Introduction
Gliomas are the most common primary tumors of the
central nervous system. Despite recent advances in surgical
approaches and available adjuvant therapies, a poor prognosis is
imminent due to the highly migratory and invasive nature of glioma
cells (1,2). Their active migration and invasion
ultimately lead to tumor recurrence and reduced life expectancy
(3). Nevertheless, the molecular
mechanisms underlying glioma cell migration and invasion remain
unclear. Therefore, it is vital to investigate the signaling
pathways mediating glioma pathogenesis and progression in order to
formulate efficient therapeutic regimes.
Ubiquitination is essential in many biologic
processes, while aberrant expression of the enzymes involved in the
regulation of the ubiquitination process plays a key role in
malignant transformation (4).
Protein ubiquitination is achieved in the ubiquitin-conjugating
process via the covalent attachment of enzymes E1, E2 and E3 on the
specific-target of substrate protein. The E3 ubiquitin ligases are
the key regulatory components, which recognize specific substrates
(5). Emerging evidence indicates
that the E3 ubiquitin ligase Nedd4-1 is involved in cancer
pathogenesis (6,7), and may be a target protein for cancer
therapy (8–10), as the overexpression of Nedd4-1 in
colorectal and gastric cancers has been found to promote malignant
cell proliferation (11,12). FoxM1-regulated Nedd4-1 expression
was reported to destabilize phosphatase and tensin homolog deleted
on chromosome 10 (PTEN) and to promote astrocyte transformation and
glioma formation (5). To date, PTEN
is the substrate which has received the most attention in Nedd4-1
ubiquitination (6,13,14).
PTEN is argued to be dispensable for Nedd4-1 (15,16),
which is supported by numerous studies (17–21).
In addition, Nedd4-1 has been reported in several cases of human
tumors, whereas the functional significance of Nedd4-1 in glioma
remains unclear.
Rap, the closest homolog of the small G protein Ras,
plays a variety of biological functions in human cells, such as
signal transduction, migration and proliferation (22,23).
In addition to the recognized multiple mammalian proteins as
Nedd4-1 substrates (24), the
mono-ubiquitination of Rap2a by Nedd4-1 reportedly controls neurite
development and decreases Rap2a activity in mouse neurons (24). Recent studies indicate that
upregulation of Rap2a expression is correlated with tumor formation
and progressive malignancy in human prostate and follicular thyroid
cancer (25,26), whereas our previous study indicated
that Rap2a is modestly expressed in glioma (27). Thus, further investigations are
warranted to ascertain the effects of Nedd4-1-mediated Rap2a
ubiquitination on the migration and invasion of glioma cells.
In the present study, we investigated the role of
Nedd4-1 and the involvement of intracellular pathways in glioma. We
validated that Nedd4-1 protein was overexpressed in glioma tissues
vs. that noted in non-cancerous tissues. Moreover, Nedd4-1 promoted
the migration and invasion of glioma cells, mediated by
Nedd4-1/Rap2a ubiquitination.
Materials and methods
Glioma and non-cancerous brain tissue
samples
The present study was approved by the Institutional
Review Boards of The Affiliated Hospital of Xuzhou Medical College.
Glioma tissue samples, obtained from patients undergoing resection
for glioma tumors at the Department of Neurosurgery of The
Affiliated Hospital of Xuzhou Medical College, were histologically
confirmed by experienced pathologists based on the World Health
Organization (WHO) grading system (28). Non-cancerous brain tissues were
collected from patients undergoing decompression surgery for
intracranial traumas.
Ethical approval
All procedures performed in the present study
involving human participants were in accordance with the ethical
standards of the Institutional and/or National Research Committee,
and with the 1964 Helsinki Declaration and its later amendments or
comparable ethical standards. Written informed consent was obtained
from all individual participants or their legally authorized
representatives prior to enrollment in the study.
Cell culture
Human glioma cell lines U251 and U87 (Shanghai Cell
Bank, Type Culture Collection Committee, Chinese Academy of
Sciences) were cultured in Dulbecco's modified Eagle's medium and
F-12 (DMEM/F-12) (Invitrogen, Carlsbad, CA, USA) supplemented with
10% fetal bovine serum (FBS) (Sijiqing, Hangzhou, China), followed
by incubation at 37̊C in a humidified incubator with 5%
CO2.
Plasmids and antibodies
The mammalian expression plasmid encoding
full-length human Nedd4-1 was a gift from Dr Xuejun Jiang (Memorial
Sloan-Kettering Cancer Center, New York, USA). The shNedd4-1
plasmid was constructed by Shanghai GenePhama, and the human
Nedd4-1 residues 217–549 was a gift from Dr Nile Brose (Max Planck
Institute, Göttingen, Germany). The Myc-tagged Rap2a plasmids,
including wild-type (WT-Rap2a), dominant-active (DA-Rap2a) and
dominant-negative (DN-Rap2a), were gifts from Dr Kenichi Kariya
(University of the Ryukyus, Okinawa, Japan). DA-Rap2a and DN-Rap2a
were constructed by mutating Gly 12 to Val and Ser 17 to Asn,
respectively. Hemagglutinin (HA)-tagged ubiquitin and its mutants
including K63R-ubiquitin and K48 R-ubiquitin were gifts from
Shanghai Institute of Neuroscience, Chinese Academy of Sciences.
The bacterial expression plasmid for the glutathione S-transferase
(GST)-fusion protein of Ras-binding region of RalGDS was cloned by
reverse transcription polymerase chain reaction (RT-PCR) using a
human brain cDNA library as a template. The following antibodies
were commercial products: rabbit polyclonal antibodies to Nedd4-1
and Rap2a (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA),
mouse anti-TNIK (BD Biosciences, Franklin Lakes, NJ, USA), mouse
monoclonal antibodies P4D1 and FK1 to ubiquitin (Santa Cruz
Biotechnology, Inc.), rabbit monoclonal antibodies to ubiquitin
Apu2 and Apu3, and mouse monoclonal (9E10) antibody to Myc
(Millipore, Billerica, MA, USA).
