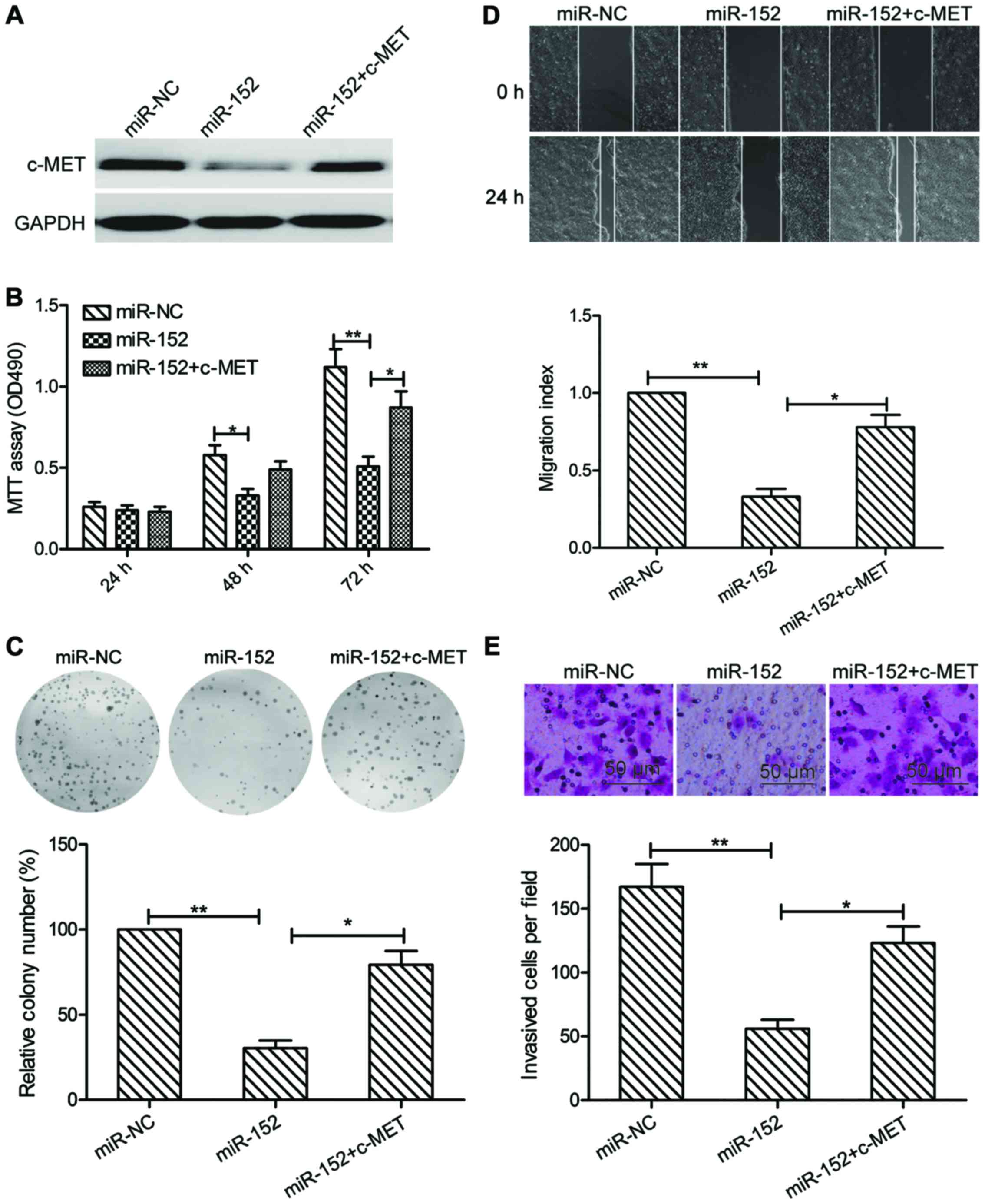
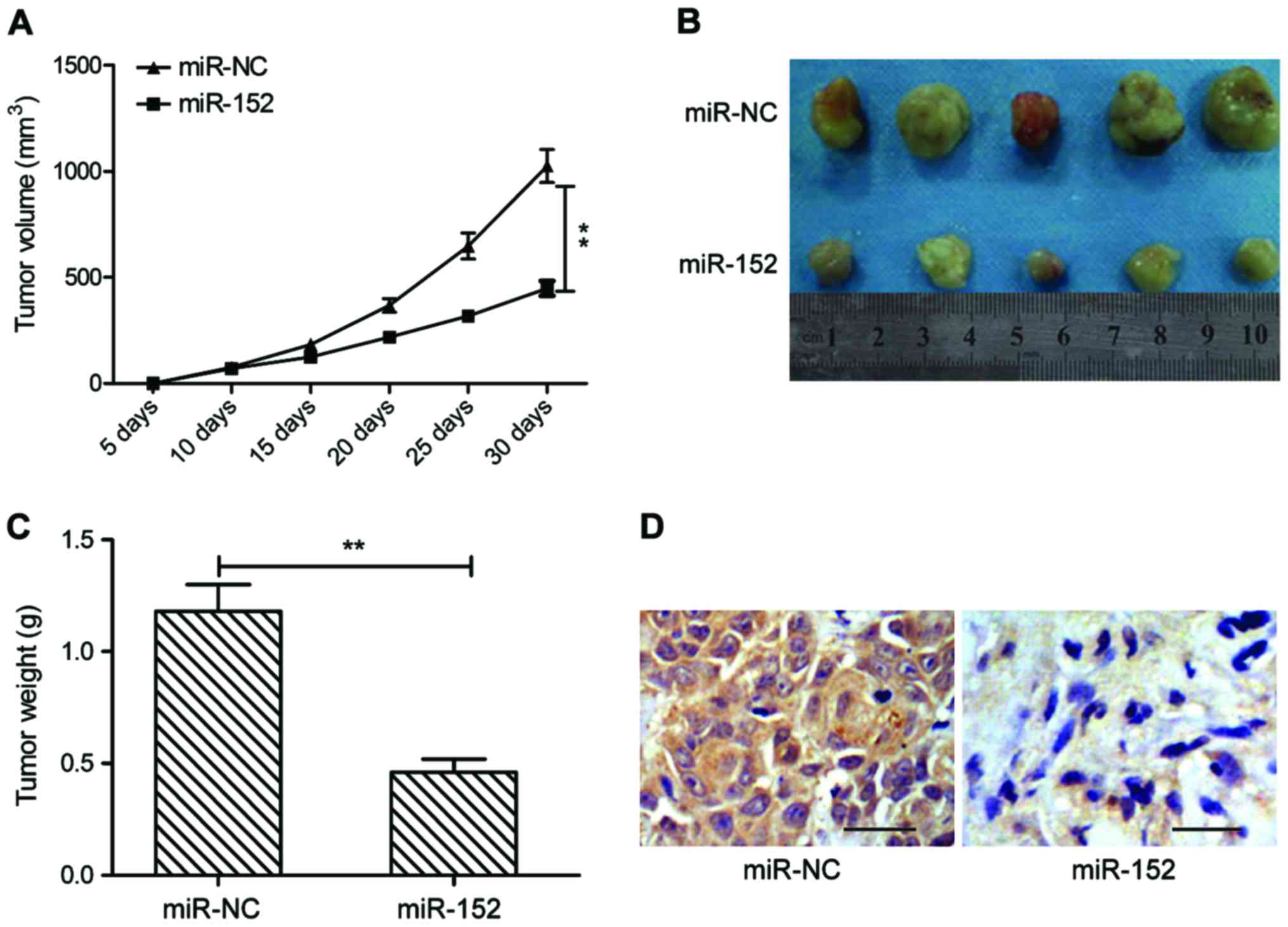
|
1
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Liao CT, Hsueh C, Lee LY, Lin CY, Fan KH,
Wang HM, Huang SF, Chen IH, Kang CJ, Ng SH, et al: Neck dissection
field and lymph node density predict prognosis in patients with
oral cavity cancer and pathological node metastases treated with
adjuvant therapy. Oral Oncol. 48:329–336. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jerjes W, Upile T, Petrie A, Riskalla A,
Hamdoon Z, Vourvachis M, Karavidas K, Jay A, Sandison A, Thomas GJ,
et al: Clinicopathological parameters, recurrence, locoregional and
distant metastasis in 115 T1-T2 oral squamous cell carcinoma
patients. Head Neck Oncol. 2:92010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Malan-Müller S, Hemmings SM and Seedat S:
Big effects of small RNAs: A review of microRNAs in anxiety. Mol
Neurobiol. 47:726–739. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
McManus MT: MicroRNAs and cancer. Semin
Cancer Biol. 13:253–258. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Farazi TA, Spitzer JI, Morozov P and
Tuschl T: miRNAs in human cancer. J Pathol. 223:102–115. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Garzon R and Marcucci G: Potential of
microRNAs for cancer diagnostics, prognostication and therapy. Curr
Opin Oncol. 24:655–659. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Prasad G, Seers C, Reynolds E and
McCullough MJ: A panel of microRNAs can be used to determine oral
squamous cell carcinoma. J Oral Pathol Med. 46:940–948.
2017.PubMed/NCBI
|
|
10
|
Ries J, Baran C, Wehrhan F, Weber M,
Neukam FW, Krautheim-Zenk A and Nkenke E: Prognostic significance
of altered miRNA expression in whole blood of OSCC patients. Oncol
Rep. 37:3467–3474. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xie G, Li W, Li R, Wu K, Zhao E, Zhang Y,
Zhang P, Shi L, Wang D, Yin Y, et al: Helicobacter pylori promote
B7-H1 expression by suppressing miR-152 and miR-200b in gastric
cancer cells. PLoS One. 12:e01688222017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li B, Xie Z and Li B: miR-152 functions as
a tumor suppressor in colorectal cancer by targeting PIK3R3. Tumour
Biol. 37:10075–10084. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen MJ, Cheng YM, Chen CC, Chen YC and
Shen CJ: miR-148a and miR-152 reduce tamoxifen resistance in
ER+ breast cancer via downregulating ALCAM. Biochem
Biophys Res Commun. 483:840–846. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang YJ, Liu XC, Du J and Zhang YJ:
miR-152 regulates metastases of non-small cell lung cancer cells by
targeting neuropilin-1. Int J Clin Exp Pathol. 8:14235–14240.
2015.PubMed/NCBI
|
|
15
|
Tang XL, Lin L, Song LN and Tang XH:
Hypoxia-inducible miR-152 suppresses the expression of WNT1 and
ERBB3, and inhibits the proliferation of cervical cancer cells. Exp
Biol Med (Maywood). 241:1429–1437. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhu C, Li J, Ding Q, Cheng G, Zhou H, Tao
L, Cai H, Li P, Cao Q, Ju X, et al: miR-152 controls migration and
invasive potential by targeting TGFα in prostate cancer cell lines.
Prostate. 73:1082–1089. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li B, Yang XX, Wang D and Ji HK:
MicroRNA-138 inhibits proliferation of cervical cancer cells by
targeting c-Met. Eur Rev Med Pharmacol Sci. 20:1109–1114.
2016.PubMed/NCBI
|
|
18
|
Chau L, Jabara JT, Lai W, Svider PF,
Warner BM, Lin HS, Raza SN and Fribley AM: Topical agents for oral
cancer chemoprevention: A systematic review of the literature. Oral
Oncol. 67:153–159. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gharat SA, Momin M and Bhavsar C: Oral
squamous cell carcinoma: Current treatment strategies and
nanotechnology-based approaches for prevention and therapy. Crit
Rev Ther Drug Carrier Syst. 33:363–400. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jasinski-Bergner S, Stoehr C, Bukur J,
Massa C, Braun J, Hüttelmaier S, Spath V, Wartenberg R, Legal W,
Taubert H, et al: Clinical relevance of miR-mediated HLA-G
regulation and the associated immune cell infiltration in renal
cell carcinoma. OncoImmunology. 4:e10088052015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu DZ, Ander BP, Tian Y, Stamova B,
Jickling GC, Davis RR and Sharp FR: Integrated analysis of mRNA and
microRNA expression in mature neurons, neural progenitor cells and
neuroblastoma cells. Gene. 495:120–127. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Huang S, Li X and Zhu H: MicroRNA-152
Targets phosphatase and tensin homolog to inhibit apoptosis and
promote cell migration of nasopharyngeal carcinoma cells. Med Sci
Monit. 22:4330–4337. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang Y, Wang D, Xie G, Yin Y, Zhao E, Tao
K and Li R: MicroRNA-152 regulates immune response via targeting
B7-H1 in gastric carcinoma. Oncotarget. 8:28125–28134.
2017.PubMed/NCBI
|
|
24
|
Ge S, Wang D, Kong Q, Gao W and Sun J:
Function of miR-152 as a tumor suppressor in human breast cancer by
targeting PIK3CA. Oncol Res. 25:1363–1371. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhou J, Zhang Y, Qi Y, Yu D, Shao Q and
Liang J: MicroRNA-152 inhibits tumor cell growth by directly
targeting RTKN in hepatocellular carcinoma. Oncol Rep.
37:1227–1234. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Skead G and Govender D: Gene of the month:
MET. J Clin Pathol. 68:405–409. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Marano L, Chiari R, Fabozzi A, De Vita F,
Boccardi V, Roviello G, Petrioli R, Marrelli D, Roviello F and
Patriti A: c-Met targeting in advanced gastric cancer: An open
challenge. Cancer Lett. 365:30–36. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Boccaccio C, Luraghi P and Comoglio PM:
MET-mediated resistance to EGFR inhibitors: An old liaison rooted
in colorectal cancer stem cells. Cancer Res. 74:3647–3651. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ho-Yen CM, Jones JL and Kermorgant S: The
clinical and functional significance of c-Met in breast cancer: A
review. Breast Cancer Res. 17:522015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mazzone M and Comoglio PM: The Met
pathway: Master switch and drug target in cancer progression. FASEB
J. 20:1611–1621. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Freudlsperger C, Alexander D, Reinert S
and Hoffmann J: Prognostic value of c-Met expression in oral
squamous cell carcinoma. Exp Ther Med. 1:69–72. 2010.PubMed/NCBI
|
|
32
|
Yasui H, Ohnishi Y, Nakajima M and Nozaki
M: Migration of oral squamous cell carcinoma cells are induced by
HGF/c-Met signalling via lamellipodia and filopodia formation.
Oncol Rep. 37:3674–3680. 2017. View Article : Google Scholar : PubMed/NCBI
|