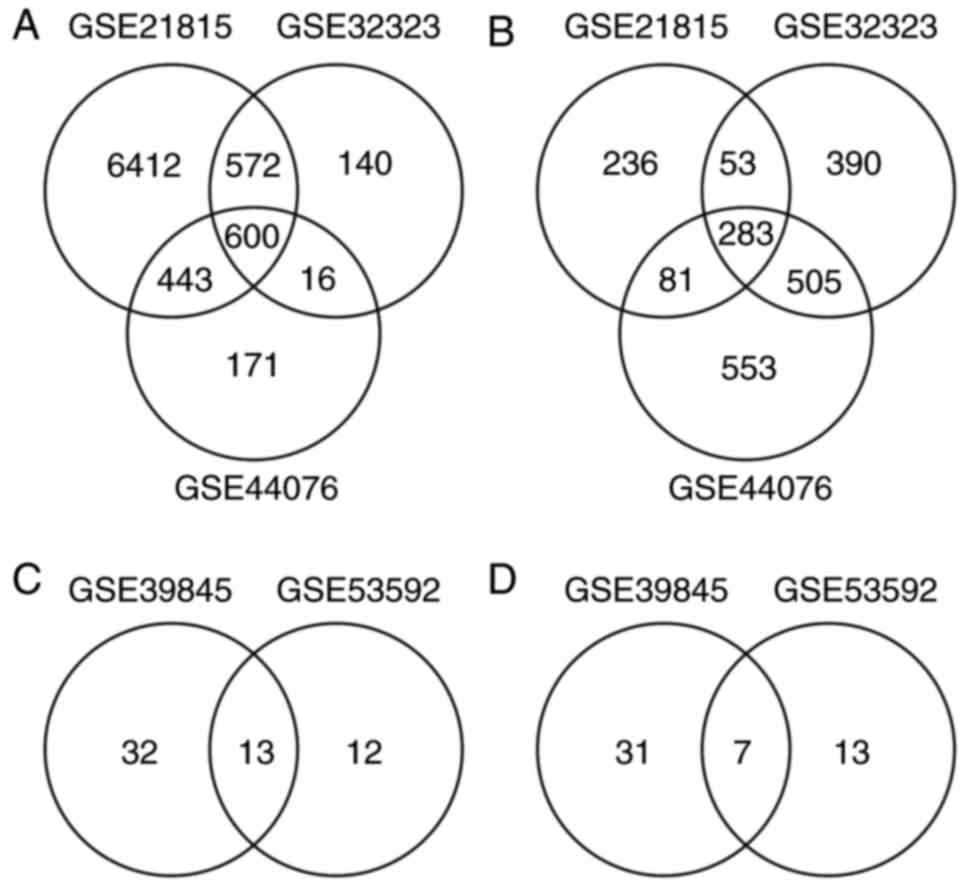
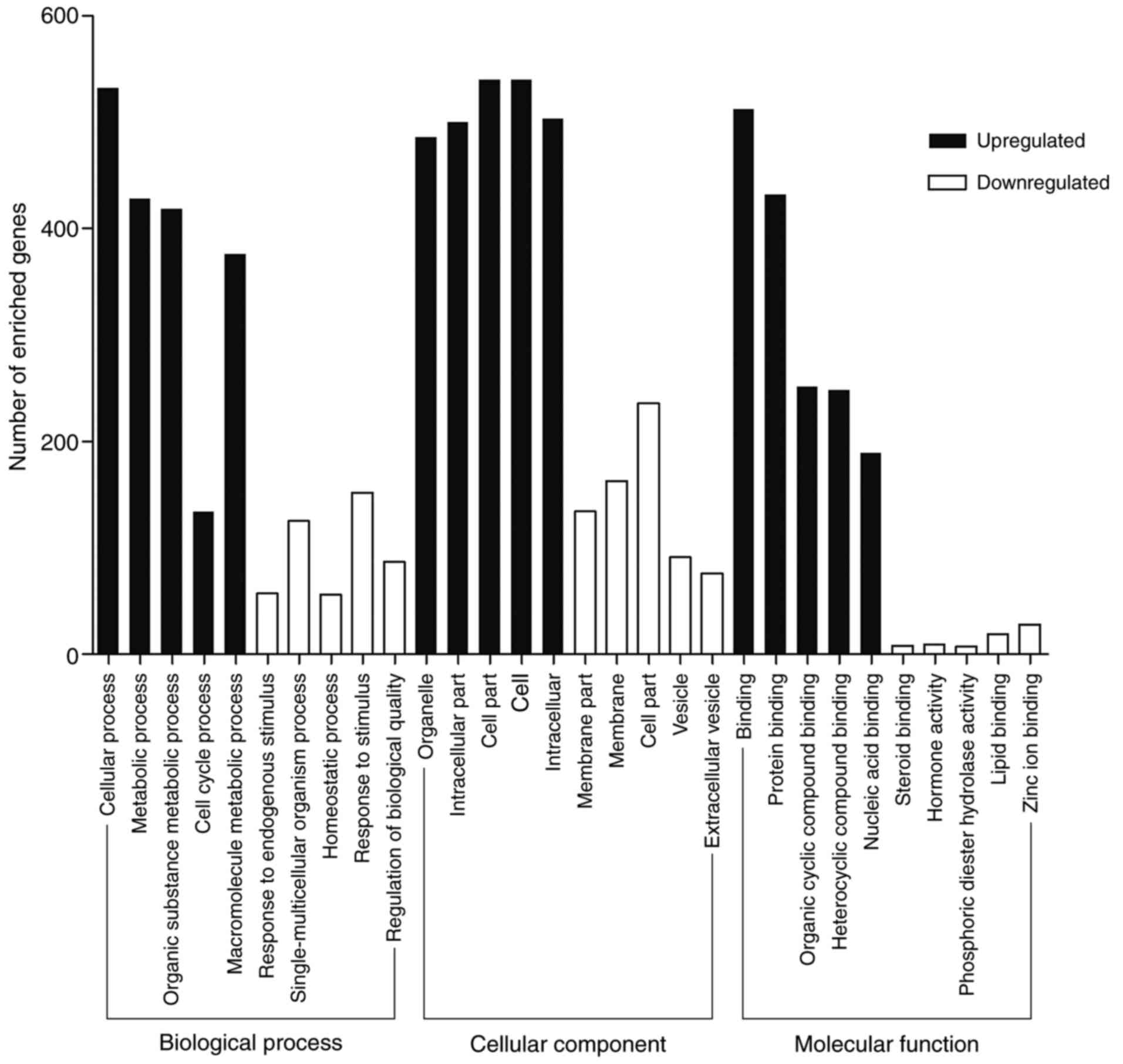
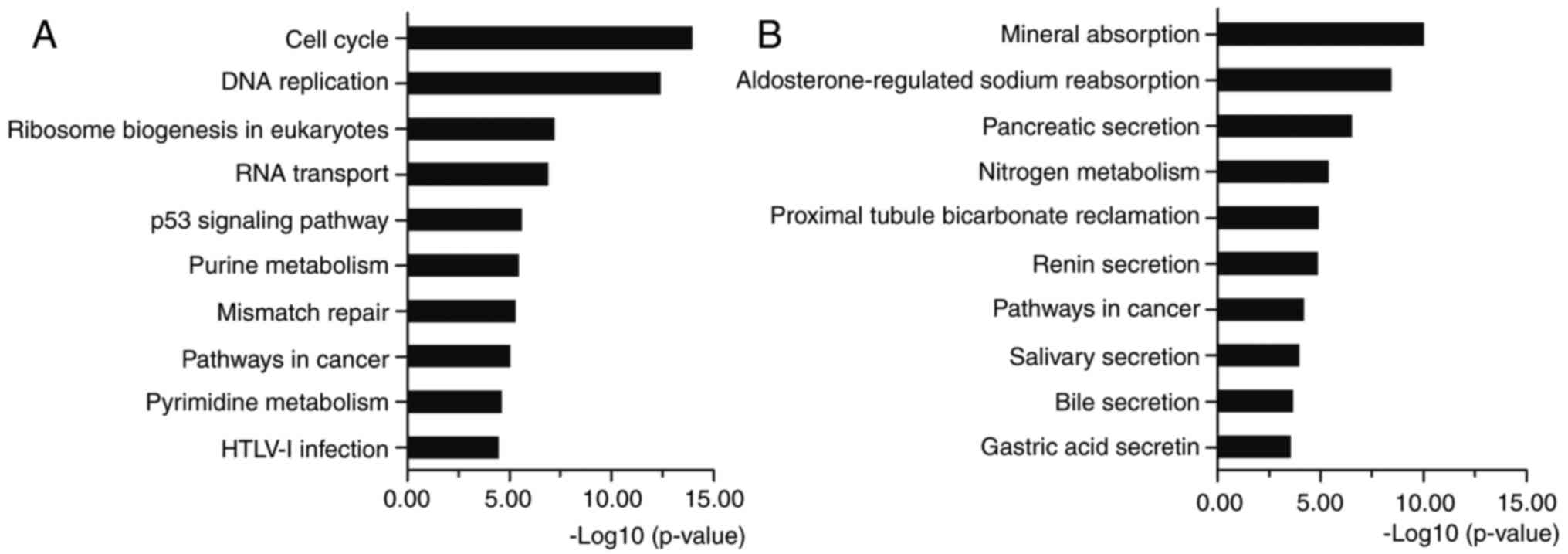