Reverse transcription-PCR
Total RNA was extracted from U251 and U87 cells
using TRIzol reagent (Tiangen, Beijing, China). The RT-PCR protocol
was performed according to the manufacturer's instructions using
reverse transcription reagents (Takara, Dalian, China). Primers
(Invitrogen, Shanghai, China) for Nedd4-1 were as follows: sense,
5′-ACTTTATCCATTACCGACAG-3′ and antisense, 5′-TGGTGGCTTCATCTTCTC-3′,
yielding a 320-bp product. Primers for the internal standard
β-actin were: sense, 5′-GCGCGGCTACAGCTTCAC-3′ and antisense,
5′-GGGGCCGGACTCGTCATA-3′, yielding a 500-bp product. PCR conditions
were as follows: the first-strand DNA template was heated at 94̊C
for 3 min before amplification, a total of 30 cycles (for Nedd4-1)
or 23 cycles (for β-actin) were used including 94̊C for 30 sec
(denaturation), 63̊C for 30 sec (annealing), 72̊C for 35 sec
(extension), and 72̊C for 7 min (extension).
Western blotting
Standard western blotting was performed using
antibodies against Nedd4-1, Rap2a, TNIK, Myc and β-actin. Cell
extracts were prepared 48 h after transfection and subjected to
10–12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis
(SDS-PAGE), followed by transference to 0.45-µm pore size
polyvinylidene fluoride membranes (Millipore) and blotted with the
antibodies as indicated.
Immunohistochemistry
Immunohistochemistry was performed as previously
described, with minor modifications (29). Paraffin-embedded tissue sections
were obtained from the Department of Pathology, The Affiliated
Hospital of Xuzhou Medical College, including 11 cases of WHO grade
II astrocytomas, 12 cases of WHO grade III anaplastic astrocytomas,
and 12 cases of WHO grade IV glioblastomas, with non-cancerous
brain tissues as controls. Slices were incubated at 4̊C overnight
with the anti-Nedd4-1 antibody (1:100 dilution; Abcam, Cambridge,
MA, USA). Subsequent to microphotography, the percentages of glioma
cells with positive staining were presented and the Nedd4-1
expression levels were thereby categorized.
Transfection assay
Cell transfection was performed using PolyJet™ In
Vitro DNA Transfection Reagent (SignaGen, Rockville, MD, USA)
when U251 cells reached 90% confluence on 10-cm plates. Plasmids
encoding shRNA-Nedd4-1, full-length Nedd4-1, HA-tagged ubiquitin,
and Myc-tagged Rap2a and its mutants were transfected in the
respective experiments. Forty-eight hours after transfection, the
cells were harvested, rinsed with phosphate-buffered saline (PBS)
and lysed in 1% SDS or 1% NP40 buffer.
In vitro scratch assay
Cell motility was examined by scratch assay as
previously described, except for minor modifications (30). Twelve hours after transfection, an
artificial gap was created on the confluent cell monolayer using a
plastic pipette tip. Migrated cells were quantified at 48 h
(magnification, ×200) in order to compare with the cell counts at
base line with a computer-aided microscopy imaging system. All
experiments were performed in triplicates.
Migration and invasion assays
The capabilities of cell migration were assessed by
Transwell chamber assay. The invasion assay was performed as
previously described with minor modifications (31). Glioma cells were harvested and
resuspended in serum-free medium at a concentration of
1×105 cells/ml, and 200 µl was added to the top chamber.
The chambers were incubated for 24 h at 37̊C in an incubator with
5% CO2. The noninvasive cells on the upper surface of
the membrane were removed by wiping with cotton-tipped swabs. The
8-µm pore-size membrane was fixed and stained, and the population
of the invaded cells was counted by light microscopy. All the
experiments were performed in triplicates.
Affinity pull-down assay with GST-Nedd4-1. The
pull-down assay was performed as previously described with minor
modifications (24). Total proteins
were extracted from a 1% NP-40 extract of U251 cells and separated
by 12% SDS-PAGE followed by western blotting using anti-TNIK
(1:500) and anti-Rap2a (1:500) antibodies.
Chemical cross-linking of the
endogenous Nedd4-1/TNIK/Rap2 complex
U251 cells in 10-cm culture dishes were incubated
with 1 mM dithiobis(succinimidyl propionate) (DSP) (Pierce,
Rockford, IL, USA) for 3 min on ice. Excess DSP was quenched with
20 mM Tris-Cl (pH 7.5 at 4̊C) for 15 min at room temperature, and
then the proteins were extracted with RIPA buffer [50 mM Tris-Cl
(pH 8.0) at 4̊C, 5 mM EDTA, 150 mM NaCl, 1% NP-40, 0.5% sodium
deoxycholate, 0.1% SDS, 0.2 mM phenylmethylsulfonyl fluoride
(PMSF), 1 µg/ml aprotinin and 0.5 µg/ml leupeptin]. Nedd4-1 was
immunoprecipitated with anti-Nedd4-1 polyclonal antibodies coupled
to protein A-sepharose beads (GE Healthcare, Milwaukee, WI, USA) at
4̊C for 14 h. The beads were rinsed with RIPA buffer, and then the
precipitated proteins were eluted and boiled in 2X loading buffer
containing 50 mM DTT, and separated by SDS-PAGE followed by western
blotting using anti-Nedd4-1, anti-TNIK and anti-Rap2 antibodies,
respectively.
GST-RalGDS pull-down assay
A total of 40 µg of GST fusion proteins of RalGDS
encoding the Ras-binding domain (RBD) was purified and immobilized
on GST-sepharose beads to pull down the GTP-bound form of Myc-Rap2a
extracted from the U251 cells transfected with Myc-Rap2a, Nedd4-1
and HA-ubiquitin. After extensive washing at 4̊C overnight with a
buffer containing 50 mM Tris-HCl (pH 7.5), 200 mM NaCl, 2.5 mM
MgCl2, 10% glycerol and 1% NP-40, the bound proteins
were eluted by boiling in 2X loading buffer followed by SDS-PAGE
and western blotting.
