector
by the In-Fusion HD Cloning kit (Takara) according to the protocol.
The recombinant vectors were then transformed into Stellar
Competent Cells (Takara) and the bacteria was spread on LB plates
containing 100 µg/ml of ampicillin. All of the plates were
incubated overnight at 37°C and individual isolated colonies were
randomly selected from each experimental plate. Plasmid DNA was
isolated using HiPure Plasmid EF Micro kit (Magen, Suzhou, China)
according to the manufacturer's instructions.
RT-PCR
First-strand cDNA from total RNA was constructed by
Transcript II All-in-One First-Strand cDNA Synthesis SuperMix kit
(Life Technologies) following the manufacturer's instructions. The
first-strand cDNA was then used as a template for PCR and the
specific primers used were as follows: TTLL12 sense primer
1, 5′-GAAGATGCCGGTGTGGTATA-3′ and TTLL12 antisense primer 1,
5′-CACGTCCGTGTAGACCTTGA-3′; TTLL12 sense primer 2,
5′-GAGACTTTGCCTACGGAGAGA-3′ and TTLL12 antisense primer 2,
5′-GCTGAGTTTCCTGTAGTCCTTGA-3′. Meanwhile, we used pUC19 vectors
inserted with TTLL12 wild-type or TTLL12 36AA plus isoform as
positive templates for PCR amplification of TTLL12 wild-type or
36AA plus isoform, respectively. PCR products were characterized by
electrophoresing on agarose gel. The desired position was
determined under UV light and PCR products were isolated by
NuceloSpin Gel and PCR Clean-Up kit.
Cloning and sequencing
PCR products were cloned into a pMD18-T Simple
Vector (Takara) and transformed into chemically competent cells.
Individual colonies were grown overnight at 37°C and plasmid DNA
was isolated using HiPure Plasmid EF Micro kit according to the
protocol, and processed to Sanger sequencing (Life
Technologies).
Bioinformatics analysis
Protein structures of the TTLL12 wild-type and new
TTLL12 36AA plus isoform were predicted by bioinformatics
softwares. The mRNA CDS and AA sequence of wild-type TTLL12 were
gained in NCBI (http://www.ncbi.nlm.nih.gov/blast). In addition, the
new transcript of TTLL12 was translated into a new AA sequence
using DNAStar EditSeq. Then, the AA sequence of the new TTLL12 36AA
plus isoform was aligned with the wild-type by NCBI protein blast
(http://blast.ncbi.nlm.nih.gov/).
Physicochemical properties of the wild-type and new isoform were
predicted by online tools ProtParam and ProtScale (http://www.expasy.ch/tools/protscale.html),
respectively. Secondary structures of the wild-type and new isoform
AA sequences were predicted by DNAStar Protean and online tool
CFSSP (http://www.biogem.org/tool/chou-fasman/). To further
investigate the function of the new isoform of the TTLL12
transcript, we also used the online prediction tool DisPort search
(http://www.disport.org/search.php) to
predict whether the additional 36 AA sequence is in a disordered
region of TTLL12.
Results
The full length of TTLL12 cDNA
amplified by 3′-RACE from total RNA of H1299 cells
To gain the full length cDNA of TTLL12, we first
generated the first-strand cDNA from total RNA of H1299 cells, and
then the first-strand cDNA was used as the template to amplify the
second-strand cDNA using 3′-RACE (Fig.
1). We aligned the 3′-RACE sequences with the consensus Homo
sapiens TTLL12 genomic sequence (wild-type) from GenBank, and
then found an additional 108-bp nucleotide sequence located at CDS
from 902 to 903 bases, between exons 5 and 6 (Fig. 2). This new isoform was called the
36AA plus isoform of TTLL12.
