Introduction
Breast cancer is the most frequently diagnosed
malignancy and the leading cause of cancer-related death among
females worldwide, with an estimated 1.7 million cases and 521,900
deaths in 2012 (1). Current breast
cancer treatment strategies include surgery and adjuvant therapy,
such as chemotherapy, radiotherapy, hormonal therapy and targeted
drugs (2). The backbone of current
chemotherapy regimens includes anthracyclines (e.g., adriamycin and
epirubicin) and taxanes (e.g., Cytoxan®) given either
sequentially or concurrently. Anti-estrogen drugs (e.g., tamoxifen)
and aromatase inhibitors (e.g., letrozole) dominate breast cancer
endocrine therapy. HER-2-positive breast cancer patients could be
treated with targeted therapy, and the main drug is trastuzumab
monoclonal antibody (Herceptin®). Chemotherapy forms an
important part of a successful treatment regimen; however, half of
the patients may fail to benefit from this, as a result of drug
resistance (3). Thus,
chemoresistance constitutes a major clinical obstacle for the
successful treatment of breast cancer.
MicroRNAs (miRNAs) are small non-coding RNAs that
negatively regulate gene expression at the post-transcriptional
level (4). Studies have shown that
miRNAs are often aberrantly expressed in human cancers and are
associated with tumorigenesis, metastasis, invasiveness and drug
resistance (5–8). For example, miR-214 is downregulated
in gastric cancer and inhibits cell migration and invasion through
targeting CSF1 (5). Most recently,
Zhang et al identified five miRNAs (miR-30b-5p, miR-96-5p,
miR-182-5p, miR-374b-5p and miR-942-5p) as candidate blood
biomarkers in breast cancer patients (9). However, studies concerning
chemoresistance-associated miRNAs in breast cancer based on human
tissues are scarce. In the present study, differentially expressed
miRNAs (DE-miRNAs) in chemoresistant and chemosensitive breast
cancer tissues were screened using miRNA expression profile of
GSE71142. The genes targeted by DE-miRNAs were predicted, and their
potential functions were analyzed by functional and pathway
enrichment analysis. Furthermore, a protein-protein interaction
(PPI) network of the predicted target genes was constructed.
Potential transcription factors that may regulate the target genes
were screened. Through these comprehensive bioinformatic analyses,
the present study aimed to explore the molecular mechanisms
underlying breast cancer chemoresistance and identify important
miRNA therapeutic targets.
Materials and methods
miRNA microarray
Breast cancer chemoresistance-associated miRNA
microarray dataset GSE71142 was downloaded from the National Center
for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO)
database (http://www.ncbi.nlm.nih.gov/geo). The dataset
GSE71142, based on the platform of GPL20717 µParaflo™ miRNA
microarray (LC Sciences, Houston, TX, USA), included five cases of
chemoresistant breast cancer tissues and five cases of
chemosensitive tissues. The chemotherapy drugs used for treating
the breast cancer patients are provided in Fig. 1B.
Screening for DE-miRNAs
Data were analyzed by subtracting the background and
then the signals were normalized using a locally-weighted
regression (LOWESS) filter (10).
The t-test analysis was conducted between chemoresistant and
chemosensitive samples, and miRNAs with P-values <0.05 and fold
change (FC) >2 were selected for cluster analysis. Mean values
of each group were used in the cluster analysis by HemI software
(Heatmap Illustrator, version 1.0) (11).
Prediction of genes targeted by
DE-miRNAs
The miRWalk2.0 database generated possible
miRNA-target interactions by gathering information from 12 types of
existing prediction software (e.g., Targetscan, miRanda and
RNAhybrid) (12). In the present
study, miRWalk2.0 was used to predict the target genes of the
DE-miRNAs. Only the common target genes predicted by at least nine
types of software were selected, which were defined as potential
target genes.
GO and pathway analysis
WEB-based GEne SeT AnaLysis Toolkit (WebGestalt)
(13) was used to perform
functional enrichment analysis including Gene Ontology (GO) and
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis,
for the potential target genes of DE-miRNAs. P<0.05 was
considered statistically significant.
Protein-protein interaction (PPI)
network and miRNA-target network construction
To evaluate the interactive relationships among
target genes, we mapped the target genes to the STRING database
(http://string-db.org) (14), and only the interactions with a
combined score >0.4 were considered as significant. The degree
of connectivity in networks was analyzed using Cytoscape software
(version 3.4.0), to obtain the significant nodes or hub proteins
(15) in the PPI networks.
Screening of potential transcription
factors
FunRich (http://www.funrich.org) (16) is a functional enrichment and
interaction network analysis tool that identifies the enriched
transcription factors for gene sets. In the present study, FunRich
was used to identify transcription factors that regulate the
DE-miRNA target genes.
Identification of breast
cancer-associated miRNAs validated in previous studies
The miRWalk2.0 database (12) provided information on experimentally
validated miRNA-gene-Human Phenotype Ontology (HPO) interactions.
In the present study, we used the Validated Targets Module of
miRWalk2.0 to identify breast cancer-associated miRNAs and
miRNA-gene interactions.
Results
Identification of DE-miRNAs and their
target genes
A total of 22 DE-miRNAs were screened out, including
10 upregulated and 12 downregulated miRNAs (Fig. 1) in chemoresistant breast cancer
tissues, compared with chemosensitive tissues. Based on FC,
miR-196a-5p, miR-4286 and miR-200b-3p were the top three most
upregulated miRNAs; and miR-4472, miR-4467 and miR-572 were the top
three most downregulated miRNAs. miRWalk2.0 was used to predict the
target genes of DE-miRNAs, generating 1,278 potential target genes,
including 1,155 genes for upregulated miRNAs and 123 genes for
downregulated miRNAs.
Functional and pathway enrichment
analyses
GO functional and KEGG pathway enrichment analyses
were performed on the aforementioned potential target genes. The
enriched GO functions for the target genes are presented in
Tables I and II, including the positive regulation of
gene expression, positive regulation of transcription
(DNA-templated) and positive regulation of cellular biosynthetic
process in the biological process (BP) category; cell junction and
nuclear transcriptional repressor complex in the cellular component
(CC) category; and transcription factor activity and enzyme binding
in the molecular function (MF) category.
 | Table I.Enriched functions for the target
genes of the upregulated miRNAs. |
Table I.
Enriched functions for the target
genes of the upregulated miRNAs.
| GO terms | P-value | FDR |
|---|
| Biological process
(BP) |
|
|
|
GO:0006366, transcription from
RNA polymerase II promoter | P<0.001 | P<0.001 |
|
GO:0009790, embryo
development | P<0.001 | P<0.001 |
|
GO:0010628, positive
regulation of gene expression | P<0.001 | P<0.001 |
|
GO:0045893, positive
regulation of transcription, DNA-templated | P<0.001 | P<0.001 |
|
GO:1903508, positive
regulation of nucleic acid-templated transcription | P<0.001 | P<0.001 |
| Cellular component
(CC) |
|
|
|
GO:0031410, cytoplasmic
vesicle | 2.354E-14 | 1.407E-11 |
|
GO:0097708, intracellular
vesicle | 2.731E-14 | 1.407E-11 |
|
GO:0098588, bounding membrane
of organelle | 1.391E-13 | 4.776E-11 |
|
GO:0030054, cell junction | 3.435E-13 | 8.845E-11 |
|
GO:0097458, neuron part | 4.348E-13 | 8.956E-11 |
| Molecular function
(MF) |
|
|
|
GO:0000981, RNA polymerase II
transcription factor activity, sequence-specific DNA binding | P<0.001 | P<0.001 |
|
GO:0000982, transcription
factor activity, RNA polymerase II core promoter proximal region
sequence-specific binding | 1.199E-14 | 1.096E-11 |
|
GO:0001228, transcriptional
activator activity, RNA polymerase II transcription regulatory
region sequence-specific binding | 4.663E-14 | 2.841E-11 |
|
GO:0019899, enzyme
binding | 1.765E-13 | 8.067E-11 |
|
GO:0044212, transcription
regulatory region DNA binding | 5.079E-13 | 1.654E-10 |
 | Table II.Enriched functions of the target genes
of the downregulated miRNAs. |
Table II.
Enriched functions of the target genes
of the downregulated miRNAs.
| GO terms | P-value | FDR |
|---|
| Biological process
(BP) |
|
|
|
GO:0001501, skeletal system
development | 5.75E-05 | 1.05E-01 |
|
GO:0051173, positive
regulation of nitrogen compound metabolic process | 6.77E-05 | 1.05E-01 |
|
GO:0045935, positive
regulation of nucleobase-containing compound metabolic process | 7.03E-05 | 1.05E-01 |
|
GO:0031328, positive
regulation of cellular biosynthetic process | 1.36E-04 | 1.05E-01 |
|
GO:2000113, negative
regulation of cellular macromolecule biosynthetic process | 1.43E-04 | 1.05E-01 |
| Cellular component
(CC) |
|
|
|
GO:0005578, proteinaceous
extracellular matrix | 3.42E-04 | 8.81E-02 |
|
GO:0030054, cell junction | 3.45E-04 | 8.81E-02 |
|
GO:0070603, SWI/SNF
superfamily-type complex | 4.87E-04 | 8.81E-02 |
|
GO:0090568, nuclear
transcriptional repressor complex | 4.90E-04 | 8.81E-02 |
|
GO:0016589, NURF complex | 5.10E-04 | 8.81E-02 |
| Molecular function
(MF) |
|
|
|
GO:0003682, chromatin
binding | 3.34E-05 | 3.18E-02 |
|
GO:0019899, enzyme
binding | 4.13E-05 | 3.18E-02 |
|
GO:0019903, protein
phosphatase binding | 5.22E-05 | 3.18E-02 |
|
GO:0003700, transcription
factor activity, sequence-specific DNA binding | 1.10E-04 | 4.08E-02 |
|
GO:0001071, nucleic acid
binding transcription factor activity | 1.12E-04 | 4.08E-02 |
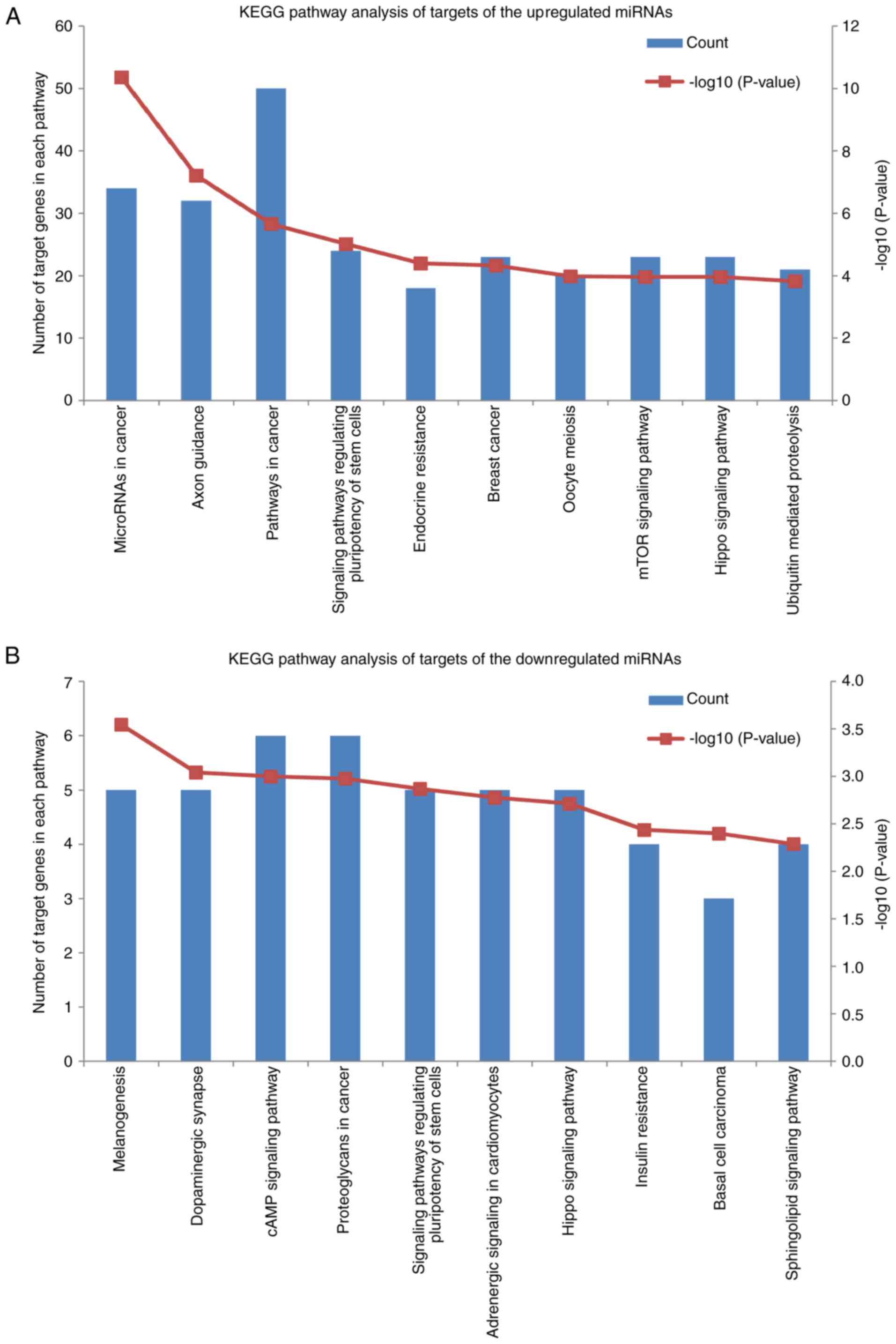
The enriched KEGG pathways for target genes of
upregulated miRNAs (Fig. 2A)
included miRNAs in cancer, pathways in cancer, signaling pathways
regulating pluripotency of stem cells, endocrine resistance, breast
cancer, mTOR signaling and Hippo signaling pathway. Of note, 23
genes (CDK6, JAG1, DVL3, E2F3, FGF9, DLL1/4, IGF1R, JUN, LRP6,
NOTCH1/2, NRAS, PIK3R1/3, MAPK1/3, FZD3/4/5/7, AXIN2 and WNT3A)
were specifically involved in the breast cancer pathway. For
downregulated miRNAs (Fig. 2B), the
enriched KEGG pathways included cAMP signaling pathway,
proteoglycans in cancer, signaling pathways regulating pluripotency
of stem cells, Hippo signaling pathway and basal cell
carcinoma.
Construction and analysis of PPI
network and miRNA-target network
Data from the STRING database showed that many of
the target genes interacted with each other. For better
visualization, the top 10 hub nodes with higher degrees were
screened (Table III). For the
upregulated miRNAs, the hub genes were NOTCH1, JUN, NRAS, MAPK1,
BCL2, MAPK3, NFKB1, ITGA2, CDK6 and IGF1R. Among these genes,
NOTCH1 showed the highest node degree (degree=103). For the
downregulated miRNAs, the hub genes were MAPK14, PRKCA, SMARCA5,
UBE2I, DVL3, WNT7B, CREB5, SLC9A1, FZD3 and NFKB1. Among these
genes, MAPK14 showed the highest node degree (degree=10).
 | Table III.Hub genes identified in the PPI
network. |
Table III.
Hub genes identified in the PPI
network.
| Upregulated
miRNAs | Downregulated
miRNAs |
|---|
|
|
|---|
| Gene symbol | Degree | Gene symbol | Degree |
|---|
| NOTCH1 | 103 | MAPK14 | 10 |
| JUN | 102 | PRKCA | 5 |
| NRAS | 83 | SMARCA5 | 4 |
| MAPK1 | 81 | UBE2I | 4 |
| BCL2 | 80 | DVL3 | 3 |
| MAPK3 | 74 | WNT7B | 3 |
| NFKB1 | 66 | CREB5 | 3 |
| ITGA2 | 64 | SLC9A1 | 2 |
| CDK6 | 52 | FZD3 | 2 |
| IGF1R | 52 | NFKB1 | 2 |
As shown in Fig. 3,
the miRNA-hub gene network was constructed. The hub target genes of
the upregulated miRNAs could be potentially regulated by miR-107,
miR-16-5p, miR-196a-5p, miR-200b-3p, miR-214-3p and miR-23a-3p
(Fig. 3A). Particularly, miR-16-5p
was predicted to target the most hub genes (n=4). The hub target
genes of the downregulated miRNAs could be potentially regulated by
miR-1275, miR-489 and miR-572 (Fig.
3B). In addition, miR-489 was predicted to target the most hub
genes (n=5).
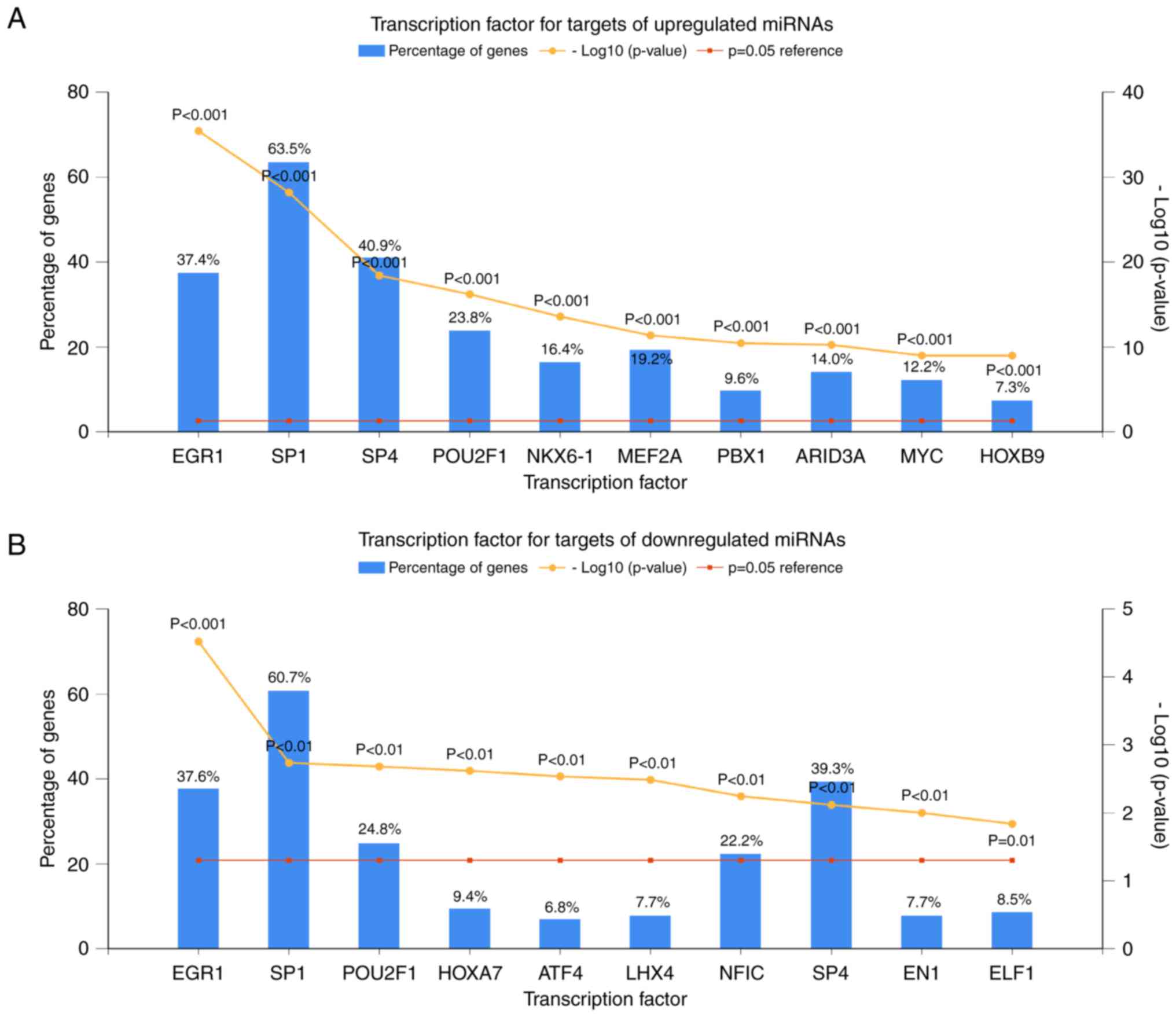
Screening of potential transcription
factors
Based on data from FunRich, the top 10 enriched
transcription factors for the target genes of the upregulated
miRNAs were EGR1, SP1, SP4, POU2F1, NKX6-1, MEF2A, PBX1, ARID3A,
MYC and HOXB9 (Fig. 4A). For the
targets of the downregulated miRNAs, the top 10 enriched
transcription factors were EGR1, SP1, POU2F1, HOXA7, ATF4, LHX4,
NFIC, SP4, EN1 and ELF1 (Fig.
4B).
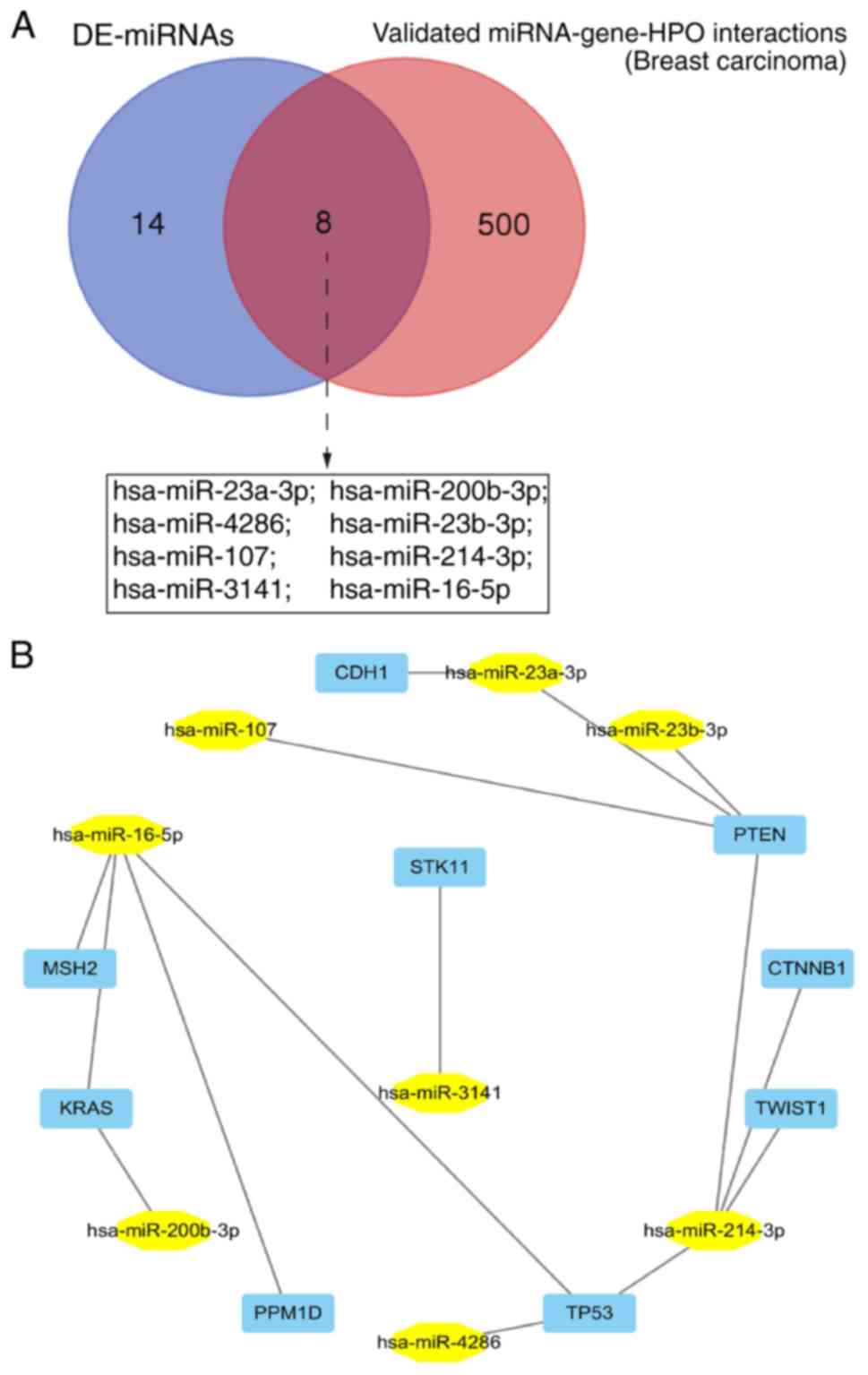
Validated breast carcinoma-associated
miRNA-gene interactions
The miRWalk2.0 database was used to search for
experimentally validated miRNA-gene-Human Phenotype Ontology (HPO)
interactions in breast carcinoma. Eight of the DE-miRNAs
(miR-23a-3p, miR-200b-3p, miR-4286, miR-23b-3p, miR-107,
miR-214-3p, miR-3141 and miR-16-5p) were previously proven to be
breast carcinoma-associated miRNAs (Fig. 5A) by targeting a series of oncogenes
or tumor suppressors such as PTEN, PPM1D, TP53, KRAS, MSH2, CTNNB1,
TWIST1, CDH1 and STK11 (Fig.
5B).
Discussion
Chemoresistance is a major limitation for breast
cancer therapy. In the present study, bioinformatics analyses were
performed to investigate microRNA (miRNA)-mediated mechanism of
breast cancer chemoresistance and to identify molecular targets. In
the present study, we identified 22 DE-miRNAs in chemoresistant
breast cancer and chemosensitive tissues based on the GEO database
GSE71142. Then, by the target gene software of miRWalk2.0, we
identified the potential target genes of these
chemoresistance-related miRNAs. The enrichment and function
analyses showed that these target genes may participate in many
important cancer-related biological processes, molecular functions
and signaling pathways.
Among the dysregulated miRNAs, miR-196a-5p
(upregulation) and miR-4472 (downregulation) were found to have the
greatest expression fold change between chemoresistant and
chemosensitive tissues. miR-196a-5p has been previously reported to
be overexpressed in triple-negative breast cancer compared with
hormone receptor-positive breast cancer by microarray (17). miR-4472 has been found to be located
in common fragile sites that are frequently affected by DNA
double-strand breaks, which may be associated with carcinogenesis
from its earliest stages (18).
These two miRNAs (miR-196a-5p and miR-4472) have not been
systemically investigated in breast cancer. This is the first
report to link these two miRNAs with breast cancer chemotherapy
resistance.
Functional enrichment analysis was performed on the
target genes of identified DE-miRNAs. The most enriched GO terms
were significantly associated with regulation processes at the BP
level, and transcription activity at the MF level, respectively.
KEGG pathway analysis was performed to check the potentially
involved pathways of the target genes. Several breast
cancer-associated pathways were identified from the top enriched
KEGG terms, namely miRNAs in cancer, pathways in cancer, signaling
pathways regulating pluripotency of stem cells, endocrine
resistance, breast cancer, mTOR signaling pathway, Hippo signaling
pathway, cAMP signaling pathway, and proteoglycans in cancer. Since
stem cells are believed to play an important role in drug
resistance (19), understanding of
target genes involved in signaling pathways regulating the
pluripotency of stem cells may help us to uncover mechanisms of
drug resistance. The mTOR pathway is pivotal not only in
tumorigenesis, but also in cancer chemotherapy and hormonal drug
sensitivity (20). Various mTOR
inhibitors have been developed to overcome breast cancer drug
resistance and increase the therapeutic efficacy of breast cancer
therapy (21). Recently, evidence
suggests that the Hippo tumor suppressor pathway may confer cells
with more aggressive traits and regulate the response of cancer
cells to chemotherapeutics (22,23).
The cAMP signaling pathway has been shown to regulate multidrug
resistance of breast cancer by modulating MDR1 transcription
(24). Enrichment of the target
genes of DE-miRNAs in these signaling pathways further supports the
potential involvement of the DE-miRNAs in breast cancer
chemoresistance.
PPI network was constructed and it was found that
NOTCH1 and mitogen-activated protein kinase 14 (MAPK14) were the
hub genes with the highest connectivity degree of 103 and 10, among
the targets of upregulated and downregulated miRNAs, respectively.
Notch1 has been shown to upregulate multidrug resistance-associated
protein 1 (MRP1) expression in breast cancer, and Notch 1 inhibitor
was found to sensitize breast cancer cells to doxorubicin and
paclitaxel (25,26). Mechanically, Notch 1 has been found
to be targeted by miR-34a to reduce breast cancer stemness and
chemoresistance (27). Cleator
et al enrolled a cohort of breast cancer patients who
received doxorubicin and cyclophosphamide (adriamycin/cytoxan, AC)
chemotherapy treatment, and found that MAPK14 was upregulated in
chemosensitive tumors (28). These
data support that NOTCH1 and MAPK14 may be candidate targets
associated with breast cancer chemoresistance.
Exploring the possible transcription factors may be
helpful in understanding the mechanisms of target genes in
DE-miRNAs. Early growth response 1 (EGR1) and SP1 are the common
transcription factors that targeted the most genes both for
upregulated and downregulated miRNAs. A recent study indicated that
EGR1 increased drug resistance of breast cancer by enhancing
multidrug resistance 1 (MDR1) expression (29). Saha et al revealed that
aberrant overexpression of SP1 in breast cancer stem cells
transcriptionally upregulated resistance-promoting genes to
decrease doxorubicin therapy sensitivity (30). Enrichment of EGR1 and SP1 by the
target genes of DE-miRNAs implies the potential roles of the target
genes and the DE-miRNAs in drug resistance.
By searching the validated miRNA-gene-human
phenotype ontology (HPO) interactions in miRWalk2.0, several
miRNA-target pairs in breast cancer were screened. For example, the
miR-214-tumor protein p53 (TP53) axis promotes apoptosis and
sensitizes breast cancer cells to doxorubicin (31). The protein phosphatase,
Mg2+/Mn2+-dependent 1D (miR-16-PPM1D)
signaling suppresses the self-renewal and proliferation of mouse
breast cancer stem cells and sensitizes MCF-7 human breast cancer
cells to doxorubicin (32).
Screening of these validated breast cancer-associated miRNAs
suggests that the current methods to identify breast cancer- and
chemoresistance-related miRNAs are credible.
In conclusion, our data provide a comprehensive
bioinformatic analysis of DE-miRNAs, which may be involved in
breast cancer chemoresistance. The target genes of DE-miRNAs are
associated with important signaling pathways in cancer, including
breast carcinogenesis, progression and drug resistance. The
DE-miRNAs such as miR-196a-5p, miR-4472, miR-16-5p and miR-489, and
the hub target genes including NOTCH1 and MAPK14 may have the
potential to be used as targets for breast cancer treatment.
However, the biological function and mechanism of these DE-miRNAs
and the target genes in breast cancer chemoresistance need further
experimental excavation and research.
Acknowledgements
We thank LC Sciences for assistance in the
microarray data analysis. This work was supported by the Shandong
Key Research and Development Plan (no. 2016GSF201128), the National
Natural Science Foundation of China (no. 81402192), the Initial
Funding for New Clinical and Practical Techniques of Qilu Hospital
of Shandong University (no. 2016–1) and the Science and Technology
Development Plan of Jinan (the medical and health science and
technology innovation plan, no. 201704091).
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kutanzi KR, Yurchenko OV, Beland FA,
Checkhun VF and Pogribny IP: MicroRNA-mediated drug resistance in
breast cancer. Clin Epigenetics. 2:171–185. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
ODriscoll L and Clynes M: Biomarkers and
multiple drug resistance in breast cancer. Curr Cancer Drug
Targets. 6:365–384. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang YW, Shi DB, Chen X, Gao C and Gao P:
Clinicopathological significance of microRNA-214 in gastric cancer
and its effect on cell biological behaviour. PLoS One.
9:e913072014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang YW, Chen X, Gao JW, Zhang H, Ma RR,
Gao ZH and Gao P: High expression of cAMP-responsive
element-binding protein 1 (CREB1) is associated with metastasis,
tumor stage and poor outcome in gastric cancer. Oncotarget.
6:10646–10657. 2015.PubMed/NCBI
|
|
7
|
Chen X, Wang YW, Xing AY, Xiang S, Shi DB,
Liu L, Li YX and Gao P: Suppression of SPIN1-mediated PI3K-Akt
pathway by miR-489 increases chemosensitivity in breast cancer. J
Pathol. 239:459–472. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang YW, Chen X, Ma R and Gao P:
Understanding the CREB1-miRNA feedback loop in human malignancies.
Tumour Biol. 37:8487–8502. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang K, Wang YW, Wang YY, Song Y, Zhu J,
Si PC and Ma R: Identification of microRNA biomarkers in the blood
of breast cancer patients based on microRNA profiling. Gene.
619:10–20. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bolstad BM, Irizarry RA, Astrand M and
Speed TP: A comparison of normalization methods for high density
oligonucleotide array data based on variance and bias.
Bioinformatics. 19:185–193. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Deng W, Wang Y, Liu Z, Cheng H and Xue Y:
HemI: A toolkit for illustrating heatmaps. PLoS One. 9:e1119882014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dweep H and Gretz N: miRWalk2.0: A
comprehensive atlas of microRNA-target interactions. Nat Methods.
12:6972015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang J, Duncan D, Shi Z and Zhang B:
WEB-based GEne SeT AnaLysis toolkit (WebGestalt): Update 2013.
Nucleic Acids Research. 41:W77–W83. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43:D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
He X and Zhang J: Why do hubs tend to be
essential in protein networks? PLoS Genet. 2:e882006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pathan M, Keerthikumar S, Ang CS, Gangoda
L, Quek CY, Williamson NA, Mouradov D, Sieber OM, Simpson RJ, Salim
A, et al: FunRich: An open access standalone functional enrichment
and interaction network analysis tool. Proteomics. 15:2597–2601.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li Z, Meng Q, Pan A, Wu X, Cui J, Wang Y
and Li L: MicroRNA-455-3p promotes invasion and migration in triple
negative breast cancer by targeting tumor suppressor EI24.
Oncotarget. 8:19455–19466. 2017.PubMed/NCBI
|
|
18
|
Georgakilas AG, Tsantoulis P, Kotsinas A,
Michalopoulos I, Townsend P and Gorgoulis VG: Are common fragile
sites merely structural domains or highly organized ‘functional’
units susceptible to oncogenic stress? Cell Mol Life Sci.
71:4519–4544. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhou C, Grottkau BE and Zou S: regulators
of stem cells proliferation in tissue regeneration. Curr Stem Cell
Res Ther. 11:177–187. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Margariti N, Fox SB, Bottini A and
Generali D: ‘Overcoming breast cancer drug resistance with mTOR
inhibitors’. Could it be a myth or a real possibility in the
short-term future? Breast Cancer Res Treat. 128:599–606.
2011.PubMed/NCBI
|
|
21
|
Carraway H and Hidalgo M: New targets for
therapy in breast cancer: Mammalian target of rapamycin (mTOR)
antagonists. Breast Cancer Res. 6:219–224. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lai D, Visser-Grieve S and Yang X: Tumour
suppressor genes in chemotherapeutic drug response. Biosci Rep.
32:361–374. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shi P, Feng J and Chen C: Hippo pathway in
mammary gland development and breast cancer. Acta Biochim Biophys
Sin. 47:53–59. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Rohlff C and Glazer RI: Regulation of
multidrug resistance through the cAMP and EGF signalling pathways.
Cell Signal. 7:431–443. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kim B, Stephen SL, Hanby AM, Horgan K,
Perry SL, Richardson J, Roundhill EA, Valleley EM, Verghese ET,
Williams BJ, et al: Chemotherapy induces Notch1-dependent MRP1
up-regulation, inhibition of which sensitizes breast cancer cells
to chemotherapy. BMC Cancer. 15:6342015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhao L, Ma Y, Gu F and Fu L: Inhibition of
Notch1 increases paclitaxel sensitivity to human breast cancer.
Chin Med J. 127:442–447. 2014.PubMed/NCBI
|
|
27
|
Park EY, Chang E, Lee EJ, Lee HW, Kang HG,
Chun KH, Woo YM, Kong HK, Ko JY, Suzuki H, et al: Targeting of
miR34a-NOTCH1 axis reduced breast cancer stemness and
chemoresistance. Cancer Res. 74:7573–7582. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cleator S, Tsimelzon A, Ashworth A,
Dowsett M, Dexter T, Powles T, Hilsenbeck S, Wong H, Osborne CK,
O'Connell P and Chang JC: Gene expression patterns for doxorubicin
(Adriamycin) and cyclophosphamide (Cytoxan) (AC) response and
resistance. Breast Cancer Res Treat. 95:229–233. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tao W, Shi JF, Zhang Q, Xue B, Sun YJ and
Li CJ: Egr-1 enhances drug resistance of breast cancer by
modulating MDR1 expression in a GGPPS-independent manner. Biomed
Pharmacother. 67:197–202. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Saha S, Mukherjee S, Mazumdar M, Manna A,
Khan P, Adhikary A, Kajal K, Jana D, Sa G, Mukherjee S, et al:
Mithramycin a sensitizes therapy-resistant breast cancer stem cells
toward genotoxic drug doxorubicin. Transl Res. 165:558–577. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang J, Su B, Gong C, Xi Q and Chao T:
miR-214 promotes apoptosis and sensitizes breast cancer cells to
doxorubicin by targeting the RFWD2-p53 cascade. Biochem Biophys Res
Commun. 478:337–342. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang X, Wan G, Mlotshwa S, Vance V,
Berger FG, Chen H and Lu X: Oncogenic Wip1 phosphatase is inhibited
by miR-16 in the DNA damage signaling pathway. Cancer Res.
70:7176–7186. 2010. View Article : Google Scholar : PubMed/NCBI
|