Introduction
Hepatocellular carcinoma (HCC) is one of the most
fatal cancers in the world while its incidence and death rate are
still increasing (1). Most studies
have demonstrated that HCC is a complex and multifarious molecular
disease (2,3). However, the molecular pathogenesis of
HCC remains elusive. Elucidation of some aberrant genes and
mechanisms in HCC is required to investigate more specific potent
targeted therapy and diagnosis methods.
MicroRNAs (miRNAs) are a type of small and
endogenous non-coding RNAs of 19–23 nucleotides that negatively
regulate gene expression by binding to the untranslated regions of
target messenger RNAs (mRNAs). Growing evidence indicates that
miRNAs play an important role in diverse biological processes, and
the aberrant expression of miRNAs contributes to tumorigenesis,
progression, diagnosis and prognosis (4–6).
miR-144-3p has been reported aberrantly and lowly expressed in
thyroid cancer targeting ZEB1 and ZEB2 could suppress the invasion
and migration capability of thyroid cancer cells (7). Moreover, as it exerts direct
regulatory roles on ZFX expression, further investigations showed
that miR-144-3p expression not only inhibits NSCLC tumor cell
growth but induces apoptosis (8).
Similar to previous studies, our previous microarray profiling also
found that miR-144-3p was downregulated in HCC (9). Though several reports revealed that
miR-144-3p can suppress proliferation of HCC by targeting AKT3 and
E2F3 (10,11), the function of miR-144-3p in tumor
angiogenesis is still unknown (12,13),
hence we investigated the role of miR-144-3p in angiogenesis and
the relationship with the survival time of HCC patients so that we
can explore new targets of anti-angiogenesis treatments in
clinic.
In this study, we found miR-144-3p not only
suppressed tumor growth in HCC, but also inhibited the ability of
angiogenesis of HCC cells. Functional studies also demonstrated
that the inhibitory effect of miR-144-3p on HCC is mainly mediated
by targeted SGK3, which reduces the activity of mTOR-VEGF pathway
in PI3K downstream signaling.
Materials and methods
Patients and tissue samples
Surgically resected paired HCC and adjacent
non-cancerous tissues were collected from 51 primary HCC patients
at the Affiliated Tumor Hospital of Guangxi Medical University
between March 2011 and May 2013. Tissue samples were immediately
frozen in liquid nitrogen until analysis. These cases selected were
based on a clear pathological diagnosis, follow-up data, and had
first undergone radical resection of HCC, and had not received
preoperative adjuvant chemotherapy, radiotherapy, targeted therapy
or immunotherapy. Informed consent was obtained from each patient,
and the study was approved by the Ethics Committee of Guangxi
Medical University, Nanning, China. The investigations were
conducted according to the Principles of Declaration of
Helsinki.
Cell culture and transfection
All cells were obtained from the Institute of
Biochemistry and Cell Biology of Chinese Academy of Science
(Shanghai, China). Human HCC cell lines (QGY-7703, SK-hep1 and
human normal liver cells (HL-7702) were maintained in RPMI-1640
with 10% fetal bovine serum (FBS) (Gibco, USA) at 37°C in a
humidified incubator containing 5% CO2. miR-144-3p
duplex mimics and negative control (NC) were from GenePharma
(Shanghai, China). Sequences used for the miR-144-3p mimics and the
corresponding control were 5′-UACAGUAUAGAUGAUGUACU-3′ and
5′-UUUGUACUACACAAAAGUACUG-3′, respectively. Sequences used for SGK3
siRNA, and NC were 5′-GCAUUGGGUUACAUUTT-3′, and
5′-UGACCUCAACUACAUGGUUTT-3′, respectively. Cells were transfected
with RNAs using INTERFERin® transfection reagent
(Polyplus Transfection, Illkirch, France) at a final concentration
of 100 nM according to the manufacturer's instructions.
RNA extraction and quantitative
RT-PCR
Total RNA, including miRNA, was extracted using
TRIzol reagent (Invitrogen, CA, USA) according to the
manufacturer's instructions. cDNAs were synthesized using ReverTra
Ace® qPCR RT kit (FSQ-101, Toyobo, Japan). MicroRNA was
reversely transcribed using First Strand cDNA Synthesis kit
ReverTra Ace-α- (FSK-100, Toyobo). Real-time PCR analyses were
performed with Thunderbird SYBR® qPCR Mix (QPS-201,
Toyobo) on an MxPro-Mx3000P Sequence Detection system (Stratagene,
USA). U6 small nuclear RNA was used as an internal normalized
reference, and fold changes were calculated by relative
quantification (2−∆∆Ct). The primers used were as
follows: miR-144-3p specific stem-loop reverse transcription
primers: 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAGTACA-3′;
miR-144-3p forward, 5′-GGGAGATCAGAAGGTGATT-3′; reverse,
5′-GTGCAGGGTCCGAGGT-3′. U6 forward, 5′-CTCGCTTCGGCAGCACA-3′;
reverse, 5′-AACGCTTCACGAATTTGCGT-3′. SGK3 forward,
5′-CCAGGAGTGAGTCTTACAG-3′; reverse, 5′-CCAGCCACATTAGGATTA-3′. All
samples were amplified in triplicate according to the
manufacturer's instructions.
Cell proliferation and colony
formation assays
Cells were seeded into 96-well plate
(5×103/well) and transfected with miR-144-3p mimics or
NC. The cell proliferation of HCC cell lines was determined by way
of Cell Counting Kit-8 assay (Dojindo, Japan) at the indicated time
points (0, 24, 48 and 72 h) according to the manufacturer's
instructions. The groups were carried out in quintuplicate wells.
For colony formation assay, cells were seeded into 6-well plates at
a low density (1×103 cells/per well) and cultured for 10
days. Then cells were fixed with 4% paraformaldehyde for 30 min and
surviving colonies (>50 cells/colony) were counted after
staining with 1% crystal violet. The experiments were carried out
in triplicate wells.
Cell cycle distribution
Forty-eight hours after transfection in 6-well
plates, QGY-7703 or SK-hep1 were harvested and washed with cold 1X
PBS. Then, cells were fixed in 70% ethanol at 4°C overnight, and
washed with PBS twice, resuspended with 100 µl RNase A, incubated
at 37°C for 30 min. Staining for DNA content was performed with 400
µl propidium iodide (KeyGen, Nanjing, China) at 4°C for 30 min in
the dark, and analyzed by a flow cytometer (Beckman Coulter EPICS
XL, USA). The experiments were carried out at least three
times.