Detection of the new TTLL12 36AA plus
isoform in both lung cancer cell lines and other cancer cell
lines
To determine whether the TTLL12 36AA plus isoform
exists in human cancers, we isolated total RNAs from 8 human lung
cancer cell lines (H1299, 95-D, SPCA-1, A549, SK-MES-1, PC-9, H2170
and Hcc-827), 2 human esophageal cancer cell lines (TE-11 and
EC-109), 2 human normal esophageal cell lines (HET-1A, HEEpiC),
human acute monocytic leukemia cell line THP-1 and human breast
cancer cell line MCF-7. The reverse transcriptase-PCR products were
then used as the template for PCR. In PCR, the plasmids harboring
the 36AA plus isoform and wild-type of the TTLL12 gene sequence
were used as the positive templates for the 36AA plus isoform and
wild-type, respectively. All the PCR products were electrophoresed
on agarose gel. We then located the position of the desired
fragment under UV light and extracted the PCR products from the
gel. Extracted PCR products were subsequently cloned into a pMD18-T
Simple Vector and transformed into chemically competent cells. All
the plasmids from the positive clone were sequenced and aligned
with the TTLL12 36AA plus isoform and TTLL12 wild-type. As a
result, we found the product of the TTLL12 36AA plus isoform in all
of the detected cell lines, and 3 lung cancer cell lines (H1299,
H2170 and Hcc-827) showed the highest proportion of the 36AA plus
isoform compared to the wild-type (Fig.
3). In addition, the sequencing results of the plasmids
extracted from the positive clone were aligned, and demonstrated
that the PCR products were from the TTLL12 36AA plus isoform
(Fig. 4). Significantly, the TTLL12
36AA plus isoform was expressed more abundantly than the wild-type
in the human cancer cell lines, particularly in the human lung
cancer cell lines, as compared with the human normal esophageal
cell lines. The findings indicate that the new TTLL12 36AA plus
isoform may play an important role in the development of various
tumors.
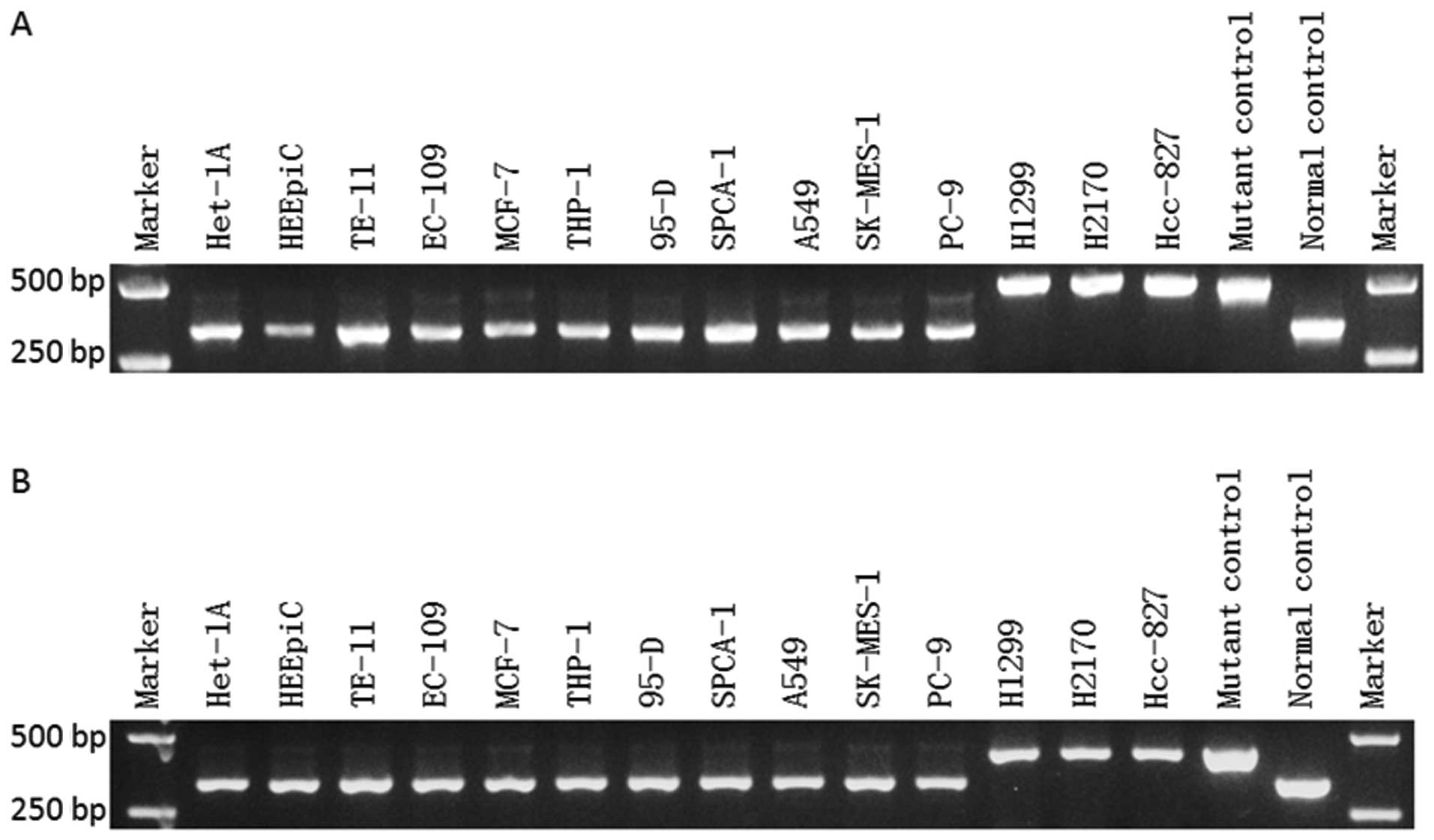 | Figure 3.Detection of the new TTLL12 36AA plus
isoform in human cancer cell lines. RT-PCR was employed in a series
of cell lines including human lung cancer cell lines (H1299, 95-D,
