Introduction
Hepatocellular carcinoma (HCC), a primary malignancy
of the liver, has become over the past years the sixth most common
cancer worldwide (1,2). The HCC is characterized by rapid cell
growth, early intrahepatic metastasis, high grade malignancy and
drug resistance, which result in poor 5-year survival rates of only
5% (3,4). The best long-term survival in patients
with HCC is achieved by liver resection (5) or liver transplantation (6,7);
however, there is no indication for resection of tumors that have
already spread to regional lymph nodes or distant organs (8–10).
Effective treatment of HCC remains a challenge because early
diagnostic markers and effective therapeutic options are still
pending, and the molecular mechanisms that contribute to
carcinogenesis and progression of HCC have not yet been fully
clarified. HCCs are discovered at a late stage of disease in most
cases and thus the diagnosis is given at a time-point when
therapeutic options are limited. Diagnostic markers for HCC would
be of upmost importance for the diagnosis of an early tumor stage,
which allows for curative treatment (11,12).
Additionally, due to the high incidence of both recurrent and
metastatic HCC, studies on the mechanisms of tumor invasion are
desperately needed (13,14).
C4.4A was found to be highly expressed in several
types of carcinomas such as non-small cell lung, colorectal and
renal cell cancers (15–17). In the majority of studies, high
C4.4A expression levels in the primary tumor correlated with poor
patient survival (18,19). Although its exact function is still
unknown, C4.4A is frequently detected in tumor metastases and in
wound healing which is pointing toward a role in cell migration
(20,21). This hypothesis was confirmed in
several studies demonstrating the engagement of C4.4A-expressing
tumor cells in migration and invasion (20,22).
Notably, a recent study with C4.4A-deficient bladder carcinoma in
C4.4A-knockout mice demonstrated unimpaired primary tumor growth,
but decreased invasion capability (23).
Most interestingly C4.4A is rarely expressed in
healthy organs, including the liver (17,24).
This suggests C4.4A as a potential diagnostic marker. Next to the
option of a diagnosis with scintigraphy, C4.4A may also be detected
in serum exosomes, as C4.4A is readily recovered in exosomes in
metastatic pancreatic carcinoma (21).
In addition, C4.4A might serve as a therapeutic
target. First preclinical results of a phase I study (NCT02134197)
with a C4.4A-directed antibody-drug conjugate (BAY1129980) in
non-small cell lung cancer showed a sufficient antitumor efficacy
in in vivo models (25).
C4.4A expression has not yet been evaluated in HCC.
Thus, the present study was designed to explore whether C4.4A could
serve as a potential diagnostic marker or therapeutic target in
patients with HCC.
Materials and methods
Human tissue and cell lines
Human tissue samples from non-inflammatory and
non-tumorous livers, colorectal liver metastases and hepatocellular
carcinoma (HCC) were collected during surgery. Tissue from lung
metastasis of HCC was kindly provided by the Tissue Bank of NCT
Heidelberg. The samples were used in accordance with the rules of
the Tissue Bank and approved by the ethics committee of the
Heidelberg University. Both human HCC cell lines, Huh7 (European
Collection of Cell Cultures) and HepG2 (Toni Lindl GmbH, Munich,
Germany), were cultured in Dulbeccos modified Eagles medium (DMEM)
supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin
and 100 µg/ml streptomycin in 5% CO2 at 37°C. Cells grew
adherently and, when confluent, were detached with trypsin for
sub-culturing. A Modular Incubator Chamber (Billups-Rothenberg,
Inc., San Diego, CA, USA) was used to culture the cells under
hypoxic conditions.
Antibodies
The following prevalidated antibodies were used:
C4.4A (rabbit anti-human, IBL; sheep anti-human, R&D Systems,
Minneapolis, MN, USA), actin (mouse anti-human; Sigma-Aldrich, St.
Louis, MO, USA), donkey anti-sheep HRP (Santa Cruz Biotechnology,
Santa Cruz, CA, USA), goat anti-mouse HRP (Santa Cruz
Biotechnology), Cy3-conjugated donkey anti-sheep IgG (Jackson
ImmunoResearch Laboratories, Inc., West Grove, PA, USA), goat
anti-rabbit APC (Jackson Immunoresearch Laboratories), APC-Annexin
V (Becton-Dickinson, San Diego, CA, USA) and PI
(Becton-Dickinson).
