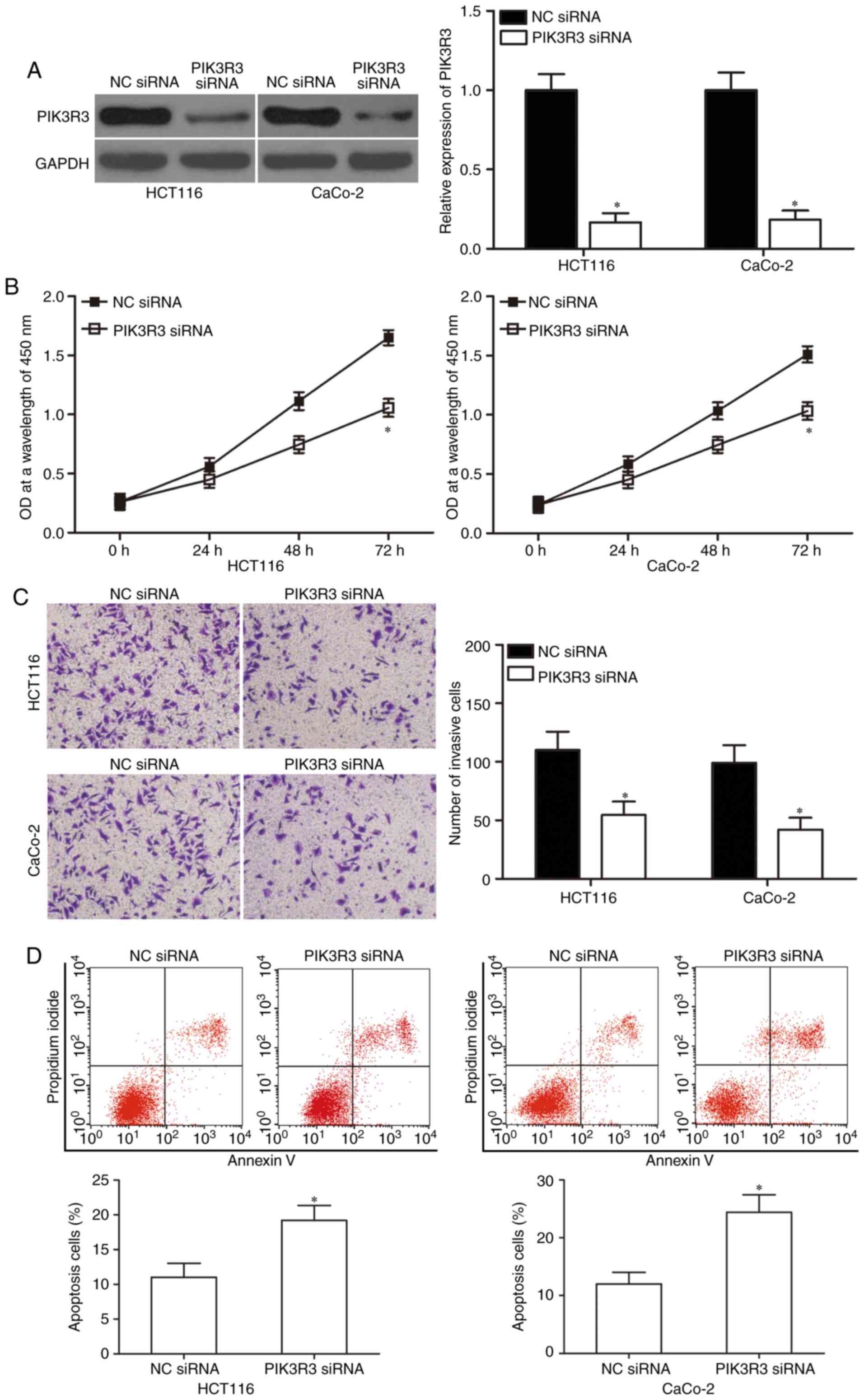
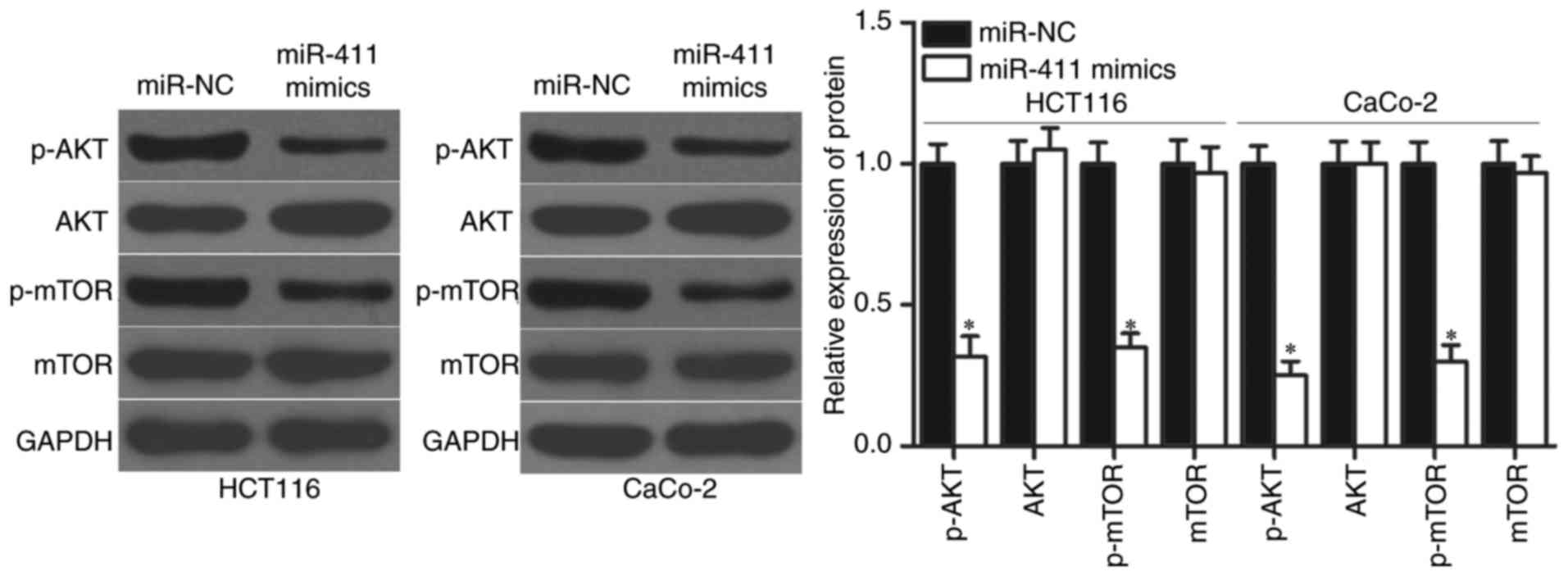
|
1
|
Haggar FA and Boushey RP: Colorectal
cancer epidemiology: Incidence, mortality, survival, and risk
factors. Clin Colon Rectal Surg. 22:191–197. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Center MM, Jemal A, Smith RA and Ward E:
Worldwide variations in colorectal cancer. CA Cancer J Clin.
59:366–378. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sung JJ, Lau JY, Goh KL and Leung WK; Asia
Pacific Working Group on Colorectal Cancer, : Increasing incidence
of colorectal cancer in Asia: Implications for screening. Lancet
Oncol. 6:871–876. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li S, Wang J, Lu Y and Fan D: Screening
and early diagnosis of colorectal cancer in China: A 12 year
retrospect (1994–2006). J Cancer Res Clin Oncol. 133:679–686. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sanz-Garcia E, Grasselli J, Argiles G,
Elez ME and Tabernero J: Current and advancing treatments for
metastatic colorectal cancer. Expert Opin Biol Ther. 16:93–110.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Meyerhardt JA and Mayer RJ: Systemic
therapy for colorectal cancer. N Engl J Med. 352:476–487. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Qiu Y, Liu Q, Chen G, Wang W, Peng K, Xiao
W and Yang H: Outcome of rectal cancer surgery in obese and
nonobese patients: A meta-analysis. World J Surg Oncol. 14:232016.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lee RC, Feinbaum RL and Ambros V: The C.
elegans heterochronic gene lin-4 encodes small RNAs with antisense
complementarity to lin-14. Cell. 75:843–854. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
OHara SP, Mott JL, Splinter PL, Gores GJ
and LaRusso NF: MicroRNAs: Key modulators of posttranscriptional
gene expression. Gastroenterology. 136:17–25. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu J: Control of protein synthesis and
mRNA degradation by microRNAs. Curr Opin Cell Biol. 20:214–221.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
He L, Thomson JM, Hemann MT,
Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe
SW, Hannon GJ, et al: A microRNA polycistron as a potential human
oncogene. Nature. 435:828–833. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang J, Lin H, Wang XY, Zhang DQ, Chen
JX, Zhuang Y and Zheng XL: Predictive value of microRNA-143 in
evaluating the prognosis of patients with hepatocellular carcinoma.
Cancer Biomark. 19:257–262. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Karatas OF, Oner M, Abay A and Diyapoglu
A: MicroRNAs in human tongue squamous cell carcinoma: From
pathogenesis to therapeutic implications. Oral Oncol. 67:124–130.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Teoh SL and Das S: The role of MicroRNAs
in diagnosis, prognosis, metastasis and resistant cases in breast
cancer. Curr Pharm Des. 23:1845–1859. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bucay N, Bhagirath D, Sekhon K, Yang T,
Fukuhara S, Majid S, Shahryari V, Tabatabai Z, Greene KL, Hashimoto
Y, et al: A novel microRNA regulator of prostate cancer
epithelial-mesenchymal transition. Cell Death Differ. 24:1263–1274.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cui L, Li Y, Lv X, Li J, Wang X, Lei Z and
Li X: Expression of MicroRNA-301a and its functional roles in
malignant melanoma. Cell Physiol Biochem. 40:230–244. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lee HW, Lee EH, Ha SY, Lee CH, Chang HK,
Chang S, Kwon KY, Hwang IS, Roh MS and Seo JW: Altered expression
of microRNA miR-21, miR-155, and let-7a and their roles in
pulmonary neuroendocrine tumors. Pathol Int. 62:583–591. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hwang HW and Mendell JT: MicroRNAs in cell
proliferation, cell death, and tumorigenesis. Br J Cancer.
94:776–780. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:pp. 2257–2261.
2006; View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tyagi N, Arora S, Deshmukh SK, Singh S,
Marimuthu S and Singh AP: Exploiting nanotechnology for the
development of MicroRNA-based cancer therapeutics. J Biomed
Nanotechnol. 12:28–42. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Barger JF and Nana-Sinkam SP: MicroRNA as
tools and therapeutics in lung cancer. Respir Med. 109:803–812.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xia K, Zhang Y, Cao S, Wu Y, Guo W, Yuan W
and Zhang S: miR-411 regulated ITCH expression and promoted cell
proliferation in human hepatocellular carcinoma cells. Biomed
Pharmacother. 70:158–163. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhao Z, Qin L and Li S: miR-411
contributes the cell proliferation of lung cancer by targeting
FOXO1. Tumour Biol. 37:5551–5560. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Guo L, Yuan J, Xie N, Wu H, Chen W, Song S
and Wang X: miRNA-411 acts as a potential tumor suppressor miRNA
via the downregulation of specificity protein 1 in breast cancer.
Mol Med Rep. 14:2975–2982. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang Y, Xu G, Liu G, Ye Y, Zhang C, Fan
C, Wang H, Cai H, Xiao R, Huang Z, et al: miR-411-5p inhibits
proliferation and metastasis of breast cancer cell via targeting
GRB2. Biochem Biophys Res Commun. 476:607–613. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang G, Yang X, Li C, Cao X, Luo X and Hu
J: PIK3R3 induces epithelial-to-mesenchymal transition and promotes
metastasis in colorectal cancer. Mol Cancer Ther. 13:1837–1847.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li B, Xie Z and Li B: miR-152 functions as
a tumor suppressor in colorectal cancer by targeting PIK3R3. Tumour
Biol. 37:10075–10084. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cao G, Dong W, Meng X, Liu H, Liao H and
Liu S: MiR-511 inhibits growth and metastasis of human
hepatocellular carcinoma cells by targeting PIK3R3. Tumour Biol.
36:4453–4459. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liu K, Li X, Cao Y, Ge Y, Wang J and Shi
B: MiR-132 inhibits cell proliferation, invasion and migration of
hepatocellular carcinoma by targeting PIK3R3. Int J Oncol.
47:1585–1593. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hwang HW and Mendell JT: MicroRNAs in cell
proliferation, cell death, and tumorigenesis. Br J Cancer. 96
Suppl:R40–R44. 2007.PubMed/NCBI
|
|
35
|
Sun M, Huang F, Yu D, Zhang Y, Xu H, Zhang
L, Li L, Dong L, Guo L and Wang S: Autoregulatory loop between
TGF-β1/miR-411-5p/SPRY4 and MAPK pathway in rhabdomyosarcoma
modulates proliferation and differentiation. Cell Death Dis.
6:e18592015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yamamoto K, Seike M, Takeuchi S, Soeno C,
Miyanaga A, Noro R, Minegishi Y, Kubota K and Gemma A: MiR-379/411
cluster regulates IL-18 and contributes to drug resistance in
malignant pleural mesothelioma. Oncol Rep. 32:2365–2372. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Oliveto S, Mancino M, Manfrini N and Biffo
S: Role of microRNAs in translation regulation and cancer. World J
Biol Chem. 8:45–56. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhu Y, Zhao H, Rao M and Xu S:
MicroRNA-365 inhibits proliferation, migration and invasion of
glioma by targeting PIK3R3. Oncol Rep. 37:2185–2192. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang L, Huang J, Yang N, Greshock J,
Liang S, Hasegawa K, Giannakakis A, Poulos N, OBrien-Jenkins A,
Katsaros D, et al: Integrative genomic analysis of
phosphatidylinositol 3-kinase family identifies PIK3R3 as a
potential therapeutic target in epithelial ovarian cancer. Clin
Cancer Res. 13:5314–5321. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhou J, Chen GB, Tang YC, Sinha RA, Wu Y,
Yap CS, Wang G, Hu J, Xia X, Tan P, et al: Genetic and
bioinformatic analyses of the expression and function of PI3K
regulatory subunit PIK3R3 in an Asian patient gastric cancer
library. BMC Med Genomics. 5:342012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Xu L, Wen Z, Zhou Y, Liu Z, Li Q, Fei G,
Luo J and Ren T: MicroRNA-7-regulated TLR9 signaling-enhanced
growth and metastatic potential of human lung cancer cells by
altering the phosphoinositide-3-kinase, regulatory subunit 3/Akt
pathway. Mol Biol Cell. 24:42–55. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Klahan S, Wu MS, Hsi E, Huang CC, Hou MF
and Chang WC: Computational analysis of mRNA expression profiles
identifies the ITG family and PIK3R3 as crucial genes for
regulating triple negative breast cancer cell migration. BioMed Res
Int. 2014:5365912014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Niemeyer BF, Parrish JK, Spoelstra NS,
Joyal T, Richer JK and Jedlicka P: Variable expression of PIK3R3
and PTEN in Ewing Sarcoma impacts oncogenic phenotypes. PLoS One.
10:e01168952015. View Article : Google Scholar : PubMed/NCBI
|