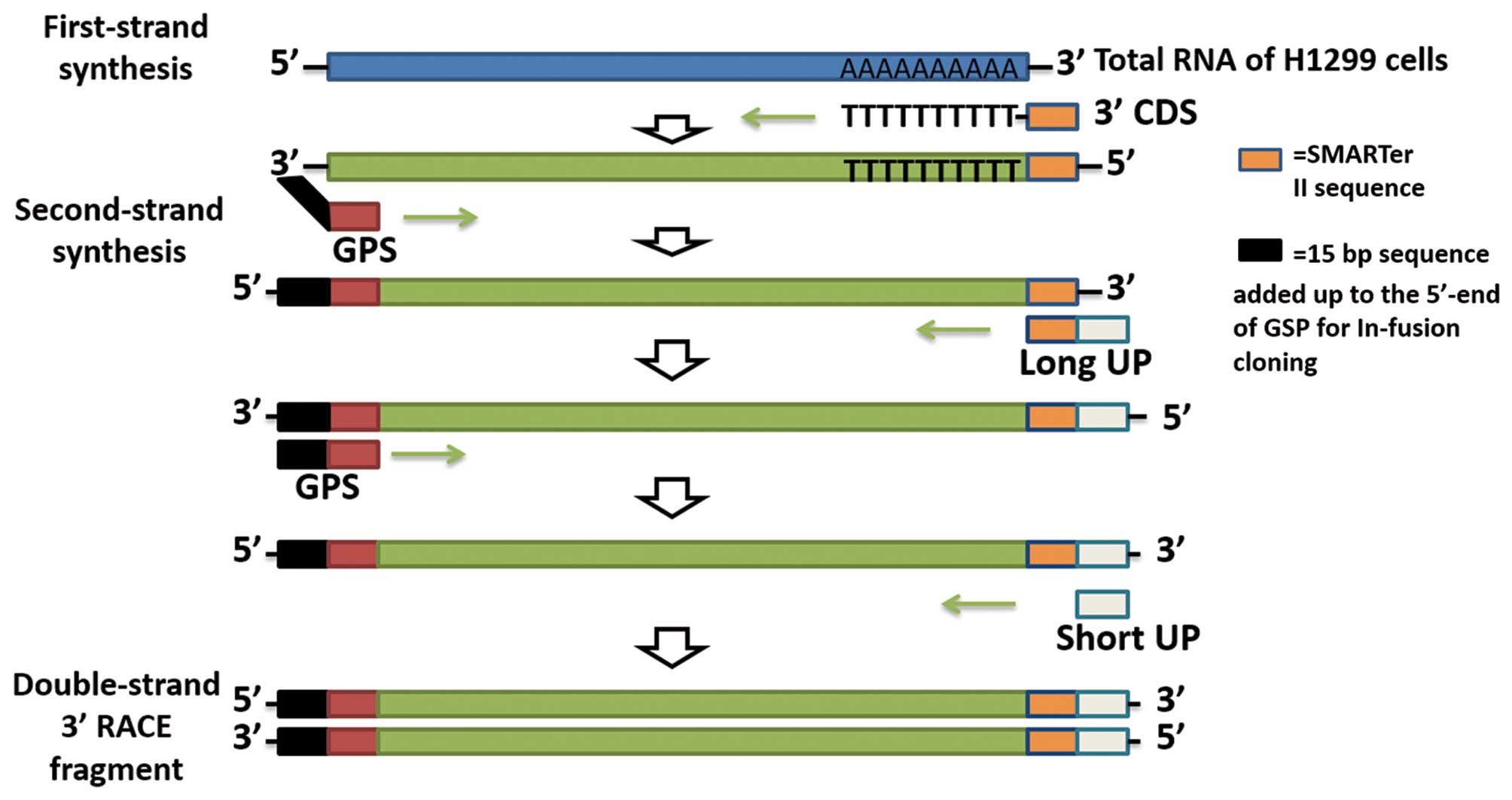
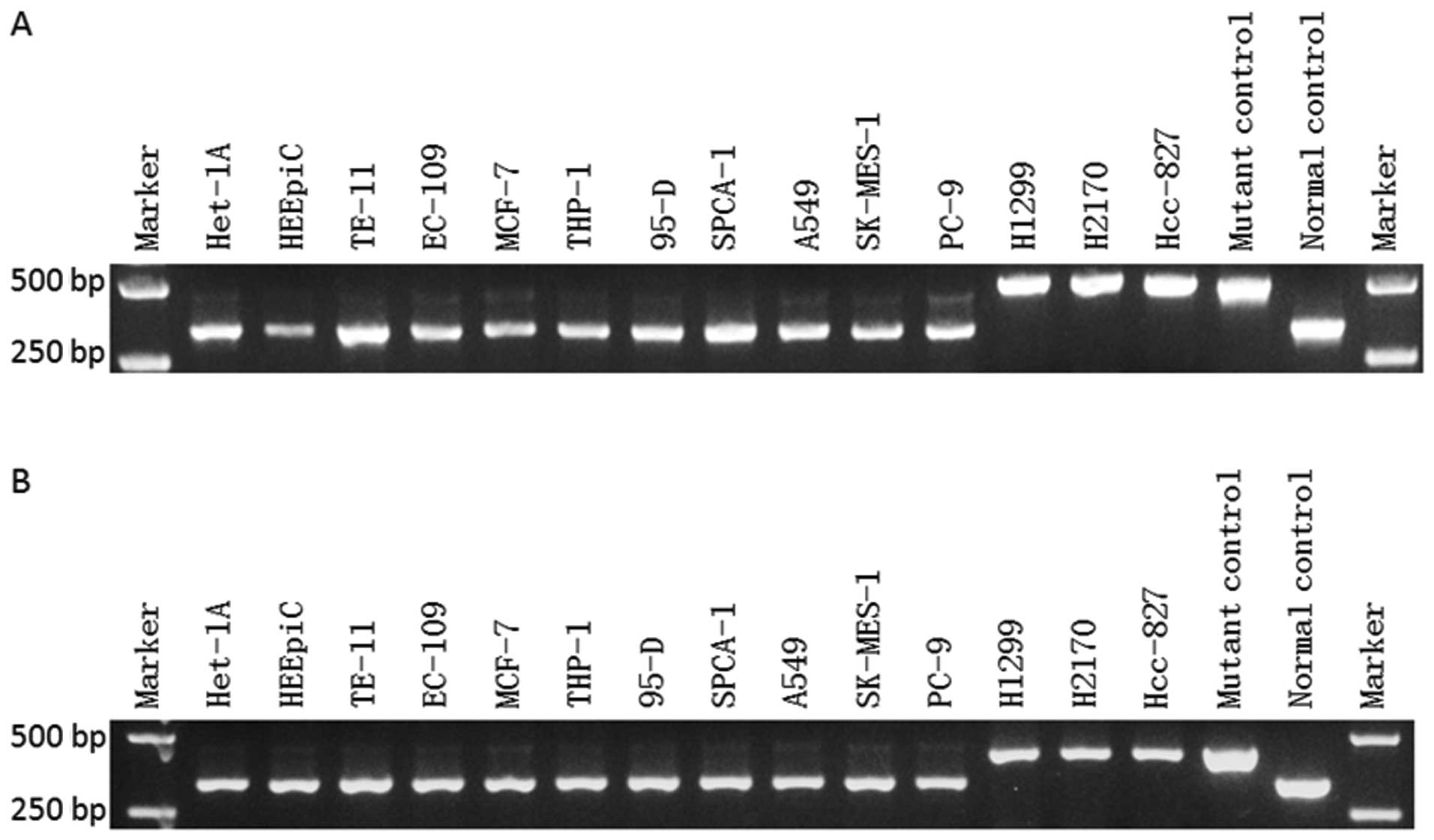
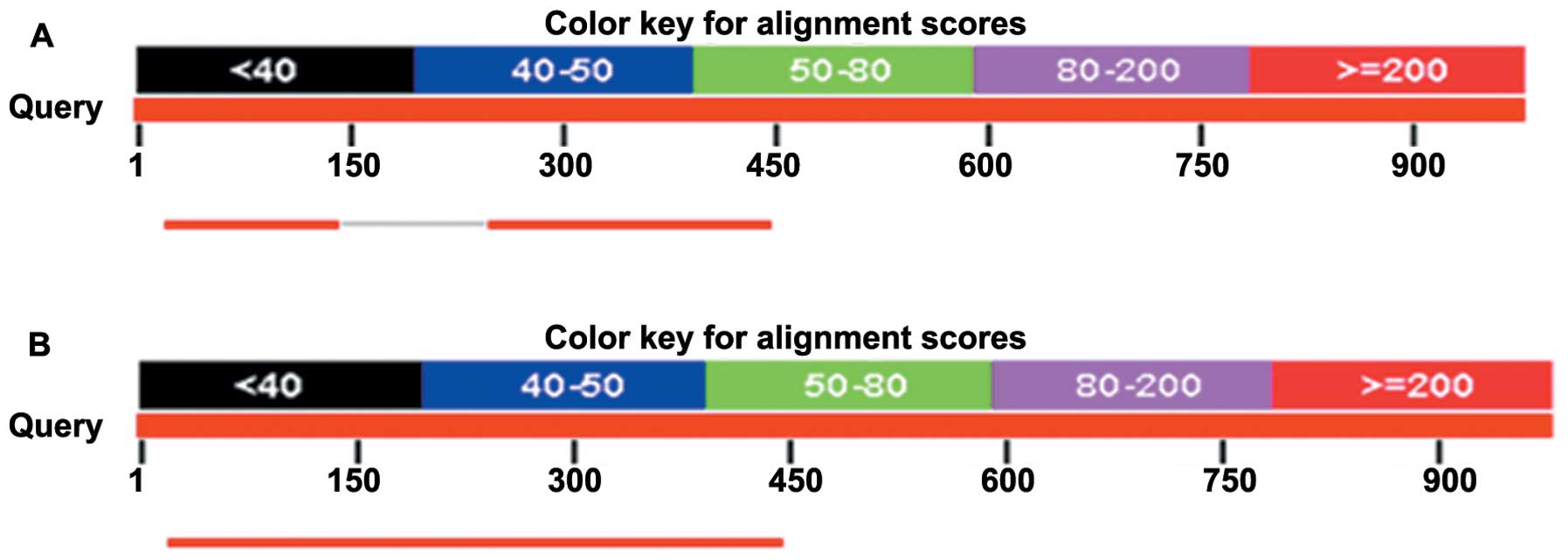
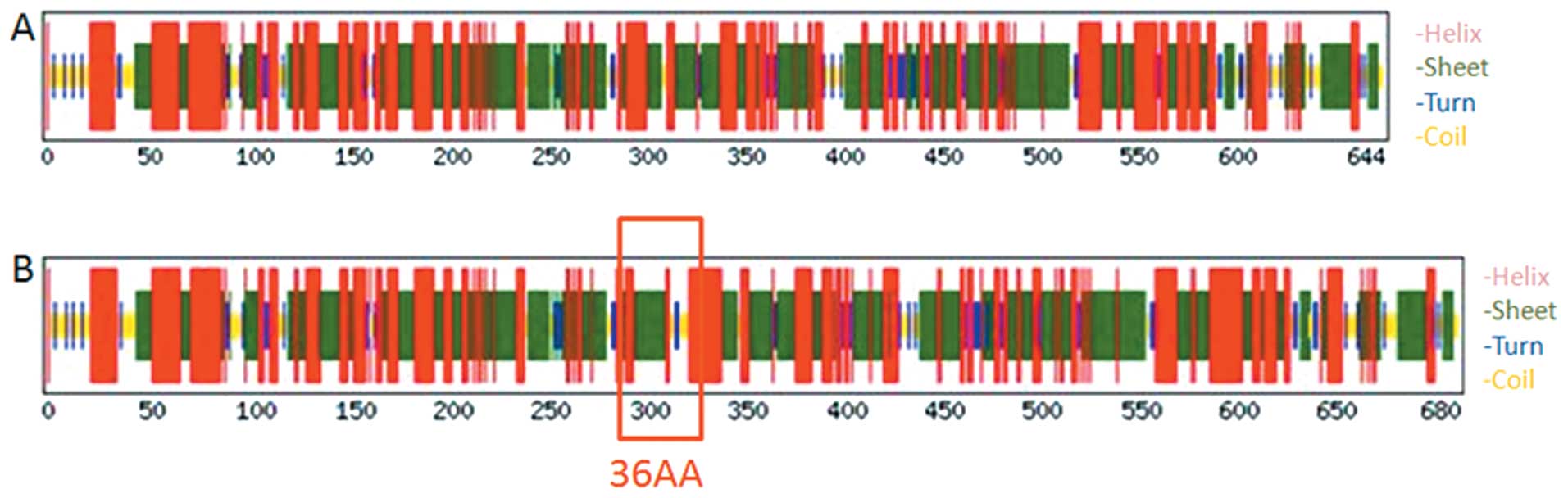
|
1
|
Janke C, Rogowski K, Wloga D, Regnard C,
Kajava AV, Strub JM, Temurak N, van Dijk J, Boucher D, van
Dorsselaer A, et al: Tubulin polyglutamylase enzymes are members of
the TTL domain protein family. Science. 308:1758–1762. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fukushima N, Furuta D, Hidaka Y, Moriyama
R and Tsujiuchi T: Post-translational modifications of tubulin in
the nervous system. J Neurochem. 109:683–693. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ersfeld K, Wehland J, Plessmann U,
Dodemont H, Gerke V and Weber K: Characterization of the