Statistical analysis
Data are presented as mean ± standard error of mean
(SEM). Statistical significance was determined using Student's
t-tests for comparisons between two groups and ANOVA for intergroup
comparisons. P-values of <0.05 were considered statistically
significant (P<0.05).
Results
Nedd4-1 expression in human glioma
tissue samples
To investigate the possible effect in glioma
pathogenesis, we determined Nedd4-1 protein expression in clinical
glioma tissues and non-cancerous brain samples. A representative
immunoblot is shown in Fig. 1A.
Analysis of the band density revealed that Nedd4-1 was
significantly upregulated in the glioma tissues vs. that observed
in the non-cancerous samples (Fig.
1B). We further verified Nedd4-1 expression by
immunohistochemistry. As shown in Fig.
1C, Nedd4-1 immunoreactivity was predominant in the cytoplasm.
As shown in Fig. 1D, the percentage
of Nedd4-1-positive cells reached 57.35±2.02% (WHO grade II) and
75.82±3.06% (WHO grade III–IV) in glioma tissues vs. the percentage
of 22.30±2.17% in non-cancerous brain samples. Thus, Nedd4-1
expression was significantly correlated with glioma malignancy.
Fig. 1 shows significant
upregulation of Nedd4-1 in glioma tissues vs. non-cancerous brain
samples. These results indicated the potential involvement of
Nedd4-1 in the pathogenesis of human glioma.
 | Figure 1.Expression profile of Nedd4-1 in
human glioma tissues. (A) Representative western blotting of the
total protein extracts isolated from human glioma (7 samples shown)
specimens and human non-cancerous (5 samples shown) brain using
rabbit Nedd4-1 antibody. β-actin served as a loading control. (B)
Relative protein levels of Nedd4-1 in glioma speimens (n=35) and
non-cancerous brain tissue (n=15). Data are expressed as the mean ±
SEM from three independent experiments; **P<0.01 compared with
non-cancerous brain tissue. (C) Representative IHC analyses of
Nedd4-1 expression in non-cancerous brain and human glioma tissues
of various (WHO) grades. a, Normal brain tissue; b, diffuse
astrocytoma, grade II; c, anaplastic astrocytoma, grade III; and d,
GBM, grade IV are shown. Scale bar, 20 µm. (D) Quantitative
analysis of the percentage of Nedd4-1-positive cells; **P<0.01
compared with non-cancerous brain tissue. Grade II, low-grade
glioma. Grade III–IV, high-grade glioma. |
Effects of Nedd4-1 on cell
migration
Given the overexpression of Nedd4-1 in glioma in
contrast to non-cancerous tissues, we employed a gain-of-function
strategy to assess the effect of Nedd4-1 on glioma cell migration.
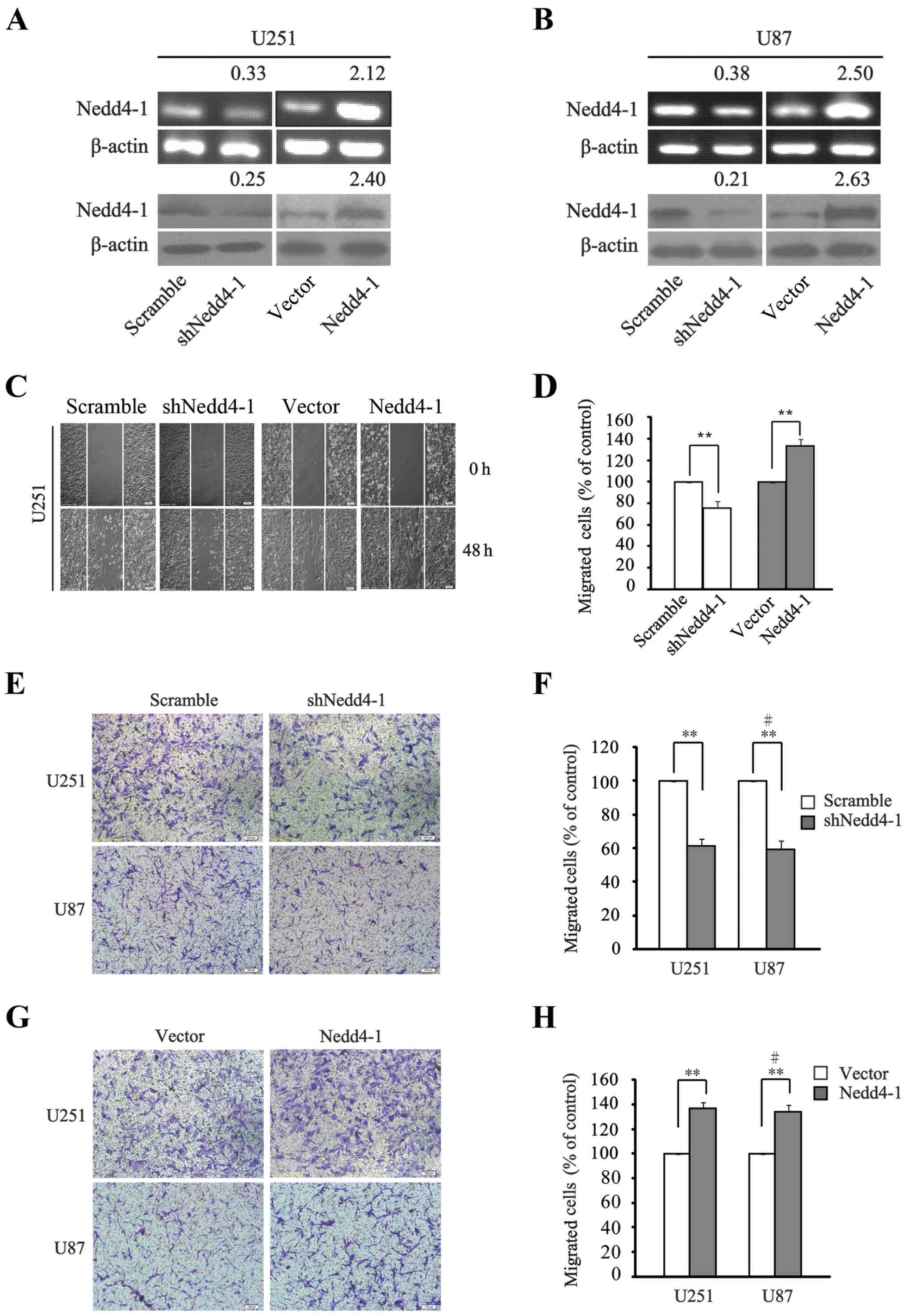
As shown in Fig. 2A and B,
exogenous shNedd4-1 and Nedd4-1 were added to interfere with or
overexpress Nedd4-1 in the U251 and U87 cell lines. The effect of
Nedd4-1 on cell migration was assessed by scratch assay. It was
shown that the migratory capacity of glioma U251 cells was
significantly reduced upon downregulation of Nedd4-1 as compared
with the scramble group. Conversely, the scratch in the
Nedd4-1-overexpressing group was rapidly replenished compared with
the vector group (Fig. 2C and D).