SPCA-1, A549, SK-MES-1, PC-9, H2170 and Hcc-827), human esophageal
cancer cell lines (TE-11 and EC-109), human normal esophageal cell
lines (HET-1A and HEEpiC), human acute monocytic leukemia cell line
THP-1, and human breast cancer cell line MCF-7. PCR was performed
using (A) TTLL12 sense primer 1 and TTLL12 antisense
primer 1, and (B) TTLL12 sense primer 2 and TTLL12
antisense primer 2. The PCR product for positive control of the
36AA plus isoform was 484 bp in size while the size of the
wild-type product was 376 bp, when the pair of primer 1 was used.
The product for positive control of the 36AA plus isoform was 434
bp in size while the wild-type was 326 bp in size as the pair of
primer 2 was used. |
Physicochemical properties of the
TTLL12 wild-type and new 36AA plus isoform
The AA sequence of the TTLL12 36AA plus isoform was
obtained using DNAStar EditSeq and translating the DNA sequences.
Then, the AA sequence of the new isoform was aligned with the
TTLL12 wild-type, and we found an additional 36AA inserted at the
location of 281AA of the TTLL12 wild-type (Fig. 5A). Next, we predicted the
physicochemical properties by ProtParam and ProtScale. The
properties of TTLL12 wild-type and 36AA plus isoform AA sequences
were predicted (Table I) as
follows: the total number of negatively charged residues (Asp +
Glu) was 93 and 96; and grand average of hydropathicity (GRAVY) was
−0.390 and −0.338. In addition, the total hydropathicities of the
two types of TTLL12 proteins are displayed in diagram (Fig. 5B and C). These results suggest that
the two types of TTLL12 proteins are hydropathical molecules, and
the 36AA plus isoform is more stable than the wild-type.
 | Table I.Physicochemical properties of the
wild-type and new 36AA plus isoform of TTLL12. |
Table I.
Physicochemical properties of the
wild-type and new 36AA plus isoform of TTLL12.
| Physicochemical
properties | Wild-type | New 36AA plus
isoform |
|---|
| Number of amino
acids | 644 | 680 |
| Molecule
formulas |
C3359H5101N905O959S28 |
C3537H5388N950O1006S29 |
| Molecular
weight | 74,403.6 | 78,245.1 |
| PI | 5.33 | 5.34 |
| Instability
index | 50.17 | 49.77 |
| Aliphatic
index | 79.18 | 82.01 |
| Negatively charged
residues (Asp + Glu) | 93 | 96 |
| Positively charged