In vitro migration assay
Migration assays were performed using the 24-well
Cell Migration with 8-µm pore size polycarbonate membrane (Corning,
NY, USA), according to the manufacturer's instructions. Briefly, 24
hours after the transfection, 5×104 cells were
resuspended in 200 µl serum-free medium and plated in the top
chamber. The lower chambers were filled with 0.6 ml of medium
containing 10% FBS. Medium with 10% FBS was added to the lower
chamber as a chemoattractant. After 24-h incubation at 37°C, the
cells on the upper surface of the membrane were removed, and the
cells on the lower surface were fixed, stained, photographed, and
counted under a microscope in five fields.
In vitro capillary tube formation
assay
HUVECs were cultured at 37°C for 24 h in a 96-well
plate coated with Matrigel (BD Biosciences, Bedford, MA, USA) in
the absence or presence of culture medium of HCC cells transfected
with NC or siRNA. The formation of capillary-like structures was
captured under an inverted phase contrast microscope. The number of
the formed tubes, which represent the degree of angiogenesis in
vitro, were scanned and quantitated in five low power
fields.
Animal studies
For mouse studies, male BALB/c-nude mice (5–6 weeks
of age) were obtained from Guangxi Province Laboratory Animal
Center (Nanning, China). HCC cells (5×106) after
transfected with miRNA-mimics or NC were suspended in 0.1 ml
phosphate-buffered saline (PBS) and then injected subcutaneously
into the backs of the mice. Tumor growth was measured over the
course of 30 days, and tumor volume was calculated according to the
formula: volume = 0.5 × length × width2. All experiments
were performed in accordance with the Guide for the Care and Use of
Laboratory Animals, with the approval of the Guangxi medical
University, China.
Luciferase reporter assay
SGK3-3′UTR-Mutant on binding sites was synthesized
by PCR. HEK293T cells were co-transfected with 80 ng of luciferase
reporter plasmid and 40 ng of pRL-TK-Renilla-luciferase
plasmid (Promega, Madison, WI, USA), and the total concentration of
RNA (100 nM) was detected with jetSI-ENDO transfection reagents
(Polyplus Transfection). With incubation for 24 h, luciferase
activity was evaluated with the Dual-Luciferase Reporter assay
system (Promega), and data were normalized for transfection
efficiency via dividing firefly luciferase activity by that of
Renilla luciferase.
Western blot analysis
Targeted prediction of miR-144-3p was analyzed by
Target Scan. Antibodies for SKG3, p-PI3K, mTOR, VEGFR2, and β-actin
were purchased from Cell Signaling Technology, and all the
antibodies were rabbit anti-human. Cells were harvested and then
lysed with RIPA buffer supplemented with 1 mmol/l PMSF (both from
Boster, Wuhan, China), and then centrifuged at 14,000 × g; at 4°C