Immunohistochemistry
Formalin-fixed, paraffin-embedded tissue sections (5
µm) were deparaffinised by three 5-min washes in Roticlear and
rehydrated using a series of ethanol/water solutions. Antigen
retrieval was achieved by boiling in 1X citrate-based, unmasking
fluid (pH 6.0) for 2×7 min in a microwave oven. Sections were
treated with 3% hydrogen peroxidase/phosphate-buffered saline (PBS)
for 5 min, followed by two washes in PBS. Immunohistochemistry was
carried out using the Sheep IgG Vectastain ABC-AP kit (Vector
Laboratories, Inc., Burlingame, CA, USA). The slides were incubated
overnight at 4°C with the primary human C4.4A antibody at a
concentration of 1 µg/ml. After three washes in PBS, the tissue
sections were incubated with biotin-conjugated, secondary antibody
(rabbit anti-sheep IgG) and Vectastain ABC-AP reagent. The sections
were developed using the Vector Red Alkaline Phosphatase substrate
kit for 30 min as specified by the manufacturer. Finally, the
sections were counterstained with Mayers hematoxylin for 10 sec,
dehydrated in ethanol and then mounted. Immunohistochemistry was
examined using a Zeiss Axiovert 40 CFL microscope.
Western blot analysis
Huh7 cells and HepG2 cells were lysed with RIPA
buffer. The suspension was put on ice for 10 min and then scraped
with a plastic cell scraper prior to ultrasonification. Finally,
the lysate was cleared by centrifugation at 4°C, 10.000 × g.
Protein lysates (7.5 µl) were separated on a 4–12% SDS-PAGE
Bis-Tris gel, transferred to a nitrocellulose membrane (Bio-Rad
Laboratories GmbH, München, Germany) and blocked in 5% non-fat
dried milk. After incubating overnight with the primary antibody,
the protein was visualized with the appropriate horseradish
peroxidase-coupled, secondary antibodies (Santa Cruz Biotechnology)
using SuperSignal West Pico Chemiluminescent substrate (Thermo
Fisher Scientific, Rockford, IL, USA).
Immunofluorescence
Cells seeded on coverslides were fixed with ice-cold
methanol. After blocking, the cells were incubated at 37°C for 45
min with primary antibody. Antibody diluent with background
reducing components from Dako (Glostrup, Denmark) was used to
dilute the primary antibodies and as a negative control. After five
washes in PBS, the cells were incubated with
fluorochrome-conjugated, secondary antibody at 37°C for 45 min
before being washed again with PBS. Nuclear staining was achieved
with DAPI (Sigma-Aldrich). Finally, coverslides were mounted in a
fluorescence mounting medium from Dako.
Flow cytometry
Cells (2×105) were fixed and
permeabilized for intracellular staining. After incubation with 30
µl primary antibody (19) at a
concentration of 2.5 µg/ml (30 min, 4°C), cells were washed two
times and incubated with dye-labeled secondary antibody (30 min,
4°C). Finally, cells were analyzed in a FACSCalibur using the
CellQuest analysis program (BD Biosciences, Heidelberg,
Germany).
siRNA transfection
For a C4.4A knockdown in human HCC cell lines, the
following predesigned and prevalidated siRNAs were purchased from
Qiagen: siC4.4A (cat. no. SI00105707) and siControl (cat. no.
SI03650318). The Huh7 and the HepG2 cell lines were transfected
with siRNA using HiPerFect Transfection reagent (Qiagen, Hilden,
Germany) according to the suppliers protocol. Transfection
efficiency was evaluated after 48 h by flow cytometry.
Cell proliferation assay
Cell proliferation was analyzed by the MTT assay.
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
was added to each well at 24, 48 and 72 h after siRNA transfection.
The plates were incubated for 4 h before addition of propanol. The
absorbance was measured in 96-well plates at 570 nm using a
microplate reader.