residues (Arg + Lys) | 65 | 68 |
| GRAVY | -0.390 | -0.338 |
Secondary structures of TTLL12
wild-type and new 36AA plus isoform
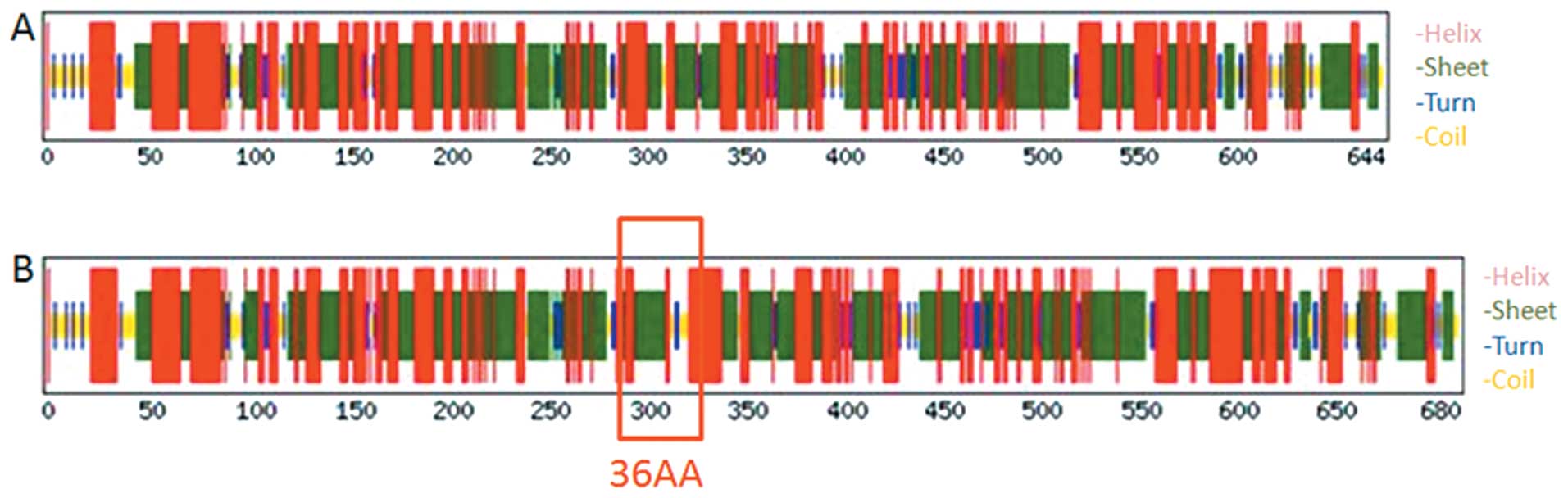
Secondary structures of the TTLL12 wild-type and
TTLL12 36AA plus isoform were predicted by DNAStar Protean and
online tool CFSSP. As a result, the total residues of α-Helix,
β-sheet and β-turn in the TTLL12 wild-type were 476, 428 and 72,
and the corresponding percentages were 73.9, 66.5 and 11.2%,
respectively (Fig. 6A). While total
residues of α-Helix, β-sheet and β-turn in the TTLL12 36AA plus
isoform were 504, 455 and 75, and the corresponding percentages
were 74.1, 66.9 and 11.0%, respectively (Fig. 6B). The results showed that the
proportions of different types of secondary structures in these two
TTLL12 proteins were slightly different (Table II).
 | Table II.Secondary structures of the TTLL12
wild-type and new 36AA plus isoform. |
Table II.
Secondary structures of the TTLL12
wild-type and new 36AA plus isoform.
|
| α-Helix | β-sheet | β-turn |
|---|
|
|
|
|
|
|---|
| TTLL12 type | Total residues | % | Total residues | % | Total residues | % |
|---|
| Wild-type | 476 | 73.9 | 428 | 66.6 | 72 | 11.2 |
| New 36AA plus
isoform | 504 | 74.1 | 455 | 66.9 | 75 | 11.0 |
The additional 36AA is involved in a
disordered region of TTLL12
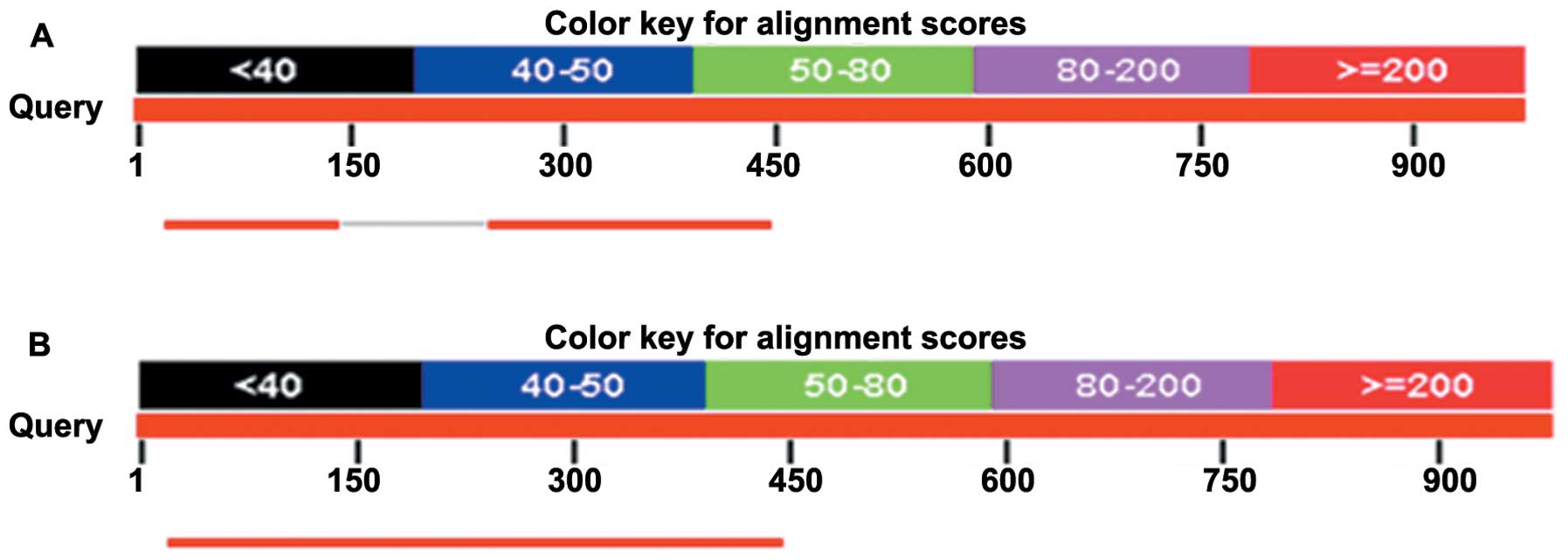
To ascertain whether the additional 36AA in the
TTLL12 36AA plus isoform is in a disordered region of TTLL12, we
firstly sought the disordered regions of the TTLL12 36AA plus
isoform by the opening accessible online resources DisProt search,
and found that the peptide of 260AA to 299AA in the TTLL12 36AA
plus isoform was highly identical to the disordered region of
Bcl-2-like protein 11 (B2L11) (Fig.
7A), suggesting this peptide could act as a disordered region
in the TTLL12 36AA plus isoform. As the additional 36AA are located
between 281AA and 282AA in the TTLL12 36AA plus isoform, we further
explored whether the additional 36AA was in the disordered region
of the protein. Using DisProt search, we found that the additional
36AA was partly identical to the disordered region of brain
natriuretic peptide-32 (BNP-32) (Fig.