We also performed cell migration assays using Transwell chambers to
investigate the migratory capacities of the U251 and U87 cells.
Likewise, the shNedd4-1 group showed significantly reduced
migration compared with that noted in the scramble group (Fig. 2E and F), whereas the migratory
behavior in the Nedd4-1-overexpressing group was evidently enhanced
compared with that noted in the vector group in both the U251 and
U87 cell lines (Fig. 2G and H).
Collectively, we demonstrated that either the downregulation or
overexpression of Nedd4-1 exerted relevant causality on the
migratory capacity of both cell lines.
Effects of Nedd4-1 on cell
invasion
In addition to the regulation of cell migration by
Nedd4-1, we investigated the effect of Nedd4-1 on cell invasion
with Matrigel-precoated Transwell chambers. The results indicated
that the invasion of U251 and U87 cells was significantly
diminished by the downregulation of Nedd4-1 vs. the scramble group
subsequent to 24 h of incubation (Fig.
3A and B), while the invasion in the Nedd4-1-overexpressing
group was markedly enhanced vs. that noted in the empty vector
group in both cell lines (Fig. 3C and
D). Thus, both downregulation and overexpression of Nedd4-1
regulated the invasive capacities of the cell lines.
Rap2a as a Nedd4-1 substrate
To verify the role of Nedd4-1 targets in glioma
cells, we selected the Nedd4-1 WW domains (32) (residue 217–547) as an affinity
matrix and U251 cell lysate (1% NP40) to pull down TNIK and Rap2a.
The results exhibited the presence of TNIK and Rap2a in the
affinity-purified complex, confirming the binding of Nedd4-1 to
either of the substrates (Fig. 4A).
We further identified that endogenous Nedd4-1, TNIK and Rap2a
constituted a complex (Fig. 4B).
U251 cell lysate was incubated with thiol-cleavable cross-linker
prior to immunoprecipitation with an anti-Nedd4-1 antibody. Under
this condition, TNIK and Rap2a were coimmunoprecipitated with
Nedd4-1.
 | Figure 4.Rap2a is a Nedd4-1 substrate. (A)
Complex of Rap2a with TNIK and GST-Nedd4-1 (residues 217–549).
Protein was extracted from U251 cells and blotted for TNIK (top)
and Rap2a (bottom) after washing GST-Nedd4-1 beads. GST beads were
used as a negative control. (B) Endogenous Rap2a, TNIK and Nedd4-1
form a complex. Nedd4-1 was immunoprecipitated using a rabbit
polyclonal anti-Nedd4-1 antibody from U251 cell extract after
treatment with a thiol-cleavable chemical cross-linker. Western
blot analyses were probed with anti-Nedd4-1, anti-TNIK and
anti-Rap2a antibodies. Rap2a and TNIK co-immunoprecipitated with
Nedd4-1, but no band was observed with the negative control IgG.
(C) Nedd4-1 selectively ubiquitinates Rap2a. Nedd4-1 (+) or (−) was
co-expressed with WT, dominant-active (DA), or dominant-negative
(DN) mutants of Myc-tagged Rap2a. Myc-Rap2a was immunoprecipitated
using a mouse monoclonal anti-Myc antibody. Immunoprecipitates were
blotted for Myc (bottom panel) and ubiquitin (P4D1, top panel).
Note that Nedd4-1 selectively ubiquitinated WT and DA mutants of
Myc-tagged Rap2a but not the DN mutant of Myc-Rap2a. (D) Mono- and
di-ubiquitination of Rap2a by Nedd4-1. Myc-Rap2a was
immunoprecipitated from U251 cells expressing Myc-Rap2a (WT) with
or without Nedd4-1. Myc-Rap2a was eluted from anti-Myc
antibody-coupled beads and immunoblotted with different monoclonal
mouse or rabbit anti-ubiquitin antibodies: P4D1, FK1, Apu3 and
Apu2. P4D1 recognizes both poly- and mono-ubiquitin-conjugated
proteins, and FK1 recognizes only poly-ubiquitin-conjugated ones.