Cell apoptosis assay
Cells were treated with 2 µg/ml of cisplatin to
induce apoptosis. At 48 h after siRNA transfection cells were
harvested and resuspended in binding buffer. After addition of
APC-Annexin V and propidium iodide (PI) the cells were incubated in
the dark for 15 min at room temperature and then analyzed by flow
cytometry (FACSCalibur; BD Biosciences).
Migration and invasion assays
Migration and invasion of HCC cells were evaluated
in Boyden chambers using a cell migration assay and a
laminin-coated cell invasion assay (Cell Biolabs, San Diego, CA,
USA). Twenty-four hours after siRNA transfection, cells were seeded
in serum-free medium in the upper chamber. The lower chamber, which
was separated by an 8 µm pore size polycarbonate-membrane,
contained medium supplemented with 10% FCS. After incubation for
12–48 h at 37°C, the non-migratory cells on the upper chamber were
removed by a cotton swab and the cells that migrated to the
undersurface of the membrane were stained with crystal-violet. The
number of migrated cells was counted within a field at ×100 under a
light microscope. For each membrane, a total of five fields were
selected at random and the numbers were averaged.
Statistical analysis
The values obtained are the mean (±SEM) of three
replicates. Statistical analysis was performed using the unpaired,
two-tailed t-test. Significance was established at a value of
P<0.05.
Results
C4.4A-expression in human liver
tissue
C4.4A-expression was evaluated in 12 samples of
non-inflammatory and non-tumorous liver, 17 HCCs and 10 pulmonary
HCC metastases. C4.4A-expression was not detected in any of the 12
samples of non-inflammatory and non-tumorous liver tissue (Fig. 1A). Since C4.4A was reported to be
expressed in liver metastasis from colorectal carcinoma (17), samples from metastatic colorectal
carcinoma (Fig. 1B) were used as
C4.4A-positive reference tissue.
Distinct C4.4A-expression was seen in 10 out of 17
(59%) HCC samples (Fig. 1C). In
addition to the presence of cytoplasmic staining, there was