7B), giving the possibility that the 36AA could endow TTLL12
with new functions by acting as part of the disordered region.
However, the biological functions of the new TTLL12 isoform in
cancers remain to be further investigated.
Discussion
The tubulin tyrosine ligase (TTLL) family is a
superfamily consisting of 14 members and they mainly catalyze
ligation of amino acids to tubulins (1–3).
TTLL12 is the least characterized and a most special member of the
TTLL family, and it plays an important role in histone and tubulin
modifications (14), mitotic
duration and chromosome ploidy (5–8). It is
the only member of the TTLL family without an assigned enzymatic
function (15–17), and is said to be a pseudo-enzyme
that has a phylogenetically conserved association of two domains
including the SET-like domain and TTL-like domain in the N- and C-
terminal of TTLL12, respectively. These two domains are related to
histone methylation (18–22) and tubulin modifications (23–25)
and then may contribute to tumorigenesis (4). The TTLL family is often suppressed in
human cancers (10) and is closely
connected with the poor prognosis of breast cancer (11), suggesting that TTLL12 could also
play roles in human cancers. It is reported that TTLL12 increases
its expression in the proliferating layer of benign human prostate
and more apparently during cancer progression to metastasis
(12), suggesting that TTLL12 may
be of importance in tumor progression.
In the present study, we found an aberrant TTLL12
transcript with an additional 108-bp nucleotide sequence inserted
between exons 5 and 6. This new isoform, named the TTLL12 36AA plus
isoform, exhibited much more abundance, as compared with the TTLL12
wild-type, in human cancer cells as to normal cells, particularly
in human lung cancer cells, suggesting it could be be considered a
new mechanism of human cancer development.
Alternatively splicing, by removing non-coding
sequences (introns) and joining a coding part (exons) (26), plays an important role in gene
expression (27–29). By the alternative splicing of
precursor mRNAs (pre-mRNAs), multiple mRNAs and proteins can be
generated from a single gene, and then the coding capacity of
genomes is expanded through such a major mechanism (27–31).