tubulin-tyrosine ligase. J Cell Biol. 120:725–732. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Brants J, Semenchenko K, Wasylyk C, Robert
A, Carles A, Zambrano A, Pradeau-Aubreton K, Birck C, Schalken JA,
Poch O, et al: Tubulin tyrosine ligase like 12, a TTLL family
member with SET- and TTL-like domains and roles in histone and
tubulin modifications and mitosis. PLoS One. 7:e512582012.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gurland G and Gundersen GG: Stable,
detyrosinated microtubules function to localize vimentin
intermediate filaments in fibroblasts. J Cell Biol. 131:1275–1290.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Peris L, Thery M, Fauré J, Saoudi Y,
Lafanechère L, Chilton JK, Gordon-Weeks P, Galjart N, Bornens M,
Wordeman L, et al: Tubulin tyrosination is a major factor affecting
the recruitment of CAP-Gly proteins at microtubule plus ends. J
Cell Biol. 174:839–849. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dunn S, Morrison EE, Liverpool TB,
Molina-París C, Cross RA, Alonso MC and Peckham M: Differential
trafficking of Kif5c on tyrosinated and detyrosinated microtubules
in live cells. J Cell Sci. 121:1085–1095. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gyoeva FK and Gelfand VI: Coalignment of
vimentin intermediate filaments with microtubules depends on
kinesin. Nature. 353:445–448. 1991. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kato C, Miyazaki K, Nakagawa A, Ohira M,
Nakamura Y, Ozaki T, Imai T and Nakagawara A: Low expression of
human tubulin tyrosine ligase and suppressed tubulin
tyrosination/detyrosination cycle are associated with impaired
neuronal differentiation in neuroblastomas with poor prognosis. Int
J Cancer. 112:365–375. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lafanechère L, Courtay-Cahen C, Kawakami
T, Jacrot M, Rüdiger M, Wehland J, Job D and Margolis RL:
Suppression of tubulin tyrosine ligase during tumor growth. J Cell
Sci. 111:171–181. 1998.PubMed/NCBI
|
|
11
|
Mialhe A, Lafanechère L, Treilleux I,
Peloux N, Dumontet C, Brémond A, Panh MH, Payan R, Wehland J,
Margolis RL, et al: Tubulin detyrosination is a frequent occurrence
in breast cancers of poor prognosis. Cancer Res. 61:5024–5027.