increased membranous C4.4A expression on the luminal side of the
glandular structures in HCCs with a pseudoglandular growth pattern
(Fig. 1D).
C4.4A is absent from healthy bronchial and alveolar
tissue (26); however, it was found
to be highly expressed in samples of pulmonary HCC metastases
(Fig. 1E). Eight out of 10 (80%)
samples were C4.4A-positive. The strongest C4.4A-expression was
noted at the invasive front of the liver tumor nodules, namely in
hepatocytes located near the fibrotic septa and next to the
surrounding lung tissue (Fig.
1F).
In summary, C4.4A was determined to be absent from
non-inflammatory and non-tumorous liver, upregulated in primary
HCCs (59%) and strongly expressed in 80% of HCC metastases.
Upregulation of C4.4A in HCC cell
lines under hypoxic conditions
Firstly, it was determined whether or not C4.4A was
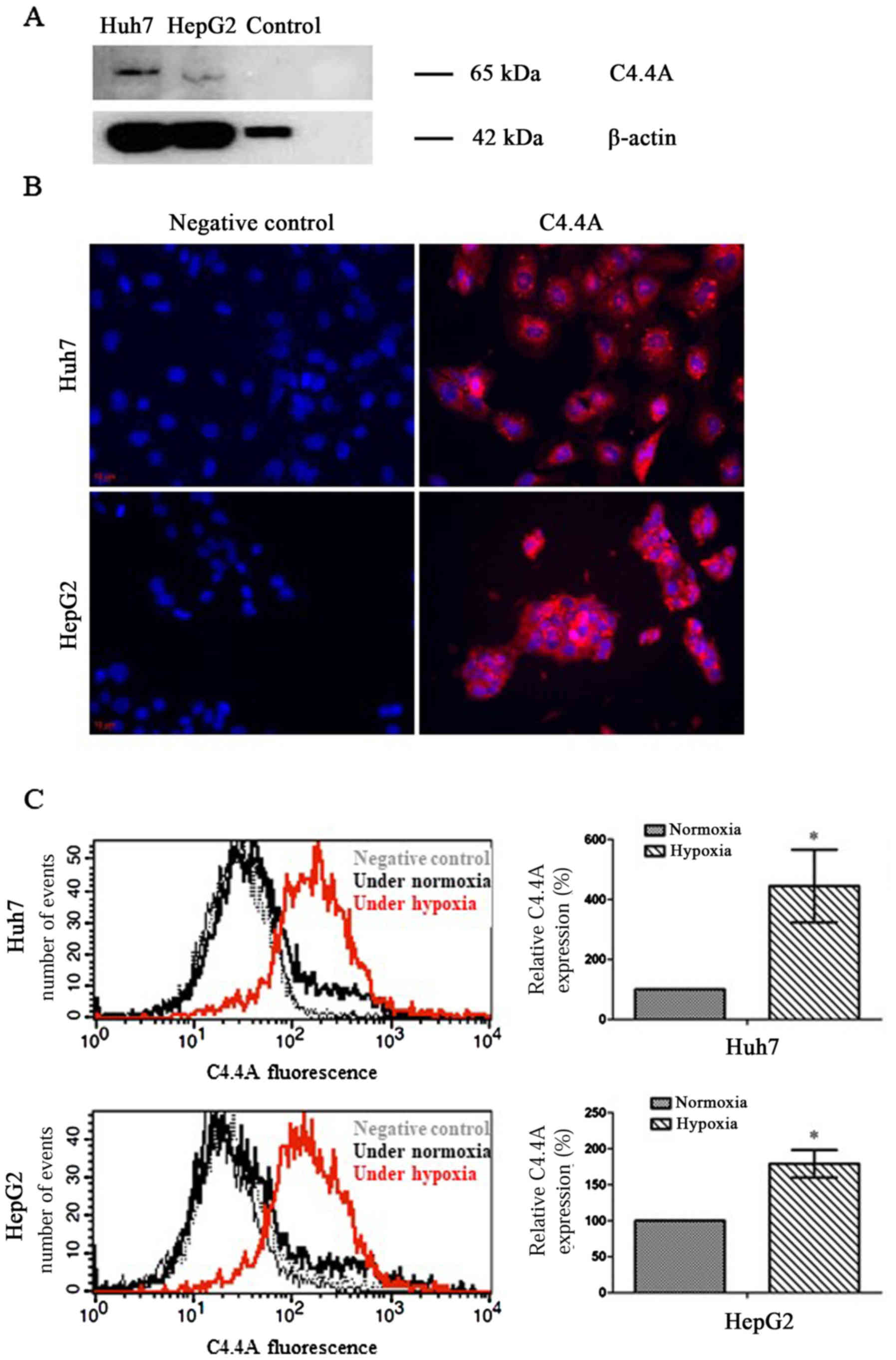
present in HCC cell lines. Western blot analysis revealed a
specific signal for C4.4A in both Huh7 and HepG2 human liver cancer
cells with a monomer band of ~65 kDa (Fig. 2A). As previously determined
(17), normal liver tissue did not
express C4.4A and as such, could be used as negative controls.
As shown in Fig. 2B,
immunofluorescence revealed that C4.4A was localized mainly within
the cytoplasmic compartment of both human cell lines. C4.4A
expression was significantly increased under hypoxic compared to
normoxic culture conditions (Fig.
2C).
Downregulation of C4.4A does not
influence HCC cell proliferation or apoptosis
C4.4A was knocked down in both Huh7 and HepG2 cells
by siRNA to examine the impact of C4.4A on HCC tumor progression.
C4.4A expression was strongly reduced in Huh7 and less pronounced,
though significant in HepG2 cells at 48 h after siC4.4A
transfection (P<0.05) (Fig.
3A).
Proliferation of both Huh7 and HepG2 cells was
evaluated by the MTT assay at 24, 48 and 72 h after transfection.
The number of viable cells increased overtime and there was no
evidence of differences between siC4.4A-treated cells and controls
(Fig. 3B).
