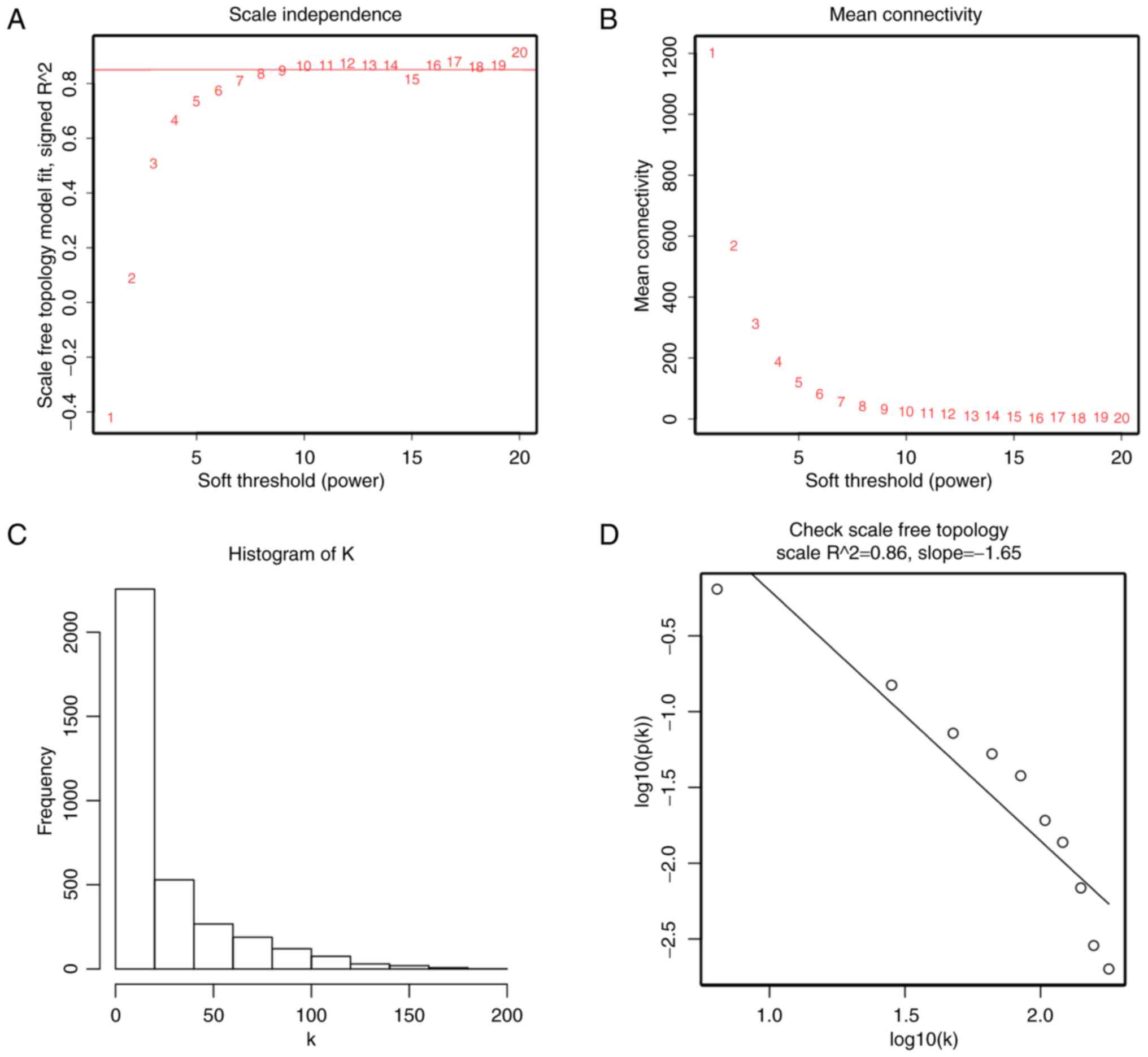
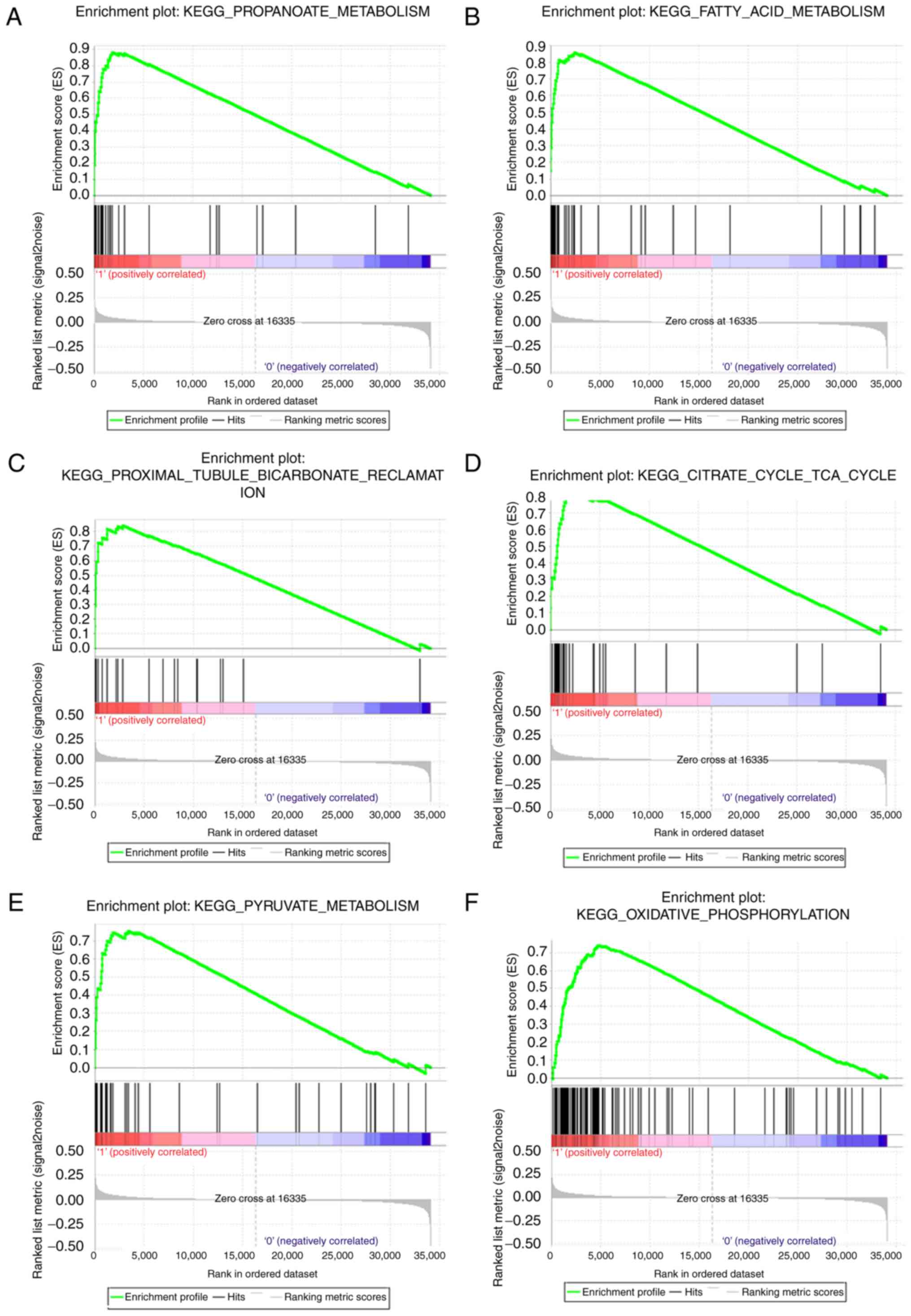
|
1
|
Cairns P: Renal cell carcinoma. Cancer
Biomark. 9:461–473. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Vera-Badillo FE, Templeton AJ, Duran I,
Ocana A, de Gouveia P, Aneja P, Knox JJ, Tannock IF, Escudier B and
Amir E: Systemic therapy for non-clear cell renal cell carcinomas:
A systematic review and meta-analysis. Eur Urol. 67:740–749. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Heng DY, Mackenzie MJ, Vaishampayan UN,
Bjarnason GA, Knox JJ, Tan MH, Wood L, Wang Y, Kollmannsberger C,
North S, et al: Primary anti-vascular endothelial growth factor
(VEGF)-refractory metastatic renal cell carcinoma: Clinical
characteristics, risk factors, and subsequent therapy. Ann Oncol.
23:1549–1555. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dahinden C, Ingold B, Wild P, Boysen G,
Luu VD, Montani M, Kristiansen G, Sulser T, Bühlmann P, Moch H, et
al: Mining tissue microarray data to uncover combinations of
biomarker expression patterns that improve intermediate staging and
grading of clear cell renal cell cancer. Clin Cancer Res. 16:88–98.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gerlinger M, Horswell S, Larkin J, Rowan
AJ, Salm MP, Varela I, Fisher R, McGranahan N, Matthews N, Santos
CR, et al: Genomic architecture and evolution of clear cell renal