2001.PubMed/NCBI
|
|
12
|
Wasylyk C, Zambrano A, Zhao C, Brants J,
Abecassis J, Schalken JA, Rogatsch H, Schaefer G, Pycha A, Klocker
H, et al: Tubulin tyrosine ligase like 12 links to prostate cancer
through tubulin posttranslational modification and chromosome
ploidy. Int J Cancer. 127:2542–2553. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Soucek K, Kamaid A, Phung AD, Kubala L,
Bulinski JC, Harper RW and Eiserich JP: Normal and prostate cancer
cells display distinct molecular profiles of α-tubulin
posttranslational modifications. Prostate. 66:954–965. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Szyk A, Deaconescu AM, Piszczek G and
Roll-Mecak A: Tubulin tyrosine ligase structure reveals adaptation
of an ancient fold to bind and modify tubulin. Nat Struct Mol Biol.
18:1250–1258. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ikegami K and Setou M: TTLL10 can perform
tubulin glycylation when co-expressed with TTLL8. FEBS Lett.
583:1957–1963. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wloga D, Webster DM, Rogowski K, Bré MH,
Levilliers N, Jerka-Dziadosz M, Janke C, Dougan ST and Gaertig J:
TTLL3 is a tubulin glycine ligase that regulates the assembly of
cilia. Dev Cell. 16:867–876. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rogowski K, Juge F, van Dijk J, Wloga D,
Strub JM, Levilliers N, Thomas D, Bré MH, Van Dorsselaer A, Gaertig
J, et al: Evolutionary divergence of enzymatic mechanisms for
posttranslational polyglycylation. Cell. 137:1076–1087. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Dillon SC, Zhang X, Trievel RC and Cheng
X: The SET-domain protein superfamily: Protein lysine
methyltransferases. Genome Biol. 6:2272005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cheng X and Zhang X: Structural dynamics
of protein lysine methylation and demethylation. Mutat Res.
618:102–115. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xu D, Bai J, Duan Q, Costa M and Dai W:
Covalent modifications of histones during mitosis and meiosis. Cell
Cycle. 8:3688–3694. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Qian C and Zhou MM: SET domain protein
lysine methyltransferases: Structure, specificity and catalysis.
Cell Mol Life Sci. 63:2755–2763. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Aravind L, Abhiman S and Iyer LM: Natural
history of the eukaryotic chromatin protein methylation system.
Prog Mol Biol Transl Sci. 101:105–176. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Garnham CP and Roll-Mecak A: The chemical
complexity of cellular microtubules: Tubulin post-translational
modification enzymes and their roles in tuning microtubule
functions. Cytoskeleton Hoboken. 69:442–463. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wloga D and Gaertig J: Post-translational
modifications of microtubules. J Cell Sci. 123:3447–3455. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Janke C and Bulinski JC:
Post-translational regulation of the microtubule cytoskeleton:
Mechanisms and functions. Nat Rev Mol Cell Biol. 12:773–786. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sharp PA: Split genes and RNA splicing.
Cell. 77:805–815. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yang X, Coulombe-Huntington J, Kang S,
Sheynkman GM, Hao T, Richardson A, Sun S, Yang F, Shen YA, Murray
RR, et al: Widespread expansion of protein interaction capabilities
by alternative splicing. Cell. 164:805–817. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Du Q, Li C, Li D and Lu S: Genome-wide
analysis, molecular cloning and expression profiling reveal
tissue-specifically expressed, feedback-regulated,
stress-responsive and alternatively spliced novel genes involved in
gibberellin metabolism in Salvia miltiorrhiza. BMC Genomics.