Apu3 and Apu2 recognize K63- and K48-linked poly-ubiquitin chains,
respectively. Lysate from U251 cells overexpressing Nedd4-1 was
also blotted with the antibodies described above to show that the
antibody titers are comparable. Asterisks in (C and D) indicate
bands corresponding to Myc-Rap2a conjugated with two ubiquitin
moieties. Triangular symbols in (C and D) indicate the light chain
of the anti-Myc used for immunoprecipitation. (E) The function of
Rap2a by Nedd4-1 ubiquitination. WT Myc-Rap2a expressed in U251
cells with or without overexpressed Nedd4-1 was precipitated using
anti-Myc antibodies. GTP-bound Rap2a was assessed with GST-RalGDS
pull-down assays compared with 20% total and GST as negative
control. (F) Relative GTP-Rap2a activity. Note that GTP-bound
active Rap2a was obviously decreased in the overexpressed Nedd4-1
condition (sixth lane). Data are expressed as the means ± SEM of
three independent experiments; **P<0.01 compared without Nedd4-1
overexpression. |
Rap2a rather than TNIK was found to be ubiquitinated
by Nedd4-1 in HEK cells, which decreased the activity of the
GTP-bound form of Rap2a (24). To
ascertain the potential effect of Nedd4-1 on Rap2a ubiquitination
in glioma cells, expression plasmids for the full-length Nedd4-1,
HA-tagged ubiquitin as well as Myc-tagged WT-Rap2a, DA-G12V Rap2a
or DN-S17N Rap2a were co-transfected in the U251 cells, with
lysates extracted under denaturing conditions (100̊C, 1% SDS) and
Myc-tagged proteins immunoprecipitated from cell extracts using
anti-Myc antibodies. As shown in Fig.
4C, immunoprecipitates from WT and DA-G12V instead of
DN-S17N-expressing U251 cells showed a gradient of protein bands
visible to an anti-ubiquitin antibody (P4D1) only in the presence
of Nedd4-1 overexpression. These data demonstrated that Nedd4-1
selectively ubiquitinated WT-Rap2a and DA-Rap2a, but not
DN-Rap2a.
To investigate the exact form of ubiquitination of
Rap2a (i.e. mono-ubiquitination, di-ubiquitination or
poly-ubiquitination), expression plasmids for full-length Nedd4-1,
HA-tagged ubiquitin and Myc-tagged WT-Rap2a were co-transfected in
U251 cells. Four types of anti-ubiquitin monoclonal antibodies
which discriminate between mono-ubiquitinated and
poly-ubiquitinated proteins were employed: P4D1 for poly-ubiquitin
and mono-ubiquitin conjugates; FK1 only for poly-ubiquitin
conjugates; Apu2 and Apu3 for K48-linked and K63-linked
poly-ubiquitin chains, respectively. As shown in Fig. 4D, the escalation of ubiquitinated
Rap2a was only identified by P4D1, indicating mono-ubiquitination
of Rap2a by Nedd4-1. Apu3 recognized a single faint band
representing ubiquitinated Rap2a by Nedd4-1 via K63-linked
ubiquitination. No bands were determined by Apu2, exempting the
Rap2a ubiquitination by Nedd4-1 from K48-linked polyubiquitin
chain, which may lead to proteasome-dependent degradation. A total
lysate of Neddd4-1-expressing U251 cells was immunoblotted using
these antibodies as controls with blurred bands.
Effect of Nedd4-1 ubiquitination on
Rap2a function
To further investigate the effect of
Nedd4-1-mediated ubiquitination on Rap2a function in glioma U251
cells, we purified and immobilized a GST fusion of the
Rap2a-binding region of RalGDS, which served to pull down active
GTP-bound Rap2a. Nedd4-1 and Myc-tagged Rap2a were thereby
co-transfected in the U251 cells, and the lysate was loaded onto
GST-RalGDS or onto GST as a control. Only active GTP-bound Rap2a
was pulled down, indicating that ubiquitination blocked Rap2a
function by inhibiting the activity of the GTP-bound Rap2a
(Fig. 4E and F).
Effects of Nedd4-1-mediated Rap2a
ubiquitination on cell migration and invasion
To verify the role of Nedd4-1-mediated
mono-ubiquitination of Rap2a in cell migration and invasion,
expression plasmids for full-length Nedd4-1, HA-tagged WT-ubiquitin
and its mutants (including HA-tagged K63R-ubiquitin, HA-tagged
K48R-ubiquitin, HA-tagged ubiquitin and Myc-tagged WT-Rap2a) were
co-transfected in the U251 and U87 cell lines, with MG-132 added to
exclude the interference of multiple-ubiquitination in all groups.
We assessed migration and invasion parameters in each group of both
U251 and U87 cells. As shown in Fig. 5A
and B, Nedd4-1 and Rap2a were overexpressed in the U251 and U87
cell lines. The Transwell assay results demonstrated that the
migration of glioma cells was significantly enhanced in the
WT-ubiquitin and K48R-ubiquitin groups vs. the vector and
K63R-ubiquitin groups (Fig. 5C and
D). The invasive capacity of glioma cells was also
significantly increased in the WT-ubiquitin and K48R-ubiquitin
groups vs. the vector and K63R-ubiquitin groups in the Matrigel
Transwell assay (Fig. 5E and F).
Furthermore, as illustrated (Fig.
5G), the results of immunoprecipitation revealed the Rap2a
mono-ubiquitination of Nedd4-1 via K63-ubiquitin. These results
indicated that Nedd4-1 may regulate the migration and invasion of
glioma cells via the inhibition of GTP-Rap2a.
Discussion
In the present study, we confirmed that Nedd4-1
promotes the migration and invasion of glioma cells via the
inhibition of GTP-Rap2a, a member of the Ras family of small G
proteins. This finding clarified the role of Nedd4-1 in migration,
invasion and other important pathogenetic processes in glioma.