for 10 min. Protein concentrations of the extracts were measured
using the bicinchoninic acid (BCA) protein assay kit (KeyGen).
Equal amounts of the proteins were concentrated and separated
through SDS-PAGE, and then transferred to polyvinylidene difluoride
(PVDF) membranes (Boster). After blocking in TBST (Tris-buffered
saline with Tween-20) which contained 5% non-fat milk for 60 min,
the membranes were incubated with the primary antibody (1:1,000
dilution; β-actin, as a loading control, 1:2,500 dilution)
overnight at 4°C. The membranes were incubated with the secondary
antibodies (mouse anti-rabbit and HRP-linked antibody, 1:5,000
dilution; Cell Signaling Technology). After incubating in enhanced
chemiluminescence solution (Boster), the proteins on the membranes
were detected using Bio-Rad Universal Hood III, and analyzed by
Image Lab™ software 2.0 (Bio-Rad).
Human Protein Atlas database
The expression of SGK3 in all major tissus and
organs in the human body can be found in the Human Protein Atlas
database. The proteins were detected by proteome methods
(http://www.proteinatlas.org).
Statistical analysis
Data are presented as mean ± standard deviation (SD)
of one representative experiment. The differences in miR-144-3p
expression between HCC tissues and non-cancerous tissues of human
subjects were calculated by a two-tailed independent samples
Student's t-test. Disease-free survival (DFS) was displayed by
Kaplan-Meier survival curves, and DFS of different groups were
compared by log-rank test. Unless otherwise noted, the differences
between groups were analyzed by one-way analysis of variance
(ANOVA) when there were more than two groups. In all cases,
differences were considered statistically significant at p<0.05.
All analyses were performed using SPSS16.0 software (Chicago, IL,
USA).
Results
miR-144-3p expression in the HCC
tissues and correlation with the clinicopathological features in
HCC patients
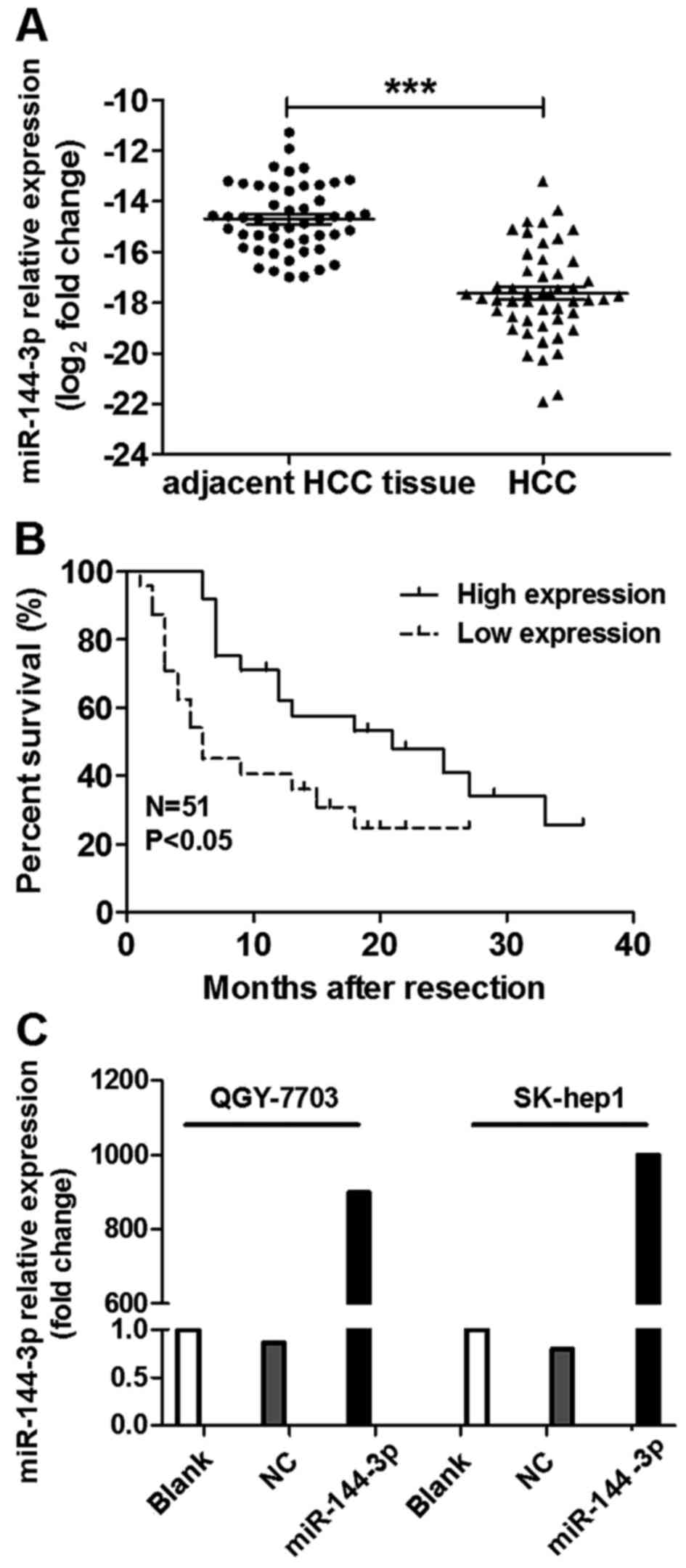
In order to verify the expression of miR-144-3p in
HCC, the levels of miR-144-3p in 51 paired HCC tissues were tested
by qRT-PCR. As shown in Fig. 1A,
miR-144-3p expression was significantly downregulated in 92% (47 of
51) of the HCC samples compared to their matched controls
(p<0.001). We further found that low miR-144-3p expression was
correlated with a shorter DFS (p<0.05) in the HCC patients as
shown in Fig. 1B. These data
suggested that miR-144-3p might be involved in tumor development
and progression in HCC.
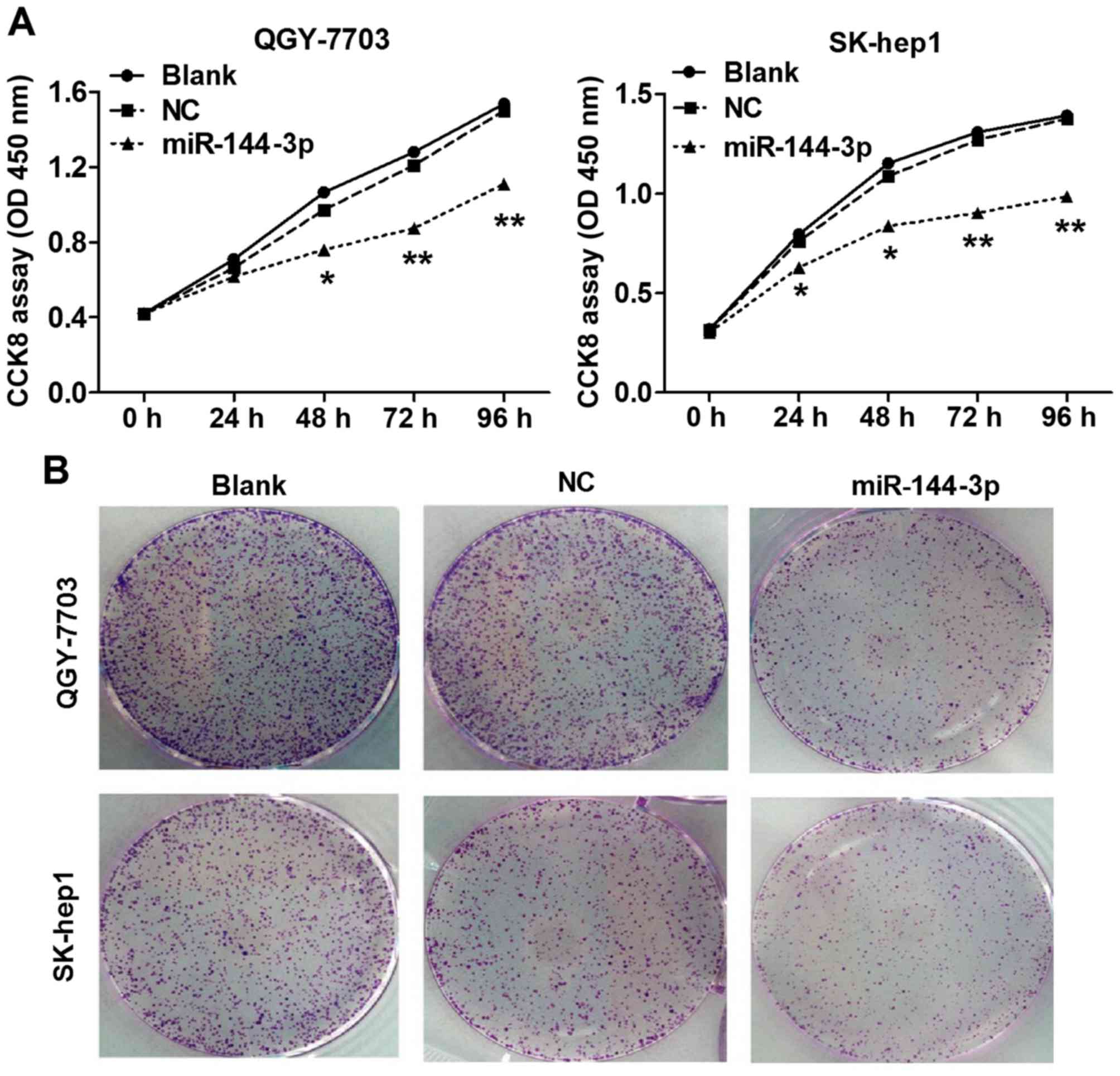
Overexpression miR-144-3p inhibits
proliferation and clonogenicity of HCC cells
To further reveal the effect of miR-144-3p
expression in HCC cells. We used miRNA mimics to rebuilt the
miR-144-3p expression, as shown in Fig.
1C. The expression of miR-144-3p in HCC cells was significantly
improved after transfection with miR-144-3p mimics. The results of
proliferation showed that improving miR-144-3p expression could
significantly suppress the proliferation of QGY-7703 or SK-hep1
cells, respectively (Fig. 2A). The
data on clonogenicity of HCC cells also found that QGY-7703 and
SK-hep1 transfected with miR-144-3p mimic for 10 days displayed
notably fewer colonies compared with NC transfection (Fig. 2B) (p<0.05).
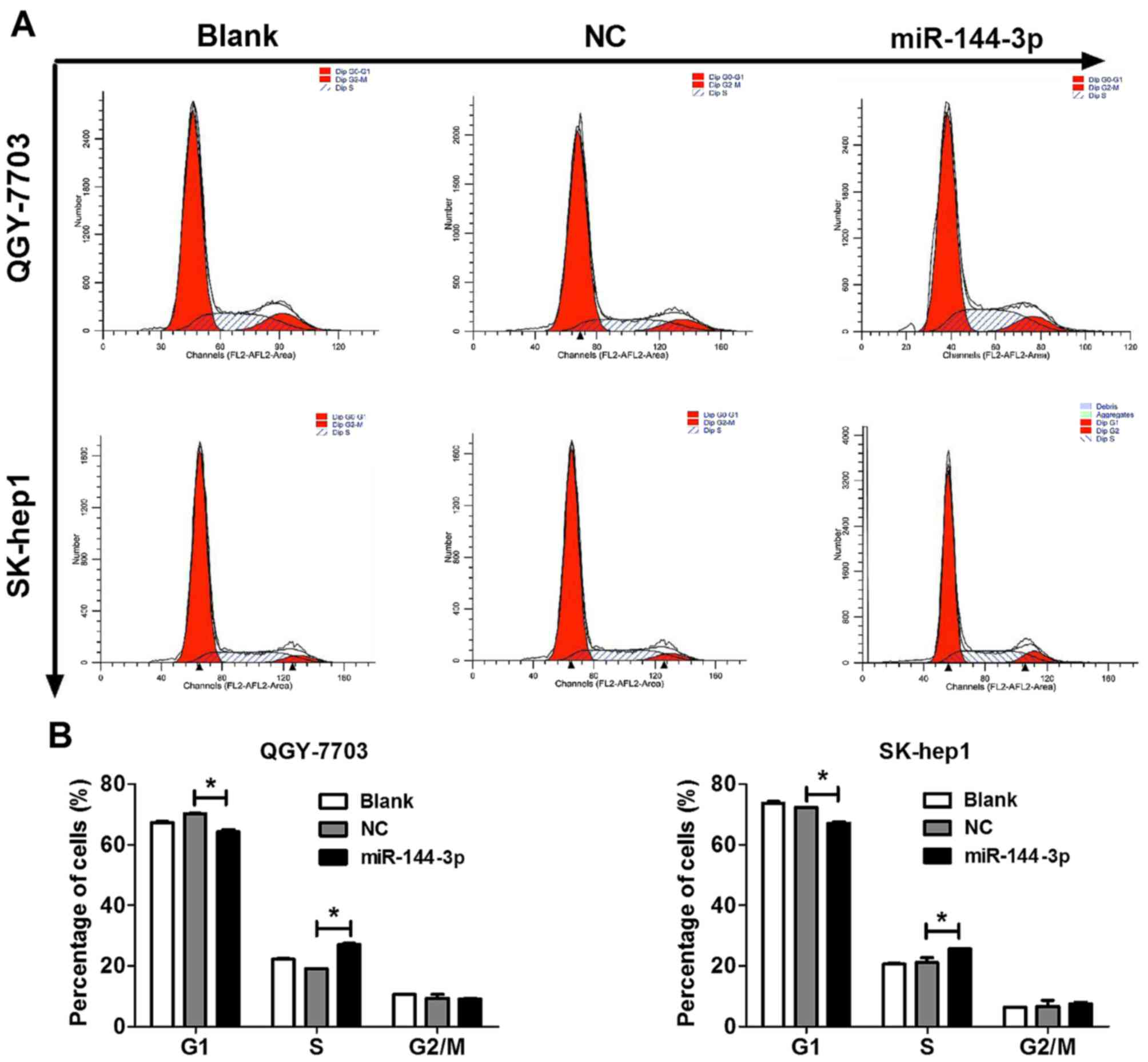
miR-144-3p suppresses HCC cell cycle
progression to inhibit tumor growth
To determine whether the antitumor role of
miR-144-3p correlates to HCC cell cycle progression, we analyzed
cell cycle of QGY-7703 and SK-hep1 HCC cells treated with the
miR-144-3p mimics for 48 h by flow cyto-metry. As observed in
Fig. 3, miR-144-3p mimics
significantly blocked G1 phase of QGY-7703 and SK-hep1 HCC cells
compared to that of NC. Therefore, the results indicated that
miR-144-3p suppressed the cell cycle progression to inhibit the
proliferation of HCC cells.
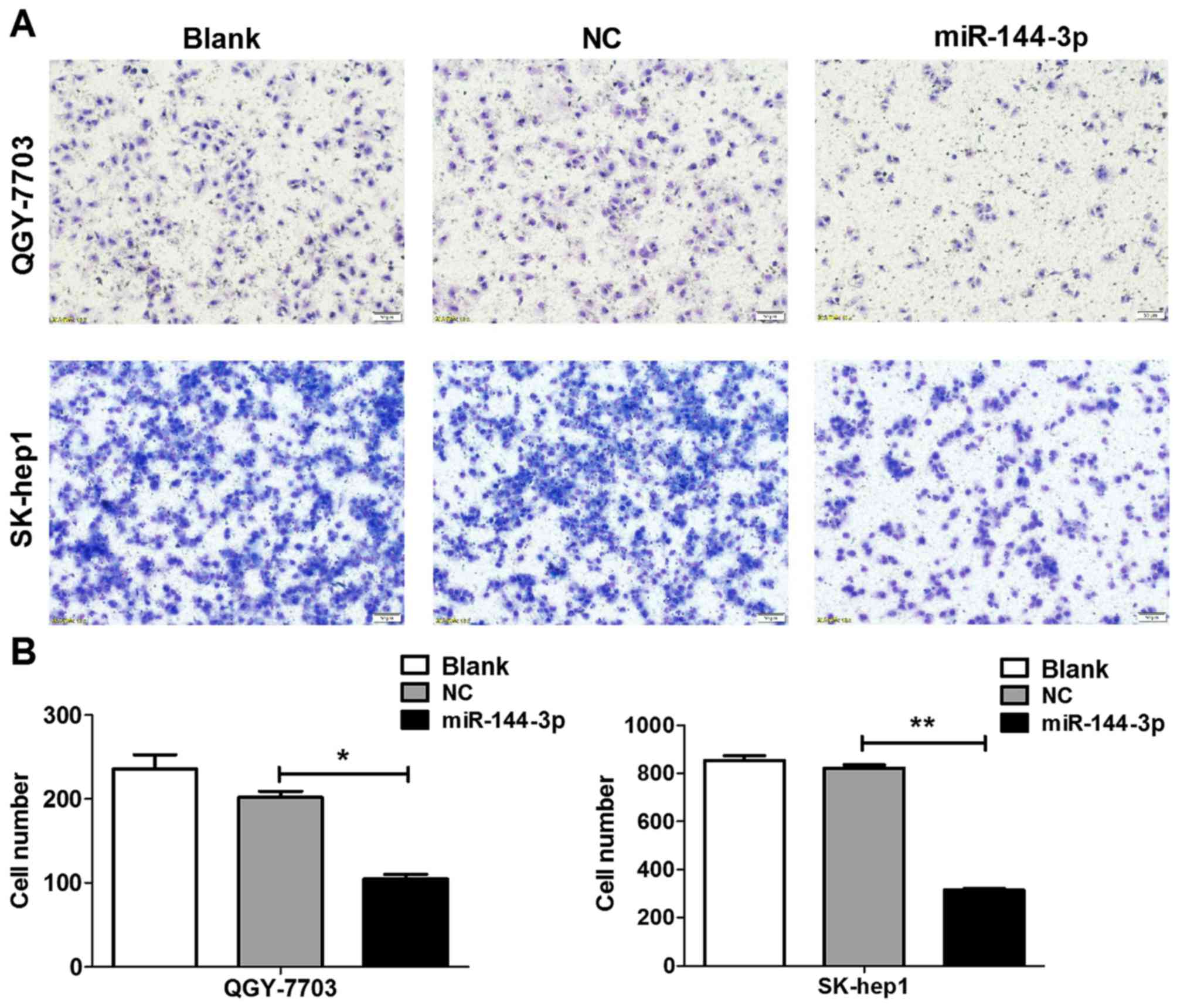
miR-144-3p overexpression inhibits HCC
cell migration
The role of miR-144-3p on HCC cell migration was
investigated. As revealed in Fig.
4, QGY-7703 and SK-hep1 cells with miR-144-3p restoration had
significantly weaker migration than control cells (P<0.05).
These observations implied that miR-144-3p could inhibit HCC
metastasis.
miR-144-3p mimics reduce the ability
of HCC cells to promote angiogenesis and tumor growth in vivo
In order to explore the function of miR-144-3p on
the angiogenesis ability of HCC, HUVECs cultured on Matrigel in
conditioned media from QGY-7703 cells and SK-hep1 cells transfected
with miR-144-3p mimics formed fewer capillary-like rings than
HUVECs cultured in conditioned media from cells transfected with NC
or siRNA as shown in the Fig. 5A.
Moreover, the animal studies also showed that overexpression
miR-144-3p suppressed the tumor growth in vivo compared to
negative control (Fig. 5B). These
results suggested that miR-144-3p could inhibit the ability of HCC
cells to promote angiogenesis and tumor growth in vivo.
miR-144-3p inhibits HCC cell growth
and angiogenesis PI3K-independently by SKG3
To further explore the mechanism of miR-144-3p in
tumor growth and angiogenesis of HCC, we found there were 1,043
putatively targeted genes of miR-144-3p in Target Scan. Then we
used the DAVID Bioinformatics Resources 6.7 database to analyze the
functional pathways (https://david-d.ncifcrf.gov), and found 8 types of
rich pathways (Table I) after
imputing these putative targets in the database, most pathways were
shown involved in angiogenesis however, also relating to tumor
growth process. Angiogenesis is a key process involved in tumor
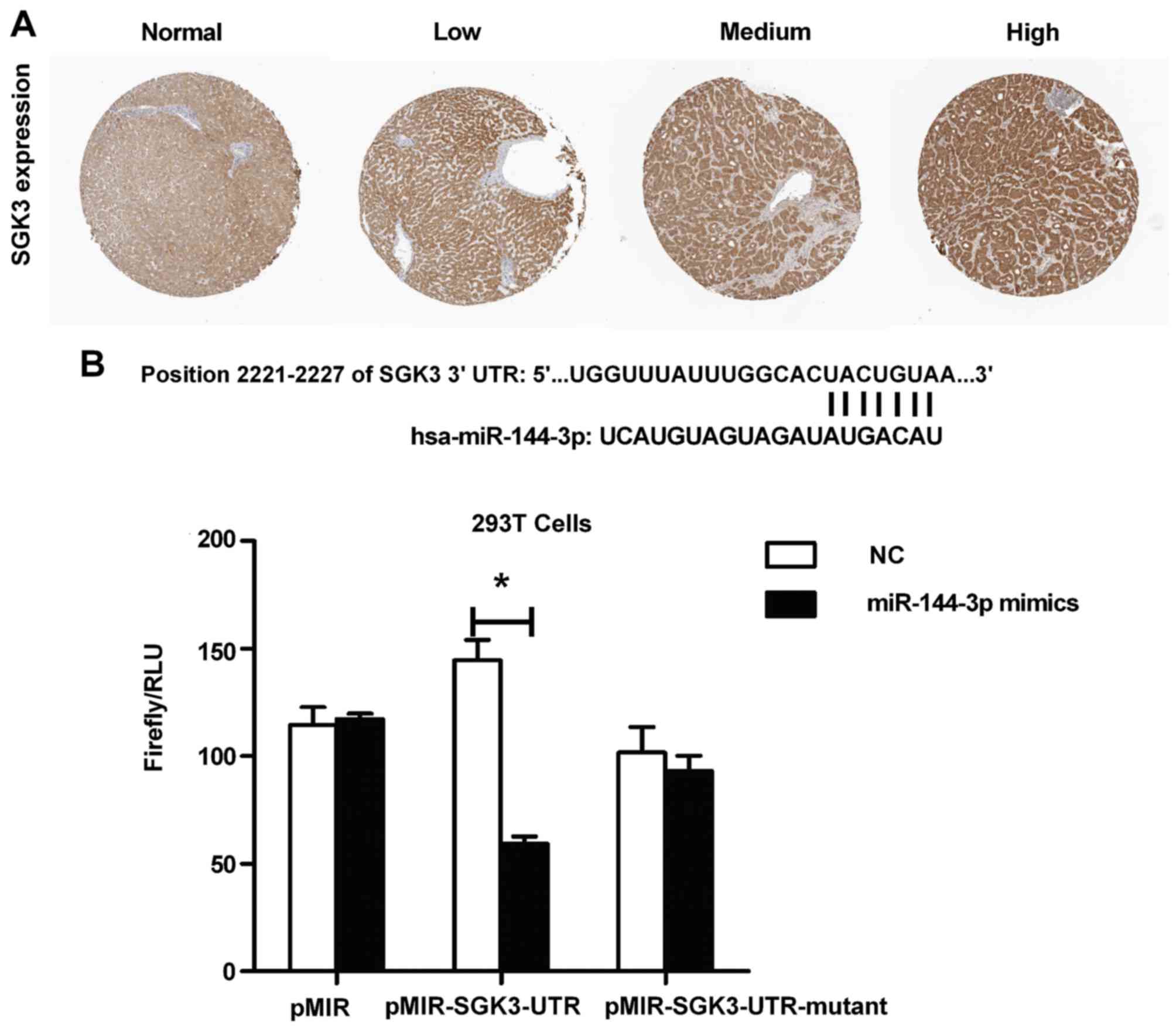
growth and metastasis. SGK3 has been shown to be highly expressed
in HCC and is hardly expressed in normal liver tissue from the
Human Protein Atlas database (Fig.
6A), so we aimed to identify the relationship between
miR-144-3p and SGK3, and the effect of miR-144-3p targeting SKG3 in
the inhibittion of PI3K/mTOR/VEGF pathway activation. As shown in
the Fig. 6B, we could find the
sequence alignment of miR-144-3p and its conserved target position
in SKG3 3′UTR where miR-144-3p might suppress gene expression
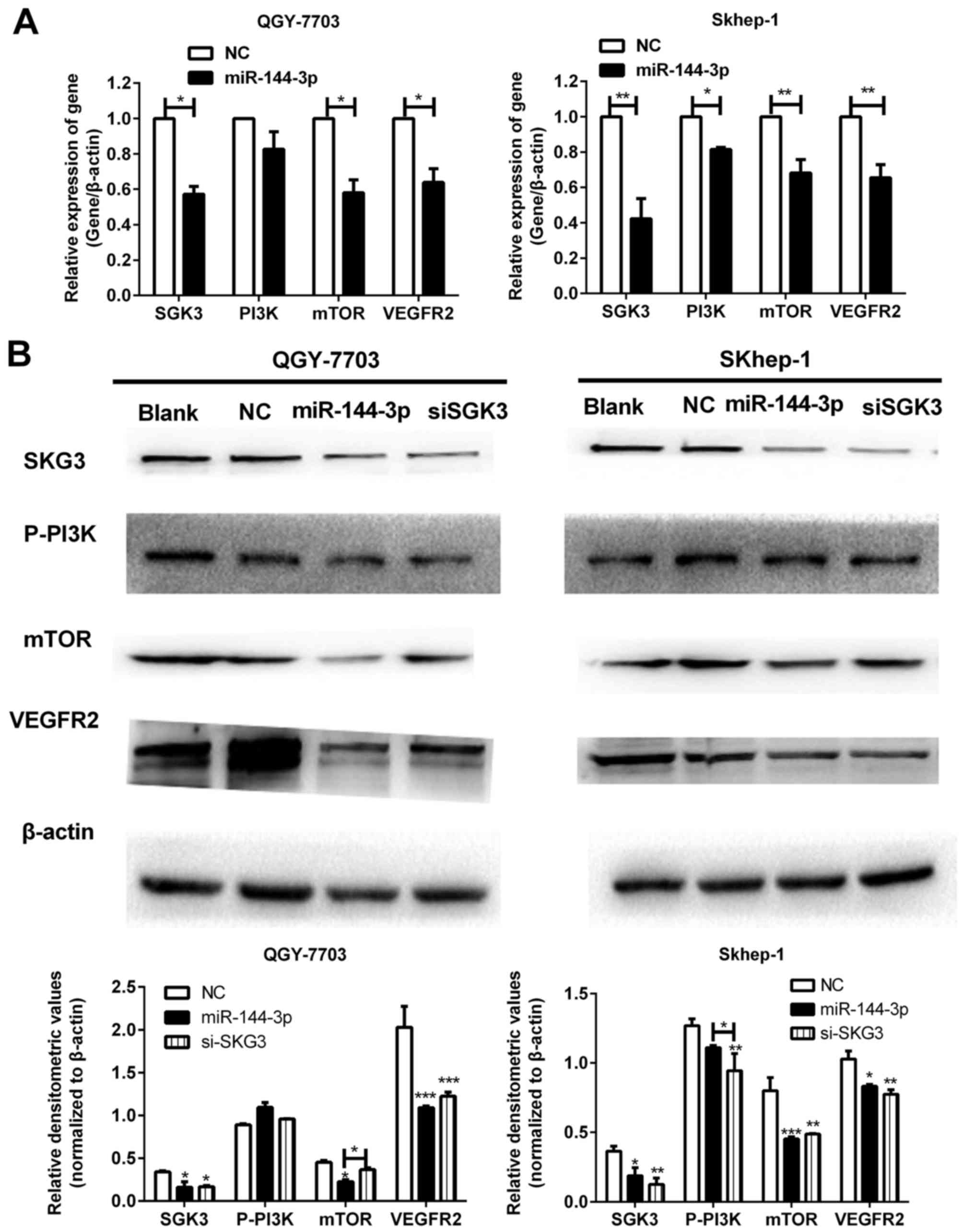
through its binding sequence. As observed in Fig. 7A, the mRNA of mTOR and VEGF2 were
significantly downregulated by miR-144-3p mimics after targeting
SGK3, but the mRNA of PI3K showed no obvious change in HCC cells
after transfected with miR-144-3p mimics. Similar to the function
of SGK3-siRNA, the results of WB found that overexpression of
miR-144-3p not only suppressed the phosphorylation of PI3K, but
also repressed the protein expression of mTOR and VEGFR2, which are
in PI3K downstream signal pathway in HCC cells (Fig. 7B). These results demonstrated that
miR-144-3p could suppress tumor growth and angiogenesis by
targeting SGK3 with PI3K/SGK/mTOR pathway.
 | Table I.Eight types of rich pathways has been
found by the DAVID Bioinformatics Resources 6.7 database
(https://david-d.ncifcrf.gov). |
Table I.
Eight types of rich pathways has been
found by the DAVID Bioinformatics Resources 6.7 database
(https://david-d.ncifcrf.gov).
| Category | Term | RT | Count | % | P-value |
|---|
| PANTHER_PATHWAY | P00021:FGF signaling
pathway | RT | 19 | 0.2 | 7.6E-3 |
|
PANTHER_PATHWAY | P00047:PDGF
signaling pathway | RT | 22 | 0.2 | 2.1E-2 |
|
PANTHER_PATHWAY | P00057:Wnt
signaling pathway | RT | 35 | 0.3 | 2.3E-2 |
|
PANTHER_PATHWAY | P00018:EGF receptor
signaling pathway | RT | 18 | 0.2 | 2.6E-2 |
|
PANTHER_PATHWAY |
P00005:Angiogenesis | RT | 25 | 0.2 | 3.0E-2 |
|
PANTHER_PATHWAY | P00034:Integrin
signaling pathway | RT | 24 | 0.2 | 4.5E-2 |
|
PANTHER_PATHWAY | P00033:Insulin/IGF
pathway-protein kinase B signaling cascade | RT | 12 | 0.1 | 5.3E-2 |
|
PANTHER_PATHWAY | P00035:Interferon-γ
signaling pathway | RT | 6 | 0.1 | 8.9E-2 |
Discussion
miRNAs have been indicated to play an important role
in diverse biological processes, including cancer. Emerging
evidence has demonstrated aberrant expression of miRNAs involving
in the tumor development and metastasis process. Thus, revealing
the potential mechanism of miRNAs in cancer can promote our
understanding in cancer biology and offer new knowledge to invent
new treatments and diagnosis methods (14).
miR-144-3p, as a tumor suppressor, has been
demonstrated in several cancer types. For instance, Navon et
al (15) using microarrays
profiled the expression of >700 miRNAs in 28 matched tumor and
normal samples representing 32 microarray measurements from 8
different tumor types (breast, colon, liver, lung, lymphoma, ovary,
prostate and testis). In addition, there are some findings that
miR-144-3p inhibits proliferation and induces apoptosis and
autophagy in lung cancer cells by targeting TIGAR (16); and miR-144-3p downregulation
increases bladder cancer cell proliferation by targeting EZH2 and
regulating Wnt signaling (17). On
the contrary, Zhang et al reported that miR-144-3p could
promote tumor cell proliferation, migration and invasion in
nasopharyngeal carcinoma through repression of PTEN (18), and Liu et al found that Myc
induced miR-144-3p/451 contributing to the acquired imatinib
resistance in chronic myelogenous leukemia K562 cells (19). In this study, we found that
miR-144-3p acted as a tumor suppressor, which is downregulated in
HCC, recovery of the expression of miR-144-3p was able to
significantly inhibit HCC growth, migrations and angiogenesis.
Also, the clinical data showed that high expression of miR-144-3p
was correlated with long disease-free survival time. This result
showed that miR-144-3p could be a prognostic marker for HCC.
To better understand the mechanisms of miR-144-3p on
HCC, we used Target-Scan database and DAVID Bioinformatics
Resources 6.7 database to analyze the potential targets and
relative function pathways. The results (Table I), showed most targets were
correlated with angiogenesis and tumor growth. As angiogenesis has
been verified to promote tumor growth, metastasis and even cause
resistance (20–26), we chose SGK3 as potential target of
miR-144-3p to investigate whether SGK3 can influence tumor
angiogenesis and growth in HCC. Encoded by chromosome 8q12.2, SGK
is known as a downstream mediator of phosphatidylinositol 3-kinase
(PI3K) oncogenic signaling. It has also been proved that SGK3 plays
a pivotal role in oncogenic progress in various cancers, including
breast cancer, ovarian cancer and hepatocellular carcinoma
(27). In physiological research
aspect, some studies reported that SGK1 is necessary for vascular
remodeling during angiogenesis (28), and ablation of SGK1 impairs
endothelial cell migration and tube formation leading to decreased
neo-angiogenesis following myocardial infarction (29). Thus, we hypothesized that miR-144-3p
might target SGK3 to inhibit tumor growth and angiogenesis.
Consistent with similar studies, we found that miR-144-3p decreased
the activation of mTOR and VEGF with no obvious change of PI3K, and
displayed more inhibitor effect on mTOR and VEGFR2 expression in
HCC cells, compared with the SGK3-siRNA groups. These results
indicated that miR-144-3p has powerful antitumor function.
In conclusion, we found miR-144-3p, similarly to a
tumor suppressor, could be a prognostic factor and inhibit tumor
growth and angiogenesis by targeting SGK3 suppressing activation of
mTOR/VEGFR2 of PI3K downstream signals. However, regardless of the
role of miR-144-3p in drug-resistance or metastasis, more in
vitro and in vivo studies are needed to fully reveal the
downregulation of miR-144-3p and its underlying mechanisms.
Acknowledgements
This study was supported by grants from National
Natural Science Foundation of China (no. 81560493), Key Project of
Natural Science Foundation of Guangxi (no. 2015GXNSFDA139024) and
the Research Fund for the College Scientific Program of Education
Department of Guangxi Province (no. ZD2014027).
Glossary
Abbreviations
Abbreviations:
|
HCC
|
hepatocellular carcinoma
|
|
SGK3
|
serum and glucocorticoid kinase 3
|
|
PI3K
|
phosphoinositide 3-kinase
|
|
miRNAs
|
microRNAs
|
|
ZEB1
|
zinc finger E-box-binding homeobox
1
|
|
ZEB2
|
zinc finger E-box-binding homeobox
2
|
|
E2F3
|
E2F transcription factor 3
|
|
ZFX
|
zinc finger X-chromosomal protein
|
|
VEGF
|
vascular endothelial growth factor
|
|
NC
|
negative control
|
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Aravalli RN, Steer CJ and Cressman EN:
Molecular mechanisms of hepatocellular carcinoma. Hepatology.
48:2047–2063. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Aravalli RN, Cressman EN and Steer CJ:
Cellular and molecular mechanisms of hepatocellular carcinoma: An
update. Arch Toxicol. 87:227–247. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kong YW, Ferland-McCollough D, Jackson TJ
and Bushell M: microRNAs in cancer management. Lancet Oncol.
13:e249–e258. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhou X, Zhang CZ, Lu SX, Chen GG, Li LZ,
Liu LL, Yi C, Fu J, Hu W, Wen JM, et al: miR-625 suppresses tumour
migration and invasion by targeting IGF2BP1 in hepatocellular
carcinoma. Oncogene. 34:965–977. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hou J, Lin L, Zhou W, Wang Z, Ding G, Dong
Q, Qin L, Wu X, Zheng Y, Yang Y, et al: Identification of miRNomes
in human liver and hepatocellular carcinoma reveals miR-199a/b-3p
as therapeutic target for hepatocellular carcinoma. Cancer Cell.
19:232–243. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Guan H, Liang W, Xie Z, Li H, Liu J, Liu
L, Xiu L and Li Y: Down-regulation of miR-144 promotes thyroid
cancer cell invasion by targeting ZEB1 and ZEB2. Endocrine.
48:566–574. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zha W, Cao L, Shen Y and Huang M: Roles of
MiR-144-ZFX pathway in growth regulation of non-small-cell lung
cancer. PLoS One. 8:e741752013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Huang XP, Hou J, Shen XY, Huang CY, Zhang
XH, Xie YA and Luo XL: MicroRNA-486-5p, which is downregulated in
hepatocellular carcinoma, suppresses tumor growth by targeting
PIK3R1. FEBS J. 282:579–594. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bruhn MA, Pearson RB, Hannan RD and
Sheppard KE: AKT-independent PI3-K signaling in cancer - emerging
role for SGK3. Cancer Manag Res. 5:281–292. 2013.PubMed/NCBI
|
|
11
|
Cao T, Li H, Hu Y, Ma D and Cai X: miR-144
suppresses the proliferation and metastasis of hepatocellular
carcinoma by targeting E2F3. Tumour Biol. 35:10759–10764. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang R, Zhao N, Li S, Fang JH, Chen MX,
Yang J, Jia WH, Yuan Y and Zhuang SM: MicroRNA-195 suppresses
angiogenesis and metastasis of hepatocellular carcinoma by
inhibiting the expression of VEGF, VAV2, and CDC42. Hepatology.
58:642–653. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yang X, Zhang XF, Lu X, Jia HL, Liang L,
Dong QZ, Ye QH and Qin LX: MicroRNA-26a suppresses angiogenesis in
human hepatocellular carcinoma by targeting hepatocyte growth
factor-cMet pathway. Hepatology. 59:1874–1885. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhu Z, Zhang X, Wang G and Zheng H: Role
of microRNAs in hepatocellular carcinoma. Hepat Mon. 14:e186722014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Navon R, Wang H, Steinfeld I, Tsalenko A,
Ben-Dor A and Yakhini Z: Novel rank-based statistical methods
reveal microRNAs with differential expression in multiple cancer
types. PLoS One. 4:e80032009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen S, Li P, Li J, Wang Y, Du Y, Chen X,
Zang W, Wang H, Chu H, Zhao G, et al: MiR-144 inhibits
proliferation and induces apoptosis and autophagy in lung cancer
cells by targeting TIGAR. Cell Physiol Biochem. 35:997–1007. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Guo Y, Ying L, Tian Y, Yang P, Zhu Y, Wang
Z, Qiu F and Lin J: miR-144 downregulation increases bladder cancer
cell proliferation by targeting EZH2 and regulating Wnt signaling.
FEBS J. 280:4531–4538. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang LY, Lee Ho-Fun V, Wong AM, Kwong DL,
Zhu YH, Dong SS, Kong KL, Chen J, Tsao SW, Guan XY, et al:
MicroRNA-144 promotes cell proliferation, migration and invasion in
nasopharyngeal carcinoma through repression of PTEN.
Carcinogenesis. 34:454–463. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu L, Wang S, Chen R, Wu Y, Zhang B,
Huang S, Zhang J, Xiao F, Wang M and Liang Y: Myc induced
miR-144/451 contributes to the acquired imatinib resistance in
chronic myelogenous leukemia cell K562. Biochem Biophys Res Commun.
425:368–373. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yuan SX, Yang F, Yang Y, Tao QF, Zhang J,
Huang G, Yang Y, Wang RY, Yang S, Huo XS, et al: Long noncoding RNA
associated with microvascular invasion in hepatocellular carcinoma
promotes angiogenesis and serves as a predictor for hepatocellular
carcinoma patients' poor recurrence-free survival after
hepatectomy. Hepatology. 56:2231–2241. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fang JH, Zhou HC, Zeng C, Yang J, Liu Y,
Huang X, Zhang JP, Guan XY and Zhuang SM: MicroRNA-29b suppresses
tumor angiogenesis, invasion, and metastasis by regulating matrix
metalloproteinase 2 expression. Hepatology. 54:1729–1740. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fernández M, Semela D, Bruix J, Colle I,
Pinzani M and Bosch J: Angiogenesis in liver disease. J Hepatol.
50:604–620. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Thomas MB, Jaffe D, Choti MM, Belghiti J,
Curley S, Fong Y, Gores G, Kerlan R, Merle P, O'Neil B, et al:
Hepatocellular carcinoma: Consensus recommendations of the National
Cancer Institute Clinical Trials Planning Meeting. J Clin Oncol.
28:3994–4005. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hao CY: Angiogenesis blockade as therapy
for hepatocellular carcinoma: Progress and challenges. J
Gastroenterol Hepatol. 26:4–6. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang L, Wang JN, Tang JM, Kong X, Yang
JY, Zheng F, Guo LY, Huang YZ, Zhang L, Tian L, et al: VEGF is
essential for the growth and migration of human hepatocellular
carcinoma cells. Mol Biol Rep. 39:5085–5093. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kerbel RS: Tumor angiogenesis. N Engl J
Med. 358:2039–2049. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hou M, Lai Y, He S, He W, Shen H and Ke Z:
SGK3 (CISK) may induce tumor angiogenesis (Hypothesis). Oncol Lett.
10:23–26. 2015.PubMed/NCBI
|
|
28
|
Catela C, Kratsios P, Hede M, Lang F and
Rosenthal N: Serum and glucocorticoid-inducible kinase 1 (SGK1) is
necessary for vascular remodeling during angiogenesis. Dev Dyn.
239:2149–2160. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zarrinpashneh E, Poggioli T, Sarathchandra
P, Lexow J, Monassier L, Terracciano C, Lang F, Damilano F, Zhou
JQ, Rosenzweig A, et al: Ablation of SGK1 impairs endothelial cell
migration and tube formation leading to decreased neo-angiogenesis
following myocardial infarction. PLoS One. 8:e802682013. View Article : Google Scholar : PubMed/NCBI
|