Downregulation of C4.4A did not affect the viability
of either Huh7 or HepG2 cells as demonstrated by Annexin V and PI
staining in samples taken at 48 h after transfection. Both the
control and the C4.4A knockdown Huh7 cells showed ~0.4% Annexin
V-positive, ~0.9% Annexin V- and PI-positive and ~1.9% necrotic
(PI-positive) cells. Similar results were obtained for the control
and C4.4A knockdown HepG2 cells [~0.3% Annexin V-positive, ~1.0%
Annexin V- and PI-positive and ~2.1% necrotic (PI-positive) cells]
(Fig. 3C).
Downregulation of C4.4A reduces HCC
cell migration and invasion
It has been reported that C4.4A is involved in the
migration and invasion of colorectal and head and neck carcinoma
(20,27). Whether this also accounts for HCC
cells was evaluated in Huh7 and HepG2 cells after downregulation of
C4.4A by siRNA treatment. Transwell migration was evaluated in a
Boyden chamber assay using membranes with a 0.8 µm pore size. After
incubation for 12 h (Huh7) or 24 h (HepG2), the lower membrane site
was stained and cells in 5 fields were counted. Downregulation of
C4.4A significantly decreased the ability of both Huh7 and HepG2
cells to migrate through the membrane (Huh7, 111±10 vs. 57±8;
HepG2, 40±4 vs. 23±3 cells per field; P<0.05; Fig. 4A).
In a Transwell invasion assay, downregulation of
C4.4A significantly decreased the number of both Huh7 and HepG2
cells that penetrated through Matrigel (Huh7, 202±14 vs. 112±12;
HepG2, 102±9 vs. 62±10 cells per field; P<0.05) (Fig. 4B). These results indicate that the
migratory and invasive potential of HCC cells is significantly
reduced by a C4.4A knockdown.
Discussion
This is the first report on C4.4A expression in
liver. While in the healthy liver hepatocytes are C4.4A negative,
C4.4A expression is strongly expressed in HCC with upregulation at
the invasive front and in lung metastasis, indicating that C4.4A
apparently contributes to HCC progression. We will discuss these
findings in view of known features of C4.4A.
C4.4A was first described as a marker of a
metastasizing rat pancreatic adenocarcinoma (22,28).
Distinct to several other metastasis markers, C4.4A is rarely
expressed in non-transformed cells. Expression of the human C4.4A
was observed in placental tissue, skin, esophagus and very weakly
in subpopulations of blood leukocytes, but not in brain, lung,
liver, kidney, stomach, colon and lymphoid organs (29). C4.4A expression in the skin
epithelium becomes upregulated during wound healing (30). Consequently, it was suggested that
C4.4A is engaged in migration and/or invasion. This has been
confirmed in vivo for wounded skin epithelia as well as for
metastasizing tumor cells (21,27,31).
In vitro assays confirmed pronounced migration and invasion,
where it also was demonstrated that invasion relies on
cooperativity of C4.4A with proteases. Our studies confirm these
findings with strongly reduced migratory and invasive capacity of
C4.4A-knockdown HCC lines.
It also should be mentioned that using the
C4.4A-knockdown HCC lines there was no evidence for an engagement
of C4.4A in proliferative activity or apoptosis resistance, two
features frequently associated with metastasis-prone tumor cells.
Similar findings were reported for colon cancer cells (20), but for pancreatic cancer cells an
engagement in apoptosis induction and proliferation was reported
(32). A most likely explanation
could be that C4.4A expression is not sufficient to fulfill these
tasks, but C4.4A may act as a coactivator. It then depends on
different expression profiles of distinct tumor entities, whether
or not C4.4A supports apoptosis resistance and proliferation.
Further studies are needed to address this question.
One of the notable findings in this study has been
the extremely strong upregulation of C4.4A in HCC lines maintained
under hypoxic conditions. This is important as hypoxic conditions
are common in HCC and often result in enhanced tumor progression
and metastasis (33–35). Promoter studies revealed so far a
contribution of C/EBP beta and JunD or c-Jun, but no contribution
of HIF-1α for C4.4A transcription (36). Further promoter analyses may unravel
the cotranscription factors accounting for the highly increased
expression under hypoxic conditions. Concerning C4.4A as a
potential diagnostic marker, the strong upregulation under hypoxia,
which is frequently seen in HCC, adds to its diagnostic
potential.