Furthermore, it has been reported that aberrant alternative
splicing underlies various pathological processes, in particular
tumorigenesis and tumor progression (32–34).
The new TTLL12 36AA plus isoform may be the result of aberrant
alternative splicing. Analysis by bioinformatics software showed
that there are some differences in peptide physicochemical
properties and protein secondary structure between the TTLL12 36AA
plus isoform and the TTLL12 wild-type, suggesting that the new 36AA
plus isoform could add some distinguished functions to TTLL12.
The traditional pattern analysis of protein
structural biology indicates that the well-defined
three-dimensional structures are pivotal to biological functions of
proteins. However, it has been reported that the disordered regions
of proteins are challenging this traditional structure-function
paradigm (35,36). Disordered regions are ubiquitous in
cellular processes and human pathological conditions, but lack a
well-defined, stable three-dimensional fold (37,38).
Proteins with disordered regions often go through disorder-to-order
when binding to their partners and they can remain partially or
fully flexible in their bound state, and form fuzzy complexes
37,39–41. In
contrast, proteins with disordered regions are mainly involved in
enzymatic activity (37,42), including cell cycle regulation
(43), cell division and
differentiation (42,44), which play critical roles in
different types of cancers (45).
In the present study, we report that the additional 36AA in the
newly found TTLL12 protein isoform is a part of the disordered
region, with the identity of those disordered regions identified in
other functional proteins. Whether the 36AA is the binding site for
certain proteins, such as those participating in post-translational
modification of tubulin or other important cell process, still
needs to be investigated.
The TTLL family has been shown to be partially
co-localized with vimentin and tubulin, which causes
post-translational modifications of tubulin (5–8).
Tubulin is a well acknowledged and important target for tumor
therapy (46), but the precise
functions of TTLL12 in tubulin modification are still poorly
understood. Proteins related to post-translational modification are
targets for new cancer therapeutic agents (47,48),
thus the new 36AA plus isoform of TTLL12 protein may have the
potential to become a novel strategy for cancer treatment involving
tubulin post-translational modifications. Therefore, the functions
and molecular mechanisms of the new 36AA plus isoform in cancer
development remain to be investigated.
In conclusion, our findings unveil a novel
transcript isoform of TTLL12 with an additional 108-bp nucleotide
sequence in the CDS. This TTLL12 36AA plus isoform could shed light
on a novel mechanism of TTLL12 in human carcinogenesis and tumor
progression, which may lead to a breakthrough of new potential
targets for human cancer therapeutics.
Acknowledgements
The present study was supported by grants from the
Science and Information Technology Bureau of Guangzhou
(2011Y1-00022 and 2012-224-8, to J.T.), the Science and Information
Technology Bureau of Guangzhou Haizhu District (2012-ZD-02, to
J.T.), and the Guangzhou Medical University (B147048, to J.T.).
References
|
1
|
Janke C, Rogowski K, Wloga D, Regnard C,
Kajava AV, Strub JM, Temurak N, van Dijk J, Boucher D, van
Dorsselaer A, et al: Tubulin polyglutamylase enzymes are members of
the TTL domain protein family. Science. 308:1758–1762. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fukushima N, Furuta D, Hidaka Y, Moriyama
R and Tsujiuchi T: Post-translational modifications of tubulin in
the nervous system. J Neurochem. 109:683–693. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ersfeld K, Wehland J, Plessmann U,
Dodemont H, Gerke V and Weber K: Characterization of the
tubulin-tyrosine ligase. J Cell Biol. 120:725–732. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Brants J, Semenchenko K, Wasylyk C, Robert
A, Carles A, Zambrano A, Pradeau-Aubreton K, Birck C, Schalken JA,
Poch O, et al: Tubulin tyrosine ligase like 12, a TTLL family
member with SET- and TTL-like domains and roles in histone and
tubulin modifications and mitosis. PLoS One. 7:e512582012.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gurland G and Gundersen GG: Stable,
detyrosinated microtubules function to localize vimentin
intermediate filaments in fibroblasts. J Cell Biol. 131:1275–1290.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Peris L, Thery M, Fauré J, Saoudi Y,
Lafanechère L, Chilton JK, Gordon-Weeks P, Galjart N, Bornens M,