Nedd4-1, an E3 ligase, plays essential regulatory
roles in physiological processes, which is exemplified by its
direct binding to and ubqiuitination of activated FGFR1, thus
regulating the endocytosis and signaling pathways of FGFR1 in
neural differentiation and embryonic development (32). Nevertheless, increasing evidence
shows that Nedd4-1 is also involved in a variety of cancers such as
bladder, lung and brain carcinomas (5,6,33). In
the present study, we identified that the level of Nedd4-1 protein
in glioma tissues was approximately 2- to 3-fold higher than that
in non-cancerous tissues (Fig. 1A and
C). The downregulation of Nedd4-1 reduced the migration and
invasion of human glioma cell lines U251 and U87, while Nedd4-1
overexpression reversed these phenomena (Figs. 2C-F and 3A-D). These findings suggest a pivotal
role of Nedd4-1 in glioma pathogenesis.
Nedd4-1 reportedly serves as a ubiquitin ligase for
multiple proteins (6,12,34,35),
with several different domains including C2, two to four WW domains
and a HECT domain. Nedd4-1 is reported to bind rather than
ubiquitinate TNIK in regulating the ubiquitination of GTP-Rap2a in
neuron development. In addition, Rap2a, in lieu of Rap1a, H-Ras and
K-Ras, is a specific substrate of Nedd4-1 (24). It is unclear whether Nedd4-1
ubiquitinates Rap2a in glioma. In the present study, Nedd4-1 was
found to bind to TNIK and Rap2a, forming a ternary complex in both
the GST-Nedd4-1 pull-down assays and co-immunoprecipitation in cell
line U251 (Fig. 4A and B). These
results suggest that the ubiquitination of Rap2a by Nedd4-1 may be
involved in the regulation of migration and invasion of glioma
cells. Moreover, the fate of the ubiquitinated proteins is often
determined by the chains of ubiquitin conjugation (36). Poly-ubiquitination of substrate
proteins by K48-linked ubiquitin chains leads to degradation within
the 26S proteasome, whereas K63-linked poly-ubiquitin chains,
mono-ubiquitination and multiple mono-ubiquitination usually
regulate protein function, gene transcription, endocytosis and DNA
repair (36,37). Additionally, there is a wealth of
evidence that selective mono-ubiquitination or alternative
ubiquitin chains can regulate protein activity (38–42).
Herein, WT-Rap2a and DA-Rap2a, but not DN-Rap2a (Fig. 4C) were robustly ubiquitinated by
Nedd4-1 by K-63-linked ubiquitin chains (Fig. 4D), with inhibition of GTP-Rap2a
activity (Fig. 4E). The present
study was supported by the study of Kawabe et al (24), in which Nedd4-1 ubiquitinated Rap2a
and inhibited GTP-Rap2a activity in neurons. Based on these
findings, we speculate that Rap2a ubiquitination by Nedd4-1 may
contribute to glioma pathogenesis.
Rap proteins belong to the Ras superfamily and have
been implicated in cell cycle control, cell adhesion and cell
migration (25,43,44).
To date, the role of Rap1 in tumorigenesis and progression remains
controversial, with some researchers hypothesizing that aberrant
Rap1 activation promotes cancer cell proliferation and
tumorigenesis (45–47) and others reporting that inactivation
of DOCK4, a Rap1 activator, rendered osteosarcoma cells with a
higher invasiveness (48), and the
expression of Rap1GAP was correlated with increased invasion in
vitro in squamous cell carcinoma (49). Similar to Rap1, the function of
Rap2a remains elusive despite its history of cloning (50). In the present study, overexpression
of Nedd4-1 promoted the migration and invasion of human glioma cell
lines U251 and U87 via Rap2a ubiquitination (Fig. 5C and D). Furthermore, only
WT-ubiquitin and K48R exhibited mono-ubiquitination (Fig.5G). Taken together, these findings
confirm the hypothesis that Nedd4-1 regulates the migration and
invasion of glioma cells via Rap2a ubiquitination. Three Rap2
proteins (Rap2a/b/c) have been cloned, but we only focused on the
function of Rap2a protein. Additionally, E3 ubiquitin ligase was
also found to be involved in the regulation of cell cycle,
apoptosis and differentiation (51,52),
which may illuminate our forthcoming investigation of glioma.
In summary, our findings suggest that Nedd4-1 plays
a pivotal role in promoting the migration and invasion of glioma
cell lines U251 and U87 via the inhibition of Rap2a activity, and
may qualify as a candidate therapeutic target in glioma.
Acknowledgements
The authors are grateful to Dr Kenichi Kariya, Dr
Nile Brose and Dr Kawabe Hiroshi for their benevolent donation of
the Rap2a plasmids. We are also indebted to Mr. Pan Li from Xuzhou
Medical College, for guidance in the style and manuscript editing.
The present study was funded by the Natural Science Foundation of
Jiangsu Province of China (BK20151165), and the National Natural
Science Foundation of China (81472345).
Glossary
Abbreviations
Abbreviations:
|
Nedd4-1
|
neuronal precursor cell expressed and
developmentally downregulated protein
|
|
TNIK
|
Traf2- and Nck-interacting kinase
|
|
Ub
|
ubiquitin
|
References
|
1
|
Watkins S and Sontheimer H: Hydrodynamic
cellular volume changes enable glioma cell invasion. J Neurosci.
31:17250–17259. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bush NA, Chang SM and Berger MS: Current
and future strategies for treatment of glioma. Neurosurg Rev.
40:1–14. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Demuth T, Rennert JL, Hoelzinger DB,
Reavie LB, Nakada M, Beaudry C, Nakada S, Anderson EM, Henrichs AN,
McDonough WS, et al: Glioma cells on the run - the migratory
transcriptome of 10 human glioma cell lines. BMC Genomics.
9:542008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nakayama KI and Nakayama K: Ubiquitin
ligases: Cell-cycle control and cancer. Nat Rev Cancer. 6:369–381.
2006. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dai B, Pieper RO, Li D, Wei P, Liu M, Woo
SY, Aldape KD, Sawaya R, Xie K and Huang S: FoxM1B regulates
NEDD4-1 expression, leading to cellular transformation and full
malignant phenotype in immortalized human astrocytes. Cancer Res.