Finally, first studies with a C4.4A knockout mouse
need to be mentioned as they confirmed again the engagement of
C4.4A in the metastatic spread of tumor cells without evidence for
functional activity as an oncogene.
In brief, the C4.4A protein is absent from normal
liver, but strongly expressed in HCC and its metastasis. C4.4A
expression in HCC cells is further enhanced by hypoxic conditions.
In Huh7 and HepG2 cells, C4.4A is engaged in tumor cell migration
and invasion.
Conclusion and outlook
Metastasis associated molecules are in many
instances abundantly expressed. C4.4A is exceptional with
displaying very restricted expression in non-transformed tissue.
This also accounts for the liver, where it is highly expressed in
59% of primary HCC and in 80% of lung metastasis.
As the prognosis of patients with HCC is poor and
reliable diagnostic markers are limited, C4.4A might well serve as
a new diagnostic and/or prognostic marker. HCC develops in
cirrhotic livers, where differential diagnosis of hepatic nodular
lesions can be difficult. To differentiate HCC with MRI and CT
scanning from the so-called regenerative nodules is especially
difficult at a small size. C4.4A could be detected using a
radiolabeled antibody by scintigraphy, thereby improving the
sensitivity and specificity of non-invasive imaging and enabling
the differentiation of C4.4A-positive HCC from C4.4A-negative
regenerative nodules in cirrhotic livers. If nodular lesions are
detected in radiologic imaging, C4.4A expression in scintigraphy
would give rise to the probalility for the presence of a HCC in
contrast to regenerative nodules, that are negative for C4.4A and
comprise of non-tumorous liver tissue. C4.4A expression also occurs
in other tumor entities and wound healing, but the differentiation
to HCC is easily possible due to the topographic localisation. As a
differential diagnosis of C4.4A-positive hepatic lesions, liver
metastases from colorectal cancer need to be considered. However,
in these cases the primary tumor can be easily identified by
endoscopy, plus liver metastases have a different behavior in MRI
and CT imaging.
Alternatively, serum exosomes or exosomes in
gallbladder secretion might be used for diagnosis in the future, as
C4.4A is readily recovered in exosomes in pancreatic carcinoma
(21). However, this diagnostic
method is connected with considerable effort and is not yet a
standard procedure.
Data provided here clearly show that C4.4A is a
highly relevant marker for HCC that can be detected in tumor
samples. This is of high clinical relevance because C4.4A as a
diagnostic marker could be detected using a radiolabeled antibody
by scintigraphy. Further studies and clinical trials are warranted
for verification, especially to test the hypothesis that serum
exosomes or exosomes in gallbladder secretion could be used to
detect C4.4A.
Furthermore, rare expression of C4.4A in
non-transformed cells suggests anti-C4.4A as an adjuvant
therapeutics in cancer treatment, a first report with an anti-C4.4A
antibody-drug conjugate revealing promising results (25).
Acknowledgements
We cordially thank Elvira Mohr for technical help
with immunohistochemistry and Nadya Phillips for English language
correction.
References
|
1
|
Kang KJ and Ahn KS: Anatomical resection
of hepatocellular carcinoma: A critical review of the procedure and
its benefits on survival. World J Gastroenterol. 23:1139–1146.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Forner A, Llovet JM and Bruix J:
Hepatocellular carcinoma. Lancet. 379:1245–1255. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hoffmann K, Franz C, Xiao Z, Mohr E, Serba
S, Büchler MW and Schemmer P: Sorafenib modulates the gene
expression of multi-drug resistance mediating ATP-binding cassette
proteins in experimental hepatocellular carcinoma. Anticancer Res.
30:4503–4508. 2010.PubMed/NCBI
|
|
5
|
Hoffmann K, Müller-Bütow V, Franz C, Hinz
U, Longerich T, Büchler MW and Schemmer P: Factors predictive of
survival after stapler hepatectomy of hepatocellular carcinoma: A
multivariate, single-center analysis. Anticancer Res. 34:767–776.