cell carcinomas defined by multiregion sequencing. Nat Genet.
46:225–233. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Eckl J, Buchner A, Prinz PU, Riesenberg R,
Siegert SI, Kammerer R, Nelson PJ and Noessner E: Transcript
signature predicts tissue NK cell content and defines renal cell
carcinoma subgroups independent of TNM staging. J Mol Med (Berl).
90:55–66. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tavazoie S, Hughes JD, Campbell MJ, Cho RJ
and Church GM: Systematic determination of genetic network
architecture. Nat Genet. 22:281–285. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chou WC, Cheng AL, Brotto M and Chuang CY:
Visual gene-network analysis reveals the cancer gene co-expression
in human endometrial cancer. BMC Genomics. 15:3002014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang F, Chang Y, Li J, Wang H, Zhou R, Qi
J, Liu J and Zhao Q: Strong correlation between ASPM gene
expression and HCV cirrhosis progression identified by
co-expression analysis. Dig Liver Dis. 49:70–76. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Clarke C, Madden SF, Doolan P, Aherne ST,
Joyce H, O'Driscoll L, Gallagher WM, Hennessy BT, Moriarty M, Crown
J, et al: Correlating transcriptional networks to breast cancer
survival: A large-scale coexpression analysis. Carcinogenesis.
34:2300–2308. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Horvath S and Dong J: Geometric
interpretation of gene coexpression network analysis. PLOS Comput
Biol. 4:e10001172008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mason MJ, Fan G, Plath K, Zhou Q and
Horvath S: Signed weighted gene co-expression network analysis of
transcriptional regulation in murine embryonic stem cells. BMC
Genomics. 10:3272009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yip AM and Horvath S: Gene network
interconnectedness and the generalized topological overlap measure.
BMC Bioinformatics. 8:222007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ravasz E, Somera AL, Mongru DA, Oltvai ZN
and Barabási AL: Hierarchical organization of modularity in
metabolic networks. Science. 297:1551–1555. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43(D1): D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Franceschini A, Lin J, von Mering C and
Jensen LJ: SVD-phy: Improved prediction of protein functional
associations through singular value decomposition of phylogenetic
profiles. Bioinformatics. 32:1085–1087. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Uhlén M, Fagerberg L, Hallström BM,
Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C,
Sjöstedt E, Asplund A, et al: Proteomics. Tissue-based map of the
human proteome. Science. 347:12604192015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Dennis G Jr, Sherman BT, Hosack DA, Yang
J, Gao W, Lane HC and Lempicki RA: DAVID: Database for Annotation,
Visualization, and Integrated Discovery. Genome Biol. 4:32003.
View Article : Google Scholar
|
|
19
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xu G and Li JY: ATP5A1 and ATP5B are
highly expressed in glioblastoma tumor cells and endothelial cells
of microvascular proliferation. J Neurooncol. 126:405–413. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Seth R, Keeley J, Abu-Ali G, Crook S,
Jackson D and Ilyas M: The putative tumour modifier gene ATP5A1 is
not mutated in human colorectal cancer cell lines but expression
levels correlate with TP53 mutations and chromosomal instability. J
Clin Pathol. 62:598–603. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jonckheere AI, Renkema GH, Bras M, Van den
Heuvel LP, Hoischen A, Gilissen C, Nabuurs SB, Huynen MA, de Vries
MC, Smeitink JA, et al: A complex V ATP5A1 defect causes fatal
neonatal mitochondrial encephalopathy. Brain. 136:1544–1554. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bata P, Gyebnar J, Tarnoki DL, Tarnoki AD,
Kekesi D, Szendroi A, Fejer B, Szasz AM, Nyirady P, Karlinger K, et
al: Clear cell renal cell carcinoma and papillary renal cell
carcinoma: Differentiation of distinct histological types with
multiphase CT. Diagn Interv Radiol. 19:387–392. 2013.PubMed/NCBI
|
|
24
|
Arnold RS, Fedewa SA, Goodman M, Osunkoya
AO, Kissick HT, Morrissey C, True LD and Petros JA: Bone metastasis
in prostate cancer: Recurring mitochondrial DNA mutation reveals
selective pressure exerted by the bone microenvironment. Bone.
78:81–86. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fang Y, Huang J, Zhang J, Wang J, Qiao F,
Chen HM and Hong ZP: Detecting the somatic mutations spectrum of
Chinese lung cancer by analyzing the whole mitochondrial DNA
genomes. Mitochondrial DNA. 26:56–60. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li LH, Kang T, Chen L, Zhang W, Liao Y,
Chen J and Shi Y: Detection of mitochondrial DNA mutations by
high-throughput sequencing in the blood of breast cancer patients.
Int J Mol Med. 33:77–82. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hashimoto M, Bacman SR, Peralta S, Falk
MJ, Chomyn A, Chan DC, Williams SL and Moraes CT: MitoTALEN: A
general approach to reduce mutant mtDNA loads and restore oxidative
phosphorylation function in mitochondrial dseases. Mol Ther.
23:1592–1599. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bonora E, Porcelli AM, Gasparre G, Biondi
A, Ghelli A, Carelli V, Baracca A, Tallini G, Martinuzzi A, Lenaz
G, et al: Defective oxidative phosphorylation in thyroid oncocytic
carcinoma is associated with pathogenic mitochondrial DNA mutations
affecting complexes I and III. Cancer Res. 66:6087–6096. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cheng L, Lin H, Hu Y, Wang J and Yang Z:
Gene function prediction based on the Gene Ontology hierarchical
structure. PLoS One. 9:e1071872014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Du J, Yuan Z, Ma Z, Song J, Xie X and Chen
Y: KEGG-PATH: Kyoto encyclopedia of genes and genomes-based pathway
analysis using a path analysis model. Mol Biosyst. 10:2441–2447.
2014. View Article : Google Scholar : PubMed/NCBI
|