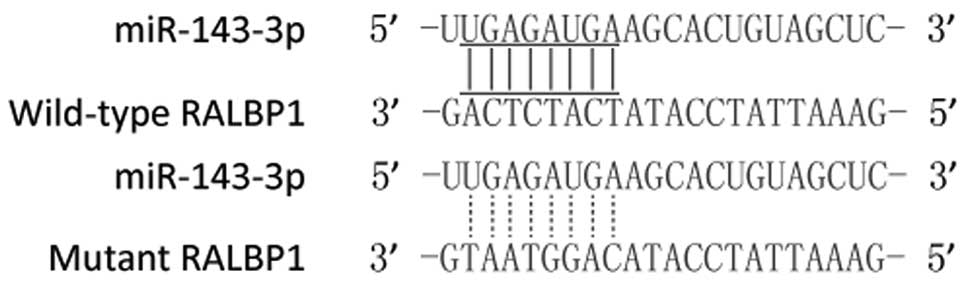
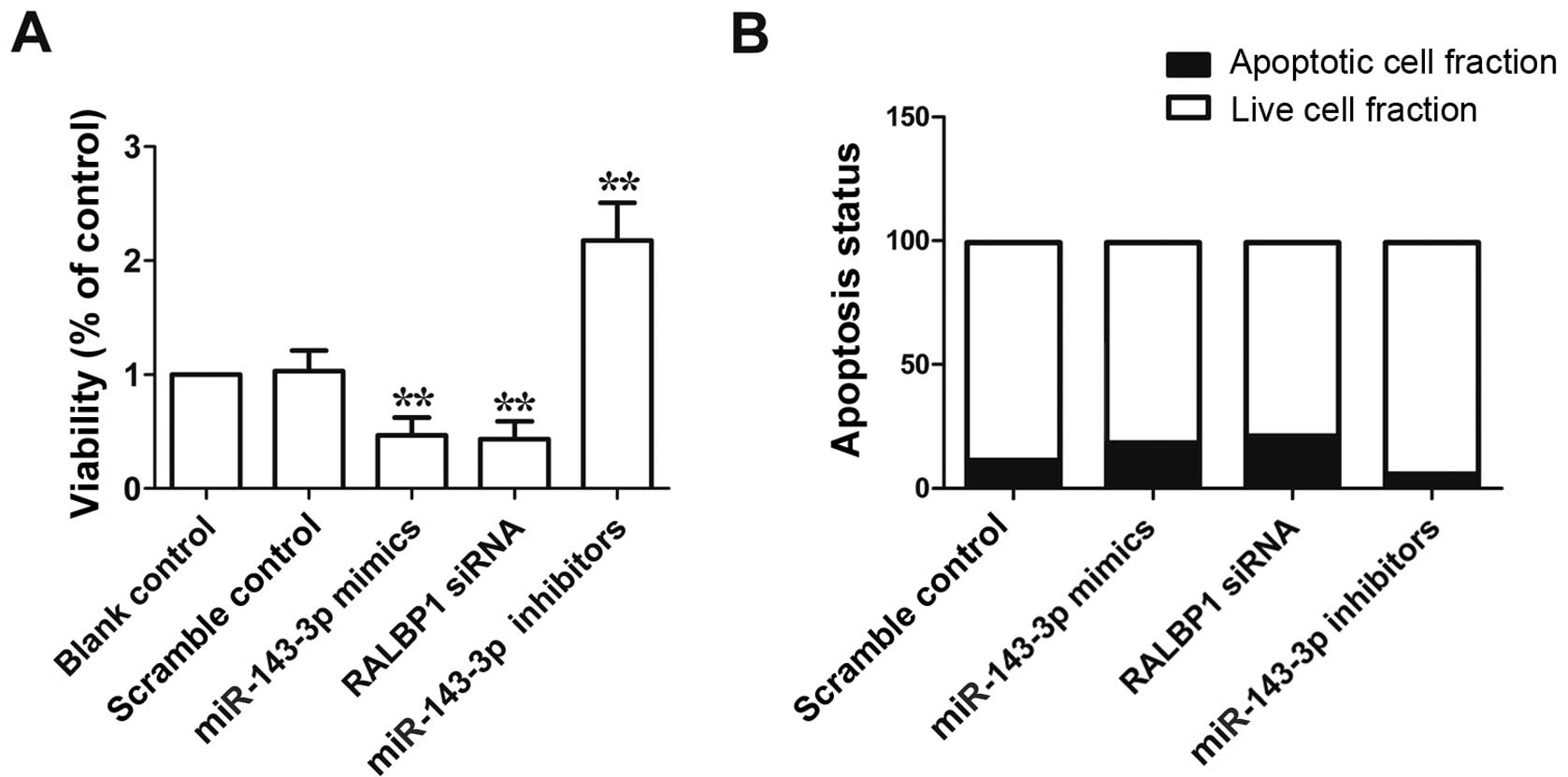
|
1
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Estimating the world cancer burden: Globocan 2000. Int J Cancer.