2014.PubMed/NCBI
|
|
6
|
Lin S, Hoffmann K and Schemmer P:
Treatment of hepatocellular carcinoma: A systematic review. Liver
Cancer. 1:144–158. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hoffmann K, Hinz U, Hillebrand N, Radeleff
BA, Ganten TM, Schirmacher P, Schmidt J, Büchler MW and Schemmer P:
Risk factors of survival after liver transplantation for HCC: A
multivariate single-center analysis. Clin Transplant. 25:E541–E551.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Clark HP, Carson WF, Kavanagh PV, Ho CP,
Shen P and Zagoria RJ: Staging and current treatment of
hepatocellular carcinoma. Radiographics. 25 Suppl 1:S3–S23. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bruix J and Sherman M: American
Association for the Study of Liver Diseases: Management of
hepatocellular carcinoma: An update. Hepatology. 53:1020–1022.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Schemmer P, Friess H, Hinz U, Mehrabi A,
Kraus TW, Zgraggen K, Schmidt J, Uhl W and Büchler MW: Stapler
hepatectomy is a safe dissection technique: Analysis of 300
patients. World J Surg. 30:419–430. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Scaggiante B, Kazemi M, Pozzato G, Dapas
B, Farra R, Grassi M, Zanconati F and Grassi G: Novel
hepatocellular carcinoma molecules with prognostic and therapeutic
potentials. World J Gastroenterol. 20:1268–1288. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Neumann O, Kesselmeier M, Geffers R,
Pellegrino R, Radlwimmer B, Hoffmann K, Ehemann V, Schemmer P,
Schirmacher P, Bermejo J Lorenzo, et al: Methylome analysis and
integrative profiling of human HCCs identify novel protumorigenic
factors. Hepatology. 56:1817–1827. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Forner A, Llovet JM and Bruix J:
Hepatocellular carcinoma. Lancet. 379:1245–1255. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liang B, Jia C, Huang Y, He H, Li J, Liao
H, Liu X, Liu X, Bai X and Yang D: TPX2 level correlates with
hepatocellular carcinoma cell proliferation, apoptosis, and EMT.
Dig Dis Sci. 60:2360–2372. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fletcher GC, Patel S, Tyson K, Adam PJ,
Schenker M, Loader JA, Daviet L, Legrain P, Parekh R, Harris AL, et
al: hAG-2 and hAG-3, human homologues of genes involved in
differentiation, are associated with oestrogen receptor-positive
breast tumours and interact with metastasis gene C4.4a and
dystroglycan. Br J Cancer. 88:579–585. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jacobsen B, Santoni-Rugiu E, Illemann M,
Kriegbaum MC, Laerum OD and Ploug M: Expression of C4.4A in
precursor lesions of pulmonary adenocarcinoma and squamous cell
carcinoma. Int J Cancer. 130:2734–2739. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Paret C, Hildebrand D, Weitz J,
Kopp-Schneider A, Kuhn A, Beer A, Hautmann R and Zöller M: C4.4A as
a candidate marker in the diagnosis of colorectal cancer. Br J
Cancer. 97:1146–1156. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hansen LV, Skov BG, Ploug M and Pappot H:
Tumour cell expression of C4.4A, a structural homologue of the
urokinase receptor, correlates with poor prognosis in non-small
cell lung cancer. Lung Cancer. 58:260–266. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Konishi K, Yamamoto H, Mimori K, Takemasa
I, Mizushima T, Ikeda M, Sekimoto M, Matsuura N, Takao T, Doki Y,
et al: Expression of C4.4A at the invasive front is a novel
prognostic marker for disease recurrence of colorectal cancer.
Cancer Sci. 101:2269–2277. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Oshiro R, Yamamoto H, Takahashi H, Ohtsuka
M, Wu X, Nishimura J, Takemasa I, Mizushima T, Ikeda M, Sekimoto M,
et al: C4.4A is associated with tumor budding and
epithelial-mesenchymal transition of colorectal cancer. Cancer Sci.