70:2951–2961. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang X, Trotman LC, Koppie T, Alimonti A,
Chen Z, Gao Z, Wang J, Erdjument-Bromage H, Tempst P, Cordon-Cardo
C, et al: NEDD4-1 is a proto-oncogenic ubiquitin ligase for PTEN.
Cell. 128:129–139. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen Y, van de Vijver MJ, Hibshoosh H,
Parsons R and Saal LH: PTEN and NEDD4 in human breast carcinoma.
Pathol Oncol Res. 22:41–47. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sun Y: Targeting E3 ubiquitin ligases for
cancer therapy. Cancer Biol Ther. 2:623–629. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hoeller D and Dikic I: Targeting the
ubiquitin system in cancer therapy. Nature. 458:438–444. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yang Y, Kitagaki J, Wang H, Hou DX and
Perantoni AO: Targeting the ubiquitin-proteasome system for cancer
therapy. Cancer Sci. 100:24–28. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Eide PW, Cekaite L, Danielsen SA,
Eilertsen IA, Kjenseth A, Fykerud TA, Ågesen TH, Bruun J, Rivedal
E, Lothe RA, et al: NEDD4 is overexpressed in colorectal cancer and
promotes colonic cell growth independently of the PI3K/PTEN/AKT
pathway. Cell Signal. 25:12–18. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kim SS, Yoo NJ, Jeong EG, Kim MS and Lee
SH: Expression of NEDD4-1, a PTEN regulator, in gastric and
colorectal carcinomas. APMIS. 116:779–784. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Drinjakovic J, Jung H, Campbell DS,
Strochlic L, Dwivedy A and Holt CE: E3 ligase Nedd4 promotes axon
branching by downregulating PTEN. Neuron. 65:341–357. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kwak YD, Wang B, Pan W, Xu H, Jiang X and
Liao FF: Functional interaction of phosphatase and tensin homologue
(PTEN) with the E3 ligase NEDD4-1 during neuronal response to zinc.
J Biol Chem. 285:9847–9857. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cao XR, Lill NL, Boase N, Shi PP, Croucher
DR, Shan H, Qu J, Sweezer EM, Place T, Kirby PA, et al: Nedd4
controls animal growth by regulating IGF-1 signaling. Sci Signal.
1:ra52008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fouladkou F, Landry T, Kawabe H, Neeb A,
Lu C, Brose N, Stambolic V and Rotin D: The ubiquitin ligase
Nedd4-1 is dispensable for the regulation of PTEN stability and
localization. Proc Natl Acad Sci USA. 105:8585–8590. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li J, Yen C, Liaw D, Podsypanina K, Bose
S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R, et al:
PTEN, a putative protein tyrosine phosphatase gene mutated
in human brain, breast, and prostate cancer. Science.
275:1943–1947. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Carico C, Nuño M, Mukherjee D, Elramsisy
A, Dantis J, Hu J, Rudnick J, Yu JS, Black KL, Bannykh SI, et al:
Loss of PTEN is not associated with poor survival in newly
diagnosed glioblastoma patients of the temozolomide era. PLoS One.
7:e336842012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zainuddin N, Jaafar H, Isa MN and Abdullah
JM: Malignant glioma: The involvement of loss of allelic
heterozygosity and PTEN mutations in a group of Malay patients.
Southeast Asian J Trop Med Public Health. 36:748–756.
2005.PubMed/NCBI
|
|
20
|
Wei Q, Clarke L, Scheidenhelm DK, Qian B,
Tong A, Sabha N, Karim Z, Bock NA, Reti R, Swoboda R, et al:
High-grade glioma formation results from postnatal pten loss or
mutant epidermal growth factor receptor expression in a transgenic
mouse glioma model. Cancer Res. 66:7429–7437. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Parsa AT, Waldron JS, Panner A, Crane CA,
Parney IF, Barry JJ, Cachola KE, Murray JC, Tihan T, Jensen MC, et
al: Loss of tumor suppressor PTEN function increases B7-H1
expression and immunoresistance in glioma. Nat Med. 13:84–88. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Itoh M, Nelson CM, Myers CA and Bissell
MJ: Rap1 integrates tissue polarity, lumen formation, and
tumorigenic potential in human breast epithelial cells. Cancer Res.
67:4759–4766. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kooistra MR, Dubé N and Bos JL: Rap1: A
key regulator in cell-cell junction formation. J Cell Sci.
120:17–22. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kawabe H, Neeb A, Dimova K, Young SM Jr,
Takeda M, Katsurabayashi S, Mitkovski M, Malakhova OA, Zhang DE,
Umikawa M, et al: Regulation of Rap2A by the ubiquitin ligase
Nedd4-1 controls neurite development. Neuron. 65:358–372. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bigler D, Gioeli D, Conaway MR, Weber MJ
and Theodorescu D: Rap2 regulates androgen sensitivity in human
prostate cancer cells. Prostate. 67:1590–1599. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Prabakaran I, Grau JR, Lewis R, Fraker DL
and Guvakova MA: Rap2A is upregulated in invasive cells
dissected from follicular thyroid cancer. J Thyroid Res.
2011:9798402011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang L, Zhan W, Xie S, Hu J, Shi Q, Zhou
X, Wu Y, Wang S, Fei Z and Yu R: Over-expression of Rap2a inhibits
glioma migration and invasion by down-regulating p-AKT. Cell Biol
Int. 38:326–334. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Louis DN, Ohgaki H, Wiestler OD, Cavenee
WK, Burger PC, Jouvet A, Scheithauer BW and Kleihues P: The 2007
WHO classification of tumours of the central nervous system. Acta
Neuropathol. 114:97–109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhou X, Hua L, Zhang W, Zhu M, Shi Q, Li
F, Zhang L, Song C and Yu R: FRK controls migration and invasion of
human glioma cells by regulating JNK/c-Jun signaling. J Neurooncol.