94:153–156. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kaku T, Ogawa S, Kawano Y, Ohishi Y,
Kobayashi H, Hirakawa T and Nakano H: Histological classification
of ovarian cancer. Med Electron Microsc. 36:9–17. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bodemann BO and White MA: Ral GTPases and
cancer: Linchpin support of the tumorigenic platform. Nat Rev
Cancer. 8:133–140. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lim KH, Baines AT, Fiordalisi JJ,
Shipitsin M, Feig LA, Cox AD, Der CJ and Counter CM: Activation of
RalA is critical for Ras-induced tumorigenesis of human cells.
Cancer Cell. 7:533–545. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Smith SC, Oxford G, Baras AS, Owens C,
Havaleshko D, Brautigan DL, Safo MK and Theodorescu D: Expression
of ral GTPases, their effectors, and activators in human bladder
cancer. Clin Cancer Res. 13:3803–3813. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Singhal SS, Awasthi YC and Awasthi S:
Regression of melanoma in a murine model by RLIP76 depletion.
Cancer Res. 66:2354–2360. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jiang H, Lu Z, Luo JQ, Wolfman A and
Foster DA: Ras mediates the activation of phospholipase D by v-Src.
J Biol Chem. 270:6006–6009. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Reinhart BJ, Weinstein EG, Rhoades MW,
Bartel B and Bartel DP: MicroRNAs in plants. Genes Dev.
16:1616–1626. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kim VN and Nam JW: Genomics of microRNA.
Trends Genet. 22:165–173. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Esteller M: Non-coding RNAs in human
disease. Nat Rev Genet. 12:861–874. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shahab SW, Matyunina LV, Mezencev R,
Walker LD, Bowen NJ, Benigno BB and McDonald JF: Evidence for the
complexity of microRNA-mediated regulation in ovarian cancer: A
systems approach. PLoS One. 6:e225082011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Permuth-Wey J, Kim D, Tsai YY, Lin HY,
Chen YA, Barnholtz-Sloan J, Birrer MJ, Bloom G, Chanock SJ, Chen Z,
et al: Ovarian Cancer Association Consortium: LIN28B polymorphisms
influence susceptibility to epithelial ovarian cancer. Cancer Res.
71:3896–3903. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nam EJ, Yoon H, Kim SW, Kim H, Kim YT, Kim
JH, Kim JW and Kim S: MicroRNA expression profiles in serous
ovarian carcinoma. Clin Cancer Res. 14:2690–2695. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hudson ME, Pozdnyakova I, Haines K, Mor G
and Snyder M: Identification of differentially expressed proteins
in ovarian cancer using high-density protein microarrays. Proc Natl
Acad Sci USA. 104:17494–17499. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nakagawa M, Tsuzuki S, Honma K, Taguchi O
and Seto M: Synergistic effect of Bcl2, Myc and Ccnd1 transforms
mouse primary B cells into malignant cells. Haematologica.