103:1155–1164. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ngora H, Galli UM, Miyazaki K and Zöller
M: Membrane-bound and exosomal metastasis-associated C4.4A promotes
migration by associating with the α6β4
integrin and MT1-MMP. Neoplasia. 14:95–107. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Rösel M, Claas C, Seiter S, Herlevsen M
and Zöller M: Cloning and functional characterization of a new
phosphatidyl-inositol anchored molecule of a metastasizing rat
pancreatic tumor. Oncogene. 17:1989–2002. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kriegbaum MC, Jacobsen B, Füchtbauer A,
Hansen GH, Christensen IJ, Rundsten CF, Persson M, Engelholm LH,
Madsen AN, Di Meo I, et al: C4.4A gene ablation is compatible with
normal epidermal development and causes modest overt phenotypes.
Sci Rep. 6:258332016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kriegbaum MC, Jacobsen B, Hald A and Ploug
M: Expression of C4.4A, a structural uPAR homolog, reflects
squamous epithelial differentiation in the adult mouse and during
embryogenesis. J Histochem Cytochem. 59:188–201. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Willuda J, Linden L, Lerchen HG, Kopitz C,
Stelte-Ludwig B, Pena C, Lange C, Golfier S, Kneip C, Carrigan PE,
et al: Preclinical anti-tumor ffficacy of BAY 1129980-a novel
auristatin-based anti-C4.4A (LYPD3) antibody-drug conjugate for the
treatment of non-small cell lung cancer. Mol Cancer Ther.
16:893–904. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jacobsen B, Kriegbaum MC, Santoni-Rugiu E
and Ploug M: C4.4A as a biomarker in pulmonary adenocarcinoma and
squamous cell carcinoma. World J Clin Oncol. 5:621–632. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu JF, Mao L, Bu LL, Ma SR, Huang CF,
Zhang WF and Sun ZJ: C4.4A as a biomarker of head and neck squamous
cell carcinoma and correlated with epithelial mesenchymal
transition. Am J Cancer Res. 5:3505–3515. 2015.PubMed/NCBI
|
|
28
|
Matzku S, Wenzel A, Liu S and Zöller M:
Antigenic differences between metastatic and nonmetastatic BSp73
rat tumor variants characterized by monoclonal antibodies. Cancer
Res. 49:1294–1299. 1989.PubMed/NCBI
|
|
29
|
Würfel J, Seiter S, Stassar M, Claas A,
Kläs R, Rösel M, Marhaba R, Savelyeva L, Schwab M, Matzku S, et al:
Cloning of the human homologue of the metastasis-associated rat
C4.4A. Gene. 262:35–41. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hansen LV, Gårdsvoll H, Nielsen BS, Lund
LR, Danø K, Jensen ON and Ploug M: Structural analysis and tissue
localization of human C4.4A: A protein homologue of the urokinase
receptor. Biochem J. 380:845–857. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Thuma F, Ngora H and Zöller M: The
metastasis-associated molecule C4.4A promotes tissue invasion and
anchorage independence by associating with the alpha6beta4
integrin. Mol Oncol. 7:917–928. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Arumugam T, Deng D, Bover L, Wang H,
Logsdon CD and Ramachandran V: New blocking antibodies against
novel AGR2-C4.4A pathway reduce growth and metastasis of pancreatic
tumors and increase survival in mice. Mol Cancer Ther. 14:941–951.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wong CC, Kai AK and Ng IO: The impact of
hypoxia in hepatocellular carcinoma metastasis. Front Med. 8:33–41.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Salnikov AV, Liu L, Platen M, Gladkich J,
Salnikova O, Ryschich E, Mattern J, Moldenhauer G, Werner J,
Schemmer P, et al: Hypoxia induces EMT in low and highly aggressive
pancreatic tumor cells but only cells with cancer stem cell
characteristics acquire pronounced migratory potential. PLoS One.
7:e463912012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hu F, Deng X, Yang X, Jin H, Gu D, Lv X,
Wang C, Zhang Y, Huo X, Shen Q, et al: Hypoxia upregulates
Rab11-family interacting protein 4 through HIF-1α to promote the
metastasis of hepatocellular carcinoma. Oncogene. 34:6007–6017.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Fries F, Nazarenko I, Hess J, Claas A,
Angel P and Zoller M: CEBPbeta, JunD and c-Jun contribute to the
transcriptional activation of the metastasis-associated C4.4A gene.
Int J Cancer. 120:2135–2147. 2007. View Article : Google Scholar : PubMed/NCBI
|