110:9–19. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liang CC, Park AY and Guan JL: In vitro
scratch assay: A convenient and inexpensive method for analysis of
cell migration in vitro. Nat Protoc. 2:329–333. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
van Golen KL, Risin S, Staroselsky A,
Berger D, Tainsky MA, Pathak S and Price JE: Predominance of the
metastatic phenotype in hybrids formed by fusion of mouse and human
melanoma clones. Clin Exp Metastasis. 14:95–106. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Persaud A, Alberts P, Hayes M, Guettler S,
Clarke I, Sicheri F, Dirks P, Ciruna B and Rotin D: Nedd4-1 binds
and ubiquitylates activated FGFR1 to control its endocytosis and
function. EMBO J. 30:3259–3273. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yim EK, Peng G, Dai H, Hu R, Li K, Lu Y,
Mills GB, Meric-Bernstam F, Hennessy BT, Craven RJ, et al: Rak
functions as a tumor suppressor by regulating PTEN protein
stability and function. Cancer Cell. 15:304–314. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Staub O, Dho S, Henry P, Correa J,
Ishikawa T, McGlade J and Rotin D: WW domains of Nedd4 bind to the
proline-rich PY motifs in the epithelial Na+ channel
deleted in Liddle's syndrome. EMBO J. 15:2371–2380. 1996.PubMed/NCBI
|
|
35
|
Murdaca J, Treins C, Monthouël-Kartmann
MN, Pontier-Bres R, Kumar S, Van Obberghen E and Giorgetti-Peraldi
S: Grb10 prevents Nedd4-mediated vascular endothelial growth factor
receptor-2 degradation. J Biol Chem. 279:26754–26761. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Shearwin-Whyatt L, Dalton HE, Foot N and
Kumar S: Regulation of functional diversity within the Nedd4 family
by accessory and adaptor proteins. BioEssays. 28:617–628. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Haglund K and Dikic I: Ubiquitylation and
cell signaling. EMBO J. 24:3353–3359. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lai Z, Ferry KV, Diamond MA, Wee KE, Kim
YB, Ma J, Yang T, Benfield PA, Copeland RA and Auger KR: Human mdm2
mediates multiple mono-ubiquitination of p53 by a mechanism
requiring enzyme isomerization. J Biol Chem. 276:31357–31367. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Dong L and Xu CW: Carbohydrates induce
mono-ubiquitination of H2B in yeast. J Biol Chem. 279:1577–1580.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Brenkman AB, de Keizer PL, van den Broek
NJ, Jochemsen AG and Burgering BM: Mdm2 induces mono-ubiquitination
of FOXO4. PLoS One. 3:e28192008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Torres MP, Lee MJ, Ding F, Purbeck C,
Kuhlman B, Dokholyan NV and Dohlman HG: G protein
mono-ubiquitination by the Rsp5 ubiquitin ligase. J Biol Chem.
284:8940–8950. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wu K, Yan H, Fang L, Wang X, Pfleger C,
Jiang X, Huang L and Pan ZQ: Mono-ubiquitination drives nuclear
export of the human DCN1-like protein hDCNL1. J Biol Chem.
286:34060–34070. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
McLeod SJ, Shum AJ, Lee RL, Takei F and
Gold MR: The Rap GTPases regulate integrin-mediated adhesion, cell
spreading, actin polymerization, and Pyk2 tyrosine phosphorylation
in B lymphocytes. J Biol Chem. 279:12009–12019. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Price LS, Hajdo-Milasinovic A, Zhao J,
Zwartkruis FJ, Collard JG and Bos JL: Rap1 regulates
E-cadherin-mediated cell-cell adhesion. J Biol Chem.
279:35127–35132. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhang L, Chenwei L, Mahmood R, van Golen
K, Greenson J, Li G, D'Silva NJ, Li X, Burant CF, Logsdon CD, et
al: Identification of a putative tumor suppressor gene
Rap1GAP in pancreatic cancer. Cancer Res. 66:898–906. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Gao L, Feng Y, Bowers R, Becker-Hapak M,
Gardner J, Council L, Linette G, Zhao H and Cornelius LA:
Ras-associated protein-1 regulates extracellular signal-regulated
kinase activation and migration in melanoma cells: Two processes
important to melanoma tumorigenesis and metastasis. Cancer Res.
66:7880–7888. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ishida D, Kometani K, Yang H, Kakugawa K,
Masuda K, Iwai K, Suzuki M, Itohara S, Nakahata T, Hiai H, et al:
Myeloproliferative stem cell disorders by deregulated Rap1
activation in SPA-1-deficient mice. Cancer Cell. 4:55–65.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Yajnik V, Paulding C, Sordella R,
McClatchey AI, Saito M, Wahrer DC, Reynolds P, Bell DW, Lake R, van
den Heuvel S, et al: DOCK4, a GTPase activator, is disrupted
during tumorigenesis. Cell. 112:673–684. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Mitra RS, Goto M, Lee JS, Maldonado D,
Taylor JM, Pan Q, Carey TE, Bradford CR, Prince ME, Cordell KG, et
al: Rap1GAP promotes invasion via induction of matrix
metalloproteinase 9 secretion, which is associated with poor
survival in low N-stage squamous cell carcinoma. Cancer Res.
68:3959–3969. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Pizon V, Chardin P, Lerosey I, Olofsson B
and Tavitian A: Human cDNAs rap1 and rap2 homologous to the
Drosophila gene Dras3 encode proteins closely related to ras
in the ‘effector’ region. Oncogene. 3:201–204. 1988.PubMed/NCBI
|
|
51
|
Chantry A: WWP2 ubiquitin ligase and its
isoforms: New biological insight and promising disease targets.
Cell Cycle. 10:2437–2439. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Tuoc TC and Stoykova A: Roles of the
ubiquitin-proteosome system in neurogenesis. Cell Cycle.
9:3174–3180. 2010. View Article : Google Scholar : PubMed/NCBI
|