Wordeman L, et al: Tubulin tyrosination is a major factor affecting
the recruitment of CAP-Gly proteins at microtubule plus ends. J
Cell Biol. 174:839–849. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dunn S, Morrison EE, Liverpool TB,
Molina-París C, Cross RA, Alonso MC and Peckham M: Differential
trafficking of Kif5c on tyrosinated and detyrosinated microtubules
in live cells. J Cell Sci. 121:1085–1095. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gyoeva FK and Gelfand VI: Coalignment of
vimentin intermediate filaments with microtubules depends on
kinesin. Nature. 353:445–448. 1991. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kato C, Miyazaki K, Nakagawa A, Ohira M,
Nakamura Y, Ozaki T, Imai T and Nakagawara A: Low expression of
human tubulin tyrosine ligase and suppressed tubulin
tyrosination/detyrosination cycle are associated with impaired
neuronal differentiation in neuroblastomas with poor prognosis. Int
J Cancer. 112:365–375. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lafanechère L, Courtay-Cahen C, Kawakami
T, Jacrot M, Rüdiger M, Wehland J, Job D and Margolis RL:
Suppression of tubulin tyrosine ligase during tumor growth. J Cell
Sci. 111:171–181. 1998.PubMed/NCBI
|
|
11
|
Mialhe A, Lafanechère L, Treilleux I,
Peloux N, Dumontet C, Brémond A, Panh MH, Payan R, Wehland J,
Margolis RL, et al: Tubulin detyrosination is a frequent occurrence
in breast cancers of poor prognosis. Cancer Res. 61:5024–5027.
2001.PubMed/NCBI
|
|
12
|
Wasylyk C, Zambrano A, Zhao C, Brants J,
Abecassis J, Schalken JA, Rogatsch H, Schaefer G, Pycha A, Klocker
H, et al: Tubulin tyrosine ligase like 12 links to prostate cancer
through tubulin posttranslational modification and chromosome
ploidy. Int J Cancer. 127:2542–2553. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Soucek K, Kamaid A, Phung AD, Kubala L,
Bulinski JC, Harper RW and Eiserich JP: Normal and prostate cancer
cells display distinct molecular profiles of α-tubulin
posttranslational modifications. Prostate. 66:954–965. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Szyk A, Deaconescu AM, Piszczek G and
Roll-Mecak A: Tubulin tyrosine ligase structure reveals adaptation
of an ancient fold to bind and modify tubulin. Nat Struct Mol Biol.
18:1250–1258. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ikegami K and Setou M: TTLL10 can perform
tubulin glycylation when co-expressed with TTLL8. FEBS Lett.
583:1957–1963. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wloga D, Webster DM, Rogowski K, Bré MH,
Levilliers N, Jerka-Dziadosz M, Janke C, Dougan ST and Gaertig J:
TTLL3 is a tubulin glycine ligase that regulates the assembly of
cilia. Dev Cell. 16:867–876. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rogowski K, Juge F, van Dijk J, Wloga D,
Strub JM, Levilliers N, Thomas D, Bré MH, Van Dorsselaer A, Gaertig
J, et al: Evolutionary divergence of enzymatic mechanisms for
posttranslational polyglycylation. Cell. 137:1076–1087. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Dillon SC, Zhang X, Trievel RC and Cheng
X: The SET-domain protein superfamily: Protein lysine
methyltransferases. Genome Biol. 6:2272005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cheng X and Zhang X: Structural dynamics
of protein lysine methylation and demethylation. Mutat Res.
618:102–115. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xu D, Bai J, Duan Q, Costa M and Dai W:
Covalent modifications of histones during mitosis and meiosis. Cell
Cycle. 8:3688–3694. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Qian C and Zhou MM: SET domain protein
lysine methyltransferases: Structure, specificity and catalysis.
Cell Mol Life Sci. 63:2755–2763. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Aravind L, Abhiman S and Iyer LM: Natural
history of the eukaryotic chromatin protein methylation system.
Prog Mol Biol Transl Sci. 101:105–176. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Garnham CP and Roll-Mecak A: The chemical
complexity of cellular microtubules: Tubulin post-translational
modification enzymes and their roles in tuning microtubule
functions. Cytoskeleton Hoboken. 69:442–463. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wloga D and Gaertig J: Post-translational
modifications of microtubules. J Cell Sci. 123:3447–3455. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Janke C and Bulinski JC:
Post-translational regulation of the microtubule cytoskeleton:
Mechanisms and functions. Nat Rev Mol Cell Biol. 12:773–786. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sharp PA: Split genes and RNA splicing.