16:10872015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Rodríguez SA, Grochová D, McKenna T,
Borate B, Trivedi NS, Erdos MR and Eriksson M: Global genome
splicing analysis reveals an increased number of alternatively
spliced genes with aging. Aging Cell. 15:267–278. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Shang J, Fan X, Shangguan L, Liu H and
Zhou Y: Global gene expression profiling and alternative splicing
events during the chondrogenic differentiation of human cartilage
endplate-derived stem cells. BioMed Res Int. 2015:6049722015.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Stevens M and Oltean S: Alternative
splicing in CKD. J Am Soc Nephrol. 27:1596–1603. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sheng Z, Sun Y, Zhu R, Jiao N, Tang K, Cao
Z and Ma C: Functional cross-talking between differentially
expressed and alternatively spliced genes in human liver cancer
cells treated with berberine. PLoS One. 10:e01437422015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Adamopoulos PG, Kontos CK, Tsiakanikas P
and Scorilas A: Identification of novel alternative splice variants
of the BCL2L12 gene in human cancer cells using next-generation
sequencing methodology. Cancer Lett. 373:119–129. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Faustino NA and Cooper TA: Pre-mRNA
splicing and human disease. Genes Dev. 17:419–437. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Forman-Kay JD and Mittag T: From sequence
and forces to structure, function, and evolution of intrinsically
disordered proteins. Structure. 21:1492–1499. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Dunker AK, Bondos SE, Huang F and Oldfield
CJ: Intrinsically disordered proteins and multicellular organisms.
Semin Cell Dev Biol. 37:44–55. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wright PE and Dyson HJ: Intrinsically
unstructured proteins: Re-assessing the protein structure-function
paradigm. J Mol Biol. 293:321–331. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chouard T: Structural biology: Breaking
the protein rules. Nature. 471:151–153. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kotta-Loizou I, Tsaousis GN and Hamodrakas
SJ: Analysis of molecular recognition features (MoRFs) in membrane
proteins. Biochim Biophys Acta. 1834:798–807. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Tompa P and Fuxreiter M: Fuzzy complexes:
Polymorphism and structural disorder in protein-protein
interactions. Trends Biochem Sci. 33:2–8. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Fuxreiter M and Tompa P: Fuzzy complexes:
A more stochastic view of protein function. Adv Exp Med Biol.
725:1–14. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Xie H, Vucetic S, Iakoucheva LM, Oldfield
CJ, Dunker AK, Uversky VN and Obradovic Z: Functional anthology of
intrinsic disorder. 1. Biological processes and functions of
proteins with long disordered regions. J Proteome Res. 6:1882–1898.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yoon MK, Mitrea DM, Ou L and Kriwacki RW:
Cell cycle regulation by the intrinsically disordered proteins p21
and p27. Biochem Soc Trans. 40:981–988. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF
and Jones DT: Prediction and functional analysis of native disorder
in proteins from the three kingdoms of life. J Mol Biol.
337:635–645. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Andresen C, Helander S, Lemak A, Farès C,
Csizmok V, Carlsson J, Penn LZ, Forman-Kay JD, Arrowsmith CH,
Lundström P, et al: Transient structure and dynamics in the
disordered c-Myc transactivation domain affect Bin1 binding.
Nucleic Acids Res. 40:6353–6366. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Harrison MR, Holen KD and Liu G: Beyond
taxanes: A review of novel agents that target mitotic tubulin and
microtubules, kinases, and kinesins. Clin Adv Hematol Oncol.
7:54–64. 2009.PubMed/NCBI
|
|
47
|
Verhey KJ and Gaertig J: The tubulin code.
Cell Cycle. 6:2152–2160. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Ghoreschi K, Laurence A and O'Shea JJ:
Selectivity and therapeutic inhibition of kinases: To be or not to
be? Nat Immunol. 10:356–360. 2009. View Article : Google Scholar : PubMed/NCBI
|