96:1318–1326. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Takagi T, Iio A, Nakagawa Y, Naoe T,
Tanigawa N and Akao Y: Decreased expression of microRNA-143 and
−145 in human gastric cancers. Oncology. 77:12–21. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li X, Luo F, Li Q, Xu M, Feng D, Zhang G
and Wu W: Identification of new aberrantly expressed miRNAs in
intestinal-type gastric cancer and its clinical significance. Oncol
Rep. 26:1431–1439. 2011.PubMed/NCBI
|
|
21
|
Keely PJ, Rusyn EV, Cox AD and Parise LV:
R-Ras signals through specific integrin alpha cytoplasmic domains
to promote migration and invasion of breast epithelial cells. J
Cell Biol. 145:1077–1088. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Holly SP, Larson MK and Parise LV: The
unique N-terminus of R-ras is required for Rac activation and
precise regulation of cell migration. Mol Biol Cell. 16:2458–2469.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Radhakrishna H and Donaldson JG:
ADP-ribosylation factor 6 regulates a novel plasma membrane
recycling pathway. J Cell Biol. 139:49–61. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jullien-Flores V, Mahé Y, Mirey G,
Leprince C, Meunier-Bisceuil B, Sorkin A and Camonis JH: RLIP76, an
effector of the GTPase Ral, interacts with the AP2 complex:
Involvement of the Ral pathway in receptor endocytosis. J Cell Sci.
113:2837–2844. 2000.PubMed/NCBI
|
|
25
|
Paleotti O, Macia E, Luton F, Klein S,
Partisani M, Chardin P, Kirchhausen T and Franco M: The small
G-protein Arf6GTP recruits the AP-2 adaptor complex to membranes. J
Biol Chem. 280:21661–21666. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ceresa BP and Schmid SL: Regulation of
signal transduction by endocytosis. Curr Opin Cell Biol.
12:204–210. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Awasthi S, Singhal SS, Awasthi YC, Martin
B, Woo JH, Cunningham CC and Frankel AE: RLIP76 and cancer. Clin
Cancer Res. 14:4372–4377. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jullien-Flores V, Dorseuil O, Romero F,
Letourneur F, Saragosti S, Berger R, Tavitian A, Gacon G and
Camonis JH: Bridging Ral GTPase to Rho pathways. RLIP76, a Ral
effector with CDC42/Rac GTPase-activating protein activity. J Biol
Chem. 270:22473–22477. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Awasthi S, Cheng JZ, Singhal SS, Pandya U,
Sharma R, Singh SV, Zimniak P and Awasthi YC: Functional reassembly
of ATP-dependent xenobiotic transport by the N- and C-terminal
domains of RLIP76 and identification of ATP binding sequences.
Biochemistry. 40:4159–4168. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Goldfinger LE, Ptak C, Jeffery ED,
Shabanowitz J, Hunt DF and Ginsberg MH: RLIP76 (RalBP1) is an R-Ras
effector that mediates adhesion-dependent Rac activation and cell
migration. J Cell Biol. 174:877–888. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kashatus DF, Lim KH, Brady DC, Pershing
NL, Cox AD and Counter CM: RALA and RALBP1 regulate mitochondrial
fission at mitosis. Nat Cell Biol. 13:1108–1115. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Nakashima S, Morinaka K, Koyama S, Ikeda
M, Kishida M, Okawa K, Iwamatsu A, Kishida S and Kikuchi A: Small G
protein Ral and its downstream molecules regulate endocytosis of
EGF and insulin receptors. EMBO J. 18:3629–3642. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Singhal SS, Roth C, Leake K, Singhal J,
Yadav S and Awasthi S: Regression of prostate cancer xenografts by
RLIP76 depletion. Biochem Pharmacol. 77:1074–1083. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Singhal SS, Singhal J, Yadav S, Dwivedi S,
Boor PJ, Awasthi YC and Awasthi S: Regression of lung and colon
cancer xenografts by depleting or inhibiting RLIP76 (Ral-binding
protein 1). Cancer Res. 67:4382–4389. 2007. View Article : Google Scholar : PubMed/NCBI
|