Cell. 77:805–815. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yang X, Coulombe-Huntington J, Kang S,
Sheynkman GM, Hao T, Richardson A, Sun S, Yang F, Shen YA, Murray
RR, et al: Widespread expansion of protein interaction capabilities
by alternative splicing. Cell. 164:805–817. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Du Q, Li C, Li D and Lu S: Genome-wide
analysis, molecular cloning and expression profiling reveal
tissue-specifically expressed, feedback-regulated,
stress-responsive and alternatively spliced novel genes involved in
gibberellin metabolism in Salvia miltiorrhiza. BMC Genomics.
16:10872015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Rodríguez SA, Grochová D, McKenna T,
Borate B, Trivedi NS, Erdos MR and Eriksson M: Global genome
splicing analysis reveals an increased number of alternatively
spliced genes with aging. Aging Cell. 15:267–278. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Shang J, Fan X, Shangguan L, Liu H and
Zhou Y: Global gene expression profiling and alternative splicing
events during the chondrogenic differentiation of human cartilage
endplate-derived stem cells. BioMed Res Int. 2015:6049722015.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Stevens M and Oltean S: Alternative
splicing in CKD. J Am Soc Nephrol. 27:1596–1603. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sheng Z, Sun Y, Zhu R, Jiao N, Tang K, Cao
Z and Ma C: Functional cross-talking between differentially
expressed and alternatively spliced genes in human liver cancer
cells treated with berberine. PLoS One. 10:e01437422015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Adamopoulos PG, Kontos CK, Tsiakanikas P
and Scorilas A: Identification of novel alternative splice variants
of the BCL2L12 gene in human cancer cells using next-generation
sequencing methodology. Cancer Lett. 373:119–129. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Faustino NA and Cooper TA: Pre-mRNA
splicing and human disease. Genes Dev. 17:419–437. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Forman-Kay JD and Mittag T: From sequence
and forces to structure, function, and evolution of intrinsically
disordered proteins. Structure. 21:1492–1499. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Dunker AK, Bondos SE, Huang F and Oldfield
CJ: Intrinsically disordered proteins and multicellular organisms.
Semin Cell Dev Biol. 37:44–55. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wright PE and Dyson HJ: Intrinsically
unstructured proteins: Re-assessing the protein structure-function
paradigm. J Mol Biol. 293:321–331. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chouard T: Structural biology: Breaking
the protein rules. Nature. 471:151–153. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kotta-Loizou I, Tsaousis GN and Hamodrakas
SJ: Analysis of molecular recognition features (MoRFs) in membrane
proteins. Biochim Biophys Acta. 1834:798–807. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Tompa P and Fuxreiter M: Fuzzy complexes:
Polymorphism and structural disorder in protein-protein
interactions. Trends Biochem Sci. 33:2–8. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Fuxreiter M and Tompa P: Fuzzy complexes:
A more stochastic view of protein function. Adv Exp Med Biol.
725:1–14. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Xie H, Vucetic S, Iakoucheva LM, Oldfield
CJ, Dunker AK, Uversky VN and Obradovic Z: Functional anthology of
intrinsic disorder. 1. Biological processes and functions of
proteins with long disordered regions. J Proteome Res. 6:1882–1898.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yoon MK, Mitrea DM, Ou L and Kriwacki RW:
Cell cycle regulation by the intrinsically disordered proteins p21
and p27. Biochem Soc Trans. 40:981–988. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF
and Jones DT: Prediction and functional analysis of native disorder
in proteins from the three kingdoms of life. J Mol Biol.
337:635–645. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Andresen C, Helander S, Lemak A, Farès C,
Csizmok V, Carlsson J, Penn LZ, Forman-Kay JD, Arrowsmith CH,
Lundström P, et al: Transient structure and dynamics in the
disordered c-Myc transactivation domain affect Bin1 binding.
Nucleic Acids Res. 40:6353–6366. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Harrison MR, Holen KD and Liu G: Beyond
taxanes: A review of novel agents that target mitotic tubulin and
microtubules, kinases, and kinesins. Clin Adv Hematol Oncol.
7:54–64. 2009.PubMed/NCBI
|
|
47
|
Verhey KJ and Gaertig J: The tubulin code.
Cell Cycle. 6:2152–2160. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Ghoreschi K, Laurence A and O'Shea JJ:
Selectivity and therapeutic inhibition of kinases: To be or not to
be? Nat Immunol. 10:356–360. 2009. View Article : Google Scholar : PubMed/NCBI
|