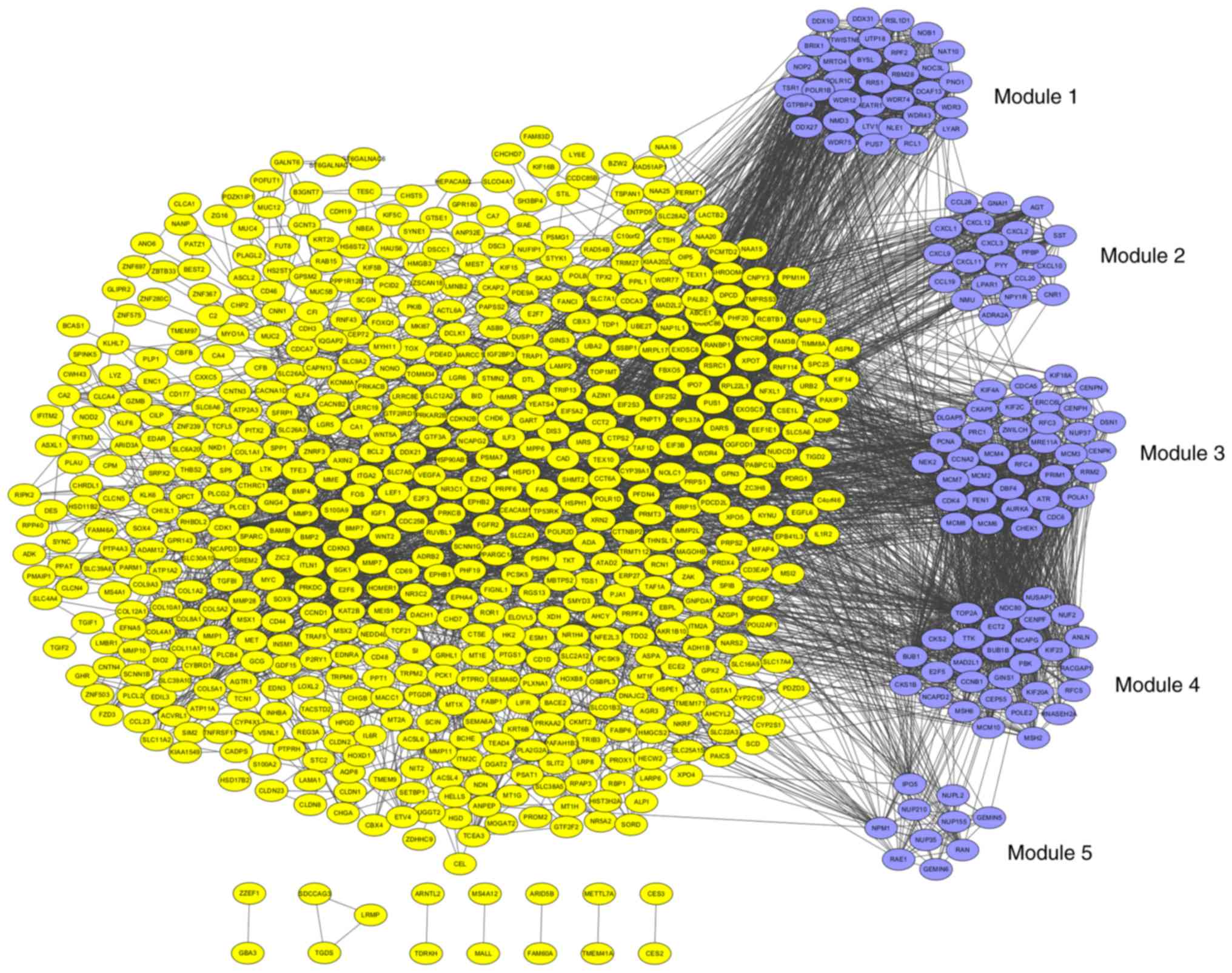
|
1
|
Haraldsdottir S, Einarsdottir HM,
Smaradottir A, Gunnlaugsson A and Halfdanarson TR: Colorectal
cancer - review. Laeknabladid. 100:75–82. 2014.(in Icelandic).
PubMed/NCBI
|
|
2
|
Fearon ER: Molecular genetics of
colorectal cancer. Annu Rev Pathol. 6:479–507. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Simon K: Colorectal cancer development and
advances in screening. Clin Interv Aging. 11:967–976. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Thomas J, Ohtsuka M, Pichler M and Ling H:
MicroRNAs: Clinical relevance in colorectal cancer. Int J Mol Sci.
16:28063–28076. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ren A, Dong Y, Tsoi H and Yu J: Detection
of miRNA as non-invasive biomarkers of colorectal cancer. Int J Mol
Sci. 16:2810–2823. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ng EK, Chong WW, Jin H, Lam EK, Shin VY,
Yu J, Poon TC, Ng SS and Sung JJ: Differential expression of
microRNAs in plasma of patients with colorectal cancer: A potential
marker for colorectal cancer screening. Gut. 58:1375–1381. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Asangani IA, Rasheed SA, Nikolova DA,
Leupold JH, Colburn NH, Post S and Allgayer H: MicroRNA-21 (miR-21)
post-transcriptionally downregulates tumor suppressor Pdcd4 and
stimulates invasion, intravasation and metastasis in colorectal
cancer. Oncogene. 27:2128–2136. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Thomas PD: The Gene Ontology and the
meaning of biological function. Methods Mol Biol. 1446:15–24. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xing Z, Chu C, Chen L and Kong X: The use
of Gene Ontology terms and KEGG pathways for analysis and
prediction of oncogenes. Biochim Biophys Acta. 1860:2725–2734.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Khunlertgit N and Yoon BJ: Incorporating
topological information for predicting robust cancer subnetwork
markers in human protein-protein interaction network. BMC
Bioinformatics. 17 Suppl 13:3512016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Clough E and Barrett T: The Gene
Expression Omnibus Database. Methods Mol Biol. 1418:93–110. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xie C, Mao X, Huang J, Ding Y, Wu J, Dong
S, Kong L, Gao G, Li CY and Wei L: KOBAS 2.0: A web server for
annotation and identification of enriched pathways and diseases.
Nucleic Acids Res. 39:(Web Server issue). W316–W322. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kanehisa M, Sato Y, Kawashima M, Furumichi
M and Tanabe M: KEGG as a reference resource for gene and protein
annotation. Nucleic Acids Res. 44(D1): D457–D462. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Killcoyne S, Carter GW, Smith J and Boyle
J: Cytoscape: A community-based framework for network modeling.
Methods Mol Biol. 563:219–239. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dashwood MR and Loizidou M: Determination
of cell-specific receptor binding using a combination of
immunohistochemistry and in vitro autoradiography: Relevance to
therapeutic receptor targeting in cancer. Methods Mol Biol.
878:137–147. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
de Lacerda TC, Costa-Silva B, Giudice FS,
Dias MV, de Oliveira GP, Teixeira BL, Dos Santos TG and Martins VR:
Prion protein binding to HOP modulates the migration and invasion
of colorectal cancer cells. Clin Exp Metastasis. 33:441–451. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tsujitani S, Saito H, Honboh T, Ataka M,
Tanida T, Makino M and Ikeguchi M: Prognostic significance of
receptor-binding cancer antigen expressed on SiSo cells (RCAS1)
expression in relation to cadherin expression in patients with
colorectal carcinoma. Dis Colon Rectum. 50:1241–1249. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liang B, Liu Z, Cao Y, Zhu C, Zuo Y, Huang
L, Wen G, Shang N, Chen Y, Yue X, et al: MC37, a new mono-carbonyl
curcumin analog, induces G2/M cell cycle arrest and
mitochondria-mediated apoptosis in human colorectal cancer cells.
Eur J Pharmacol. 796:139–148. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen X, Wu X, Ouyang W, Gu M, Gao Z, Song
M, Chen Y, Lin Y, Cao Y and Xiao H: Novel ent-Kaurane
diterpenoid from Rubus corchorifolius L. f. inhibits human
colon cancer cell growth via inducing cell cycle arrest and
apoptosis. J Agric Food Chem. 65:1566–1573. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Boyer AS, Walter D and Sørensen CS: DNA
replication and cancer: From dysfunctional replication origin
activities to therapeutic opportunities. Semin Cancer Biol.
37–38:16–25. 2016. View Article : Google Scholar
|
|
22
|
Hills SA and Diffley JF: DNA replication
and oncogene-induced replicative stress. Curr Biol. 24:R435–R444.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Stegh AH: Targeting the p53 signaling
pathway in cancer therapy - the promises, challenges and perils.
Expert Opin Ther Targets. 16:67–83. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu YH, Liu GH, Mei JJ and Wang J: The
preventive effects of hyperoside on lung cancer in vitro by
inducing apoptosis and inhibiting proliferation through caspase-3
and P53 signaling pathway. Biomed Pharmacother. 83:381–391. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen J, Wei Y, Feng Q, Ren L, He G, Chang
W, Zhu D, Yi T, Lin Q, Tang W, et al: Ribosomal protein S15A
promotes malignant transformation and predicts poor outcome in
colorectal cancer through misregulation of p53 signaling pathway.
Int J Oncol. 48:1628–1638. 2016.PubMed/NCBI
|
|
26
|
Xiao S, Zhou Y, Yi W, Luo G, Jiang B, Tian
Q, Li Y and Xue M: Fra-1 is downregulated in cervical cancer
tissues and promotes cervical cancer cell apoptosis by p53
signaling pathway in vitro. Int J Oncol. 46:1677–1684. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shaikhibrahim Z, Lindstrot A, Buettner R
and Wernert N: Analysis of laser-microdissected prostate cancer
tissues reveals potential tumor markers. Int J Mol Med. 28:605–611.
2011.PubMed/NCBI
|
|
28
|
Ferraro A, Boni T and Pintzas A: EZH2
regulates cofilin activity and colon cancer cell migration by
targeting ITGA2 gene. PLoS One. 9:e1152762014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lind GE, Kleivi K, Meling GI, Teixeira MR,
Thiis-Evensen E, Rognum TO and Lothe RA: ADAMTS1, CRABP1, and NR3C1
identified as epigenetically deregulated genes in colorectal
tumorigenesis. Cell Oncol. 28:259–272. 2006.PubMed/NCBI
|
|
30
|
Morin A, Fritsch L, Mathieu JR, Gilbert C,
Guarmit B, Firlej V, Gallou-Kabani C, Vieillefond A, Delongchamps
NB and Cabon F: Identification of CAD as an androgen receptor
interactant and an early marker of prostate tumor recurrence. FASEB
J. 26:460–467. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chang SW, Yue J, Wang BC and Zhang XL:
miR-503 inhibits cell proliferation and induces apoptosis in
colorectal cancer cells by targeting E2F3. Int J Clin Exp Pathol.
8:12853–12860. 2015.PubMed/NCBI
|
|
32
|
Fang Y, Gu X, Li Z, Xiang J and Chen Z:
miR-449b inhibits the proliferation of SW1116 colon cancer stem
cells through downregulation of CCND1 and E2F3 expression. Oncol
Rep. 30:399–406. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tong Z, Liu N, Lin L, Guo X, Yang D and
Zhang Q: miR-125a-5p inhibits cell proliferation and induces
apoptosis in colon cancer via targeting BCL2, BCL2L12 and MCL1.
Biomed Pharmacother. 75:129–136. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kaluzki I, Hrgovic I, Hailemariam-Jahn T,
Doll M, Kleemann J, Valesky EM, Kippenberger S, Kaufmann R, Zoeller
N and Meissner M: Dimethylfumarate inhibits melanoma cell
proliferation via p21 and p53 induction and bcl-2 and cyclin B1
downregulation. Tumour Biol. 37:13627–13635. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Meka Bhushann P, Jarjapu S, Vishwakarma
SK, Nanchari SR, Cingeetham A, Annamaneni S, Mukta S, Triveni B and
Satti V: Influence of BCL2-938 C>A promoter polymorphism and
BCL2 gene expression on the progression of breast cancer. Tumour
Biol. 37:6905–6912. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yang X, Gao F, Ma F, Ren Y, Chen H, Liang
X, Han S, Xiong X, Pan W, Zhou C, et al: Association of the
functional BCL-2 rs2279115 genetic variant and small cell lung
cancer. Tumour Biol. 37:1693–1698. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kvissel AK, Ramberg H, Eide T, Svindland
A, Skålhegg BS and Taskén KA: Androgen dependent regulation of
protein kinase A subunits in prostate cancer cells. Cell Signal.
19:401–409. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wong HL, Koh WP, Probst-Hensch NM, Van den
Berg D, Yu MC and Ingles SA: Insulin-like growth factor-1 promoter
polymorphisms and colorectal cancer: A functional genomics
approach. Gut. 57:1090–1096. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Feik E, Baierl A, Hieger B, Führlinger G,
Pentz A, Stättner S, Weiss W, Pulgram T, Leeb G, Mach K, et al:
Association of IGF1 and IGFBP3 polymorphisms with colorectal polyps
and colorectal cancer risk. Cancer Causes Control. 21:91–97. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Shiratsuchi I, Akagi Y, Kawahara A,
Kinugasa T, Romeo K, Yoshida T, Ryu Y, Gotanda Y, Kage M and
Shirouzu K: Expression of IGF-1 and IGF-1R and their relation to
clinicopathological factors in colorectal cancer. Anticancer Res.
31:2541–2545. 2011.PubMed/NCBI
|
|
41
|
Lang F, Perrotti N and Stournaras C:
Colorectal carcinoma cells -regulation of survival and growth by
SGK1. Int J Biochem Cell Biol. 42:1571–1575. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Xiaobo Y, Qiang L, Xiong Q, Zheng R,
Jianhua Z, Zhifeng L, Yijiang S and Zheng J: Serum and
glucocorticoid kinase 1 promoted the growth and migration of
non-small cell lung cancer cells. Gene. 576:339–346. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Conza D, Mirra P, Calì G, Tortora T,
Insabato L, Fiory F, Schenone S, Amato R, Beguinot F, Perrotti N,
et al: The SGK1 inhibitor SI113 induces autophagy, apoptosis, and
endoplasmic reticulum stress in endometrial cancer cells. J Cell
Physiol. Feb 8–2017.(Epub ahead of print). https://doi.org/10.1002/jcp.25850 View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wang J, Li H, Wang Y, Wang L, Yan X, Zhang
D, Ma X, Du Y, Liu X and Yang Y: MicroRNA-552 enhances metastatic
capacity of colorectal cancer cells by targeting a disintegrin and
metalloprotease 28. Oncotarget. 7:70194–70210. 2016.PubMed/NCBI
|
|
45
|
Zhang Q, Tang Q, Qin D, Yu L, Huang R, Lv
G, Zou Z, Jiang XC, Zou C, Liu W, et al: Role of microRNA 30a
targeting insulin receptor substrate 2 in colorectal tumorigenesis.
Mol Cell Biol. 35:988–1000. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zeng T, Sun SY, Wang Y, Zhu H and Chen L:
Network biomarkers reveal dysfunctional gene regulations during
disease progression. FEBS J. 280:5682–5695. 2013. View Article : Google Scholar : PubMed/